Abstract
Using green fluorescent protein probes and rapid acquisition of high-resolution fluorescence images, sister centromeres in budding yeast are found to be separated and oscillate between spindle poles before anaphase B spindle elongation. The rates of movement during these oscillations are similar to those of microtubule plus end dynamics. The degree of preanaphase separation varies widely, with infrequent centromere reassociations observed before anaphase. Centromeres are in a metaphase-like conformation, whereas chromosome arms are neither aligned nor separated before anaphase. Upon spindle elongation, centromere to pole movement (anaphase A) was synchronous for all centromeres and occurred coincident with or immediately after spindle pole separation (anaphase B). Chromatin proximal to the centromere is stretched poleward before and during anaphase onset. The stretched chromatin was observed to segregate to the spindle pole bodies at rates greater than centromere to pole movement, indicative of rapid elastic recoil between the chromosome arm and the centromere. These results indicate that the elastic properties of DNA play an as of yet undiscovered role in the poleward movement of chromosome arms.
Keywords: budding yeast, motility, mitosis, chromosome dynamics, centromeres
Introduction
The discovery of centromere DNA and kinetochore proteins in budding yeast has been dependent upon genetic and biochemical assays. These studies indicate that many proteins and features of mitosis in Saccharomyces cerevisiae appear to be analogous to those of mammalian tissue culture cells, making budding yeast an excellent genetic system to study the molecular mechanisms of mitosis (Maney et al. 2000; Pidoux and Allshire 2000; O'Toole and Winey 2001).
The mechanics of chromosome segregation in budding yeast has been difficult to analyze because of its small size and problems produced by the lack of chromosome condensation during mitosis. The 1.5–2-μm preanaphase spindle labeled with green fluorescent protein (GFP)–tubulin probes appears as two bright fluorescent half-spindles joined by a region of much lower fluorescence intensity in the middle of the spindle (Maddox et al. 2000). EM has shown a central spindle composed of approximately four overlapping microtubules from each pole. Also emanating from each pole are microtubules peripheral to the central spindle and extending on average 0.3 μm towards the spindle equator (Peterson and Ris 1976; Winey et al. 1995; O'Toole et al. 1999). These microtubules have been proposed to be individual kinetochore microtubules linking centromeres to the spindle poles (Peterson and Ris 1976; Winey et al. 1995; O'Toole et al. 1999). The average number of these microtubules, 16, matches the number of chromosomes, however, they do not reach the spindle equator, indicating that kinetochore proteins or centromeres must either be elongated or separated to attain a bipolar attachment (Winey et al. 1995; O'Toole et al. 1999). The medial lengths of these presumptive kinetochore microtubules decrease in anaphase 0.3–0.03 μm, as expected for centromere movement to the spindle poles (anaphase A) (O'Toole et al. 1999). However, it has not been possible to identify either centromere chromatin densities or kinetochores at the ends of these peripheral microtubules. As a result, it has been difficult to identify centromere position and their movement to the poles during anaphase.
Major advances in our understanding of how chromosomes move during mitosis in budding yeast have come from the development of live cell imaging and GFP probes for marking spindle microtubules, spindle poles, centromeres (kinetochores), and loci along chromosome arms. To determine how chromosomes move and when chromosomes separate during mitosis, Straight et al. 1997 generated fluorescent marks on chromosome arms by incorporating 256 copies (∼13.6 kb) of a truncated lacO sequence, 23 kb from the centromere in cells expressing a GFP-labeled lac repressor (lacI). This study showed that chromosomes in budding yeast oscillate from pole to pole and separate to the poles at anaphase onset. The 23-kb marker was not separated before anaphase onset in apparent conflict with the distribution of the presumptive kinetochore microtubules from EM analyses (Peterson and Ris 1976; Winey et al. 1995; O'Toole et al. 1999). This issue remained unresolved until lacO arrays were integrated 1.8–3.8 kb from the centromere (Goshima and Yanagida 2000). Unlike the 23-kb marker, the centromere proximal markers were seen in fixed preparations to be separated ≤0.8 μm before anaphase onset, indicating that the centromere proximal sister chromatin is stretched apart (Goshima and Yanagida 2000). Subsequently, live cell imaging of cells with GFP markers on individual chromosomes 1.4–2 kb from centromeres and at spindle pole bodies showed that separated sister centromeres oscillate relative to each other and to their poles before anaphase in a microtubule- and kinetochore-dependent manner (He et al. 2000; Tanaka et al. 2000). This dynamic stretching of centromere proximal chromatin reconciles the position of centromeres with EM studies. However, it raises new questions about the dynamic movements of centromeres, specifically during mitosis.
We have used live cell imaging of GFP-labeled kinetochores, chromosome loci, and spindle poles at higher temporal resolution to address centromere alignment, oscillations, and poleward movement at anaphase and poleward chromatin recoil at anaphase onset. To increase resolution, we have used a short 1.7-kb lacO repeat to mark chromatin 1.1, 12.7, and 23 kb from the centromere of individual chromosomes. The tandem repeat sequences used previously to measure chromosome movements and centromere proximal chromatin stretch were 5.6–13.6 kb in length (Straight et al. 1997; Goshima and Yanagida 2000; He et al. 2000; Tanaka et al. 2000). Thus, the actual position and movements of the centromeres may have been significantly underestimated. These changes, together with a sensitive imaging system, were used to detect the much weaker fluorescence of shorter lacO (∼1.7 kb) spots. To avoid conclusions based on observations of only one chromosome, we also marked centromeres of all chromosomes using Cse4–GFP, a kinetochore protein (Meluh et al. 1998; Chen et al. 2000).
In contrast to tissue cells, fluorescent marks on yeast chromosome arms have not been previously observed to congress to a metaphase plate (Straight et al. 1997). This may reflect underlying differences in yeast versus other eukaryotic cells and is critical for evaluating the importance and evolutionary significance of metaphase (Nicklas and Arana 1992). However, Peterson and Ris' studies (1976) indicate that the site of microtubule attachment to chromatin or centromere localization occurs within the medial portion of the spindle, and the ends of presumptive kinetochore microtubules are on average found within two bands on either side of the spindle equator close to the central spindle (Peterson and Ris 1976; Winey et al. 1995; O'Toole et al. 1999). Our imaging of Cse4-GFP–labeled centromeres shows that oscillations in position occur, but, on average, the separation between sister centromeres spans a large part of the central spindle, and the midpoint of the separated centromeres defines a metaphase plate.
With respect to timing, it has not been clear when anaphase A starts relative to the onset of anaphase B and how fast centromeres move poleward, since the chromosome arm markers do not accurately locate the position of the centromeres. Also, none of the previous studies using centromere proximal markers has addressed anaphase. Our experiments show that anaphase A begins synchronously for all centromeres at or within several minutes after the onset of anaphase B. Finally, the centromere proximal markers move poleward at ∼0.33 μm/min. In contrast, chromosome disjunction of a marker 23 kb from the centromere rapidly moves poleward, recoiling from stretching produced by poleward movement of centromeres and anaphase B separation of the spindle poles.
Materials and Methods
Plasmids
All plasmids used in this study are defined in Table and Table . The SPC72-GFP-LEU2 CEN plasmid (pXC224), NUF2-GFP-URA3 CEN plasmid (pJK6), CSE4-GFP-TRP1 plasmid (pKK1), GFP-TUB1-URA plasmid (pAFS125), pLKL60Y, and pLKL55Y were gifts from Dr. Tim Huffaker (Cornell University, Ithaca, NY) (Chen et al. 1998), Dr. Jason Kahana (University of California at San Diego, San Diego, CA) (Kahana et al. 1995), Dr. Richard Baker (University of Massachusetts Medical School, Worcester, MA) (Chen et al. 2000), Dr. Aaron Straight (Harvard University, Boston, MA) (Straight et al. 1997), and Dr. Kevin Lewis (National Institute of Environmental Health Sciences, Research Triangle Park, NC), respectively.
Table 1.
S. cerevisiae Strains
| Strain name | Relevant genotype | Source or reference |
|---|---|---|
| J178-1d | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 | K. Bloom/J. Carbon |
| 9d | MATa lys2-801 his3-200 ura3-52 leu2-3,112 | K. Bloom |
| KBY2001 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r leu2-3,112::lacO-KANr | This study |
| KBY2002 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r 1.1Kb-CEN3::lacO-KANr | This study |
| KBY2101 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r leu2-3,112::lacO-KANr TUB3-GFP-LEU2 | This study |
| KBY2102 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r 1.1Kb-CEN3::lacO-KANr pXC224 | This study |
| KBY2501 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r met14::lacO-KANr | This study |
| KBY2501.1 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r met14::lacO-HYGr 1KbCEN3::lacO-KANr | This study |
| KBY2502 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r 12.7Kb(CENXI)::lacO-KANr | This study |
| KBY2511 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r met14::lacO-KANr pXC224 | This study |
| KBY2521 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r met14::lacO-KANr pJK6 | This study |
| KBY2531 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r met14::lacO-HPH SPC29-GFP-KANr | This study |
| KBY2512 | MATa ade1 met14 ura3-52 leu2-3,112 his3-11,15 lys2Δ::lacI-GFP-NLS-NAT r 12.7Kb(CENXI)::lacO-KANrpXC224 | This study |
| KC100 | MATα ade2-101 his3-11,15 leu2-3 lys2-801 trp1 ura3-52 cse4::HIS3 pKK1 | K.C. Keith/R. Baker/ M. Fitzgerald-Hayes |
| KBY2006 | MATα ade2-101 his3-11,15 leu2-3 lys2-801 trp1 ura3-52 cse4::HIS3 pKK1 SPC29-CFP-KANr | This study |
| 9dgt1 | MATα lys2-801 his3-200 ura3-52 leu2-3,112 ura3-52::GFP-TUB1-URA3 | P.S. Maddox |
Table 2.
S. cerevisiae Plasmids
| Plasmid | Description/markers | Source or reference |
|---|---|---|
| pXC224 | SPC72–GFP fusion, pRS315/LEU2 | X. Chen/T. Huffaker |
| pJK6 | NUF2–GFP fusion, 2 μm/URA3 | J.A. Kahana/P.A. Silver |
| pKK1 | CSE4–GFP fusion, pRS314/TRP1 | K.C. Keith/R. Baker/ M. Fitzgerald-Hayes |
PCR Fragments for Integration
Integration of the lac operator into defined chromosomal loci was performed using PCR-based methods with 50 bp of homology flanking each 5′ and 3′ primer to the plasmid pLKL60Y. pLKL60Y was constructed using a BstEII–AccI fragment of an 8 mer of the lacO repeat (Robinett et al. 1996). Four copies of this repeat were then cloned into SmaI of pFA6MX4 (Wach et al. 1994) to create a 32 mer (1.2 kb in length) of the lac operator adjacent to the G418 resistance marker.
The lacI–GFP fragment was synthesized from pLKL55Y and targeted for integration into LYS2. pLKL55Y contains the lacI sequence, fused a nuclear localization signal (NLS), S65T GFP, and the NAT1r dominant drug resistance marker gene under the HIS3 promoter (Robinett et al. 1996) cloned into pRS303 (Sikorski and Hieter 1989).
A COOH-terminal fusion of SPC29–cyan fluorescent protein (CFP) was synthesized from pDH5 (Yeast Resource Center). Primers were constructed by using 50 bp of homology one codon upstream of the SPC29 stop codon and one codon downstream of the SPC29 coding region to amplify the CFP-G418 sequence.
Yeast Strains and Media
The S. cerevisiae strains and references or sources used in these experiments are listed in Table and Table . Strains with stable integrations were maintained in YEPD (2% glucose, 2% peptone, and 1% yeast extract). All strains requiring selection for maintenance of plasmids were grown on synthetic medium containing dextrose (0.67% yeast nitrogen base, 2% glucose, and the appropriate amino acids).
To induce lacI-NLS-GFP, cells were resuspended in synthetic media lacking histidine for ∼1.5 h at 30°C before adding 20 mM 3-aminotriazole (Sigma-Aldrich) for ∼30 min. All strains were grown to midlogarithmic phase growth before preparation for imaging.
Imaging
Techniques and equipment for GFP imaging have been previously described (Shaw et al. 1997; Maddox et al. 2000). For two-color imaging, excitation and emission of the fluorescent proteins was performed using the Chroma CFP–yellow fluorescent protein (YFP) narrow pass excitation and emission filters (86002 series; Chroma Technology Corp.). The CFP signal was imaged using the CFP excitation filters, whereas GFP was imaged in the YFP channel using the YFP narrow pass filters. YFP filters were used to accurately separate the GFP and CFP emission signals with a loss in the efficiency of GFP detection.
The centromere-specific histone (Cse4p) fused to GFP was used as a pan-specific marker for all centromeres (Chen et al. 2000). Centromere proximal spots were labeled with a lacO spot (Robinett et al. 1996; Straight et al. 1996) placed at ∼1.1 kb from CEN11 (centromere proximal spot). Chromosome arms were labeled ∼12.7 kb from CEN11 and ∼23 kb from CEN3 with lacO markers. To visualize the lacO elements, LacI–GFP was expressed under the control of the His3 promoter. This system facilitated the dynamic analysis of particular domains along chromosomes. Spindle pole bodies and the mitotic spindle were labeled with GFP to provide a reference point for the analysis of chromosome movement. Multiple spindle pole body markers (Spc72–GFP, Nuf2–GFP, and Spc29–GFP) and GFP–Tub1 were used for these studies to ensure that alterations in spindle architecture by different fluorescent protein fusions did not influence results. Nuf2p, reported as a spindle pole body protein (Osborne et al. 1994; Kahana et al. 1995), also localizes to the kinetochore, based on GFP fusion protein localization relative to fluorescently labeled spindle pole bodies (data not shown; Janke et al. 2001; Wigge and Kilmartin 2001). However, in strains used in this study, we observed the predominant Nuf2–GFP localization to occur at the spindle pole bodies, based on localization relative to a lacO centromere proximal marker and distance measurements of the separated Nuf2–GFP fluorescent spots (data not shown). The predominant localization of Nuf2–GFP and Spc72–GFP at the spindle pole body allowed us to accurately measure the anaphase movements of centromere proximal spots relative to the spindle pole body during anaphase onset.
Images were acquired at 0.7–60-s intervals, depending on the experiment. Fast rate analysis (0.7–5-s intervals) and single plane acquisitions were used to measure the rate of preanaphase chromosome oscillations, the rapid spot separation rates at anaphase onset, and Cse4–GFP movements. The single plane images had a depth of focus of ∼1 μm. Therefore, a slightly tilted spindle may appear in focus, however, oscillation distances and rates could be underestimated. Additionally, our measurements reflect a distance measurement from one spindle pole body to a chromosome spot. Chromosome rate measurements that are not along the spindle axis will also likely reflect an underestimate in movement rates. Time-lapse acquisition of optical Z-series of either three or five frames, separated by 0.75 μm steps, were used in all other imaging procedures. All images were acquired using 2 × 2 binning to increase camera signal-to-noise fourfold. However, preanaphase Cse4–GFP images and anaphase Tub1–GFP images were collected without binning for twofold-increased resolution.
Rate and Distance Measurement Analysis
All rate measurements were analyzed using the spindle pole body as a fiducial marker for chromosome movement along the spindle. Frame stacks were analyzed using the Metamorph Track Points function (Universal Imaging Corp.), and data sets were exported into Microsoft Excel™ for analysis. To correct for random errors in tracking, each frame stack analysis was repeated three times, and the mean for all collected data points was recorded. The rate of chromosome movement was determined in sequences where the length of the spindle did not change by a distance >0.2 μm. Linear regression plots were drawn through directed movements of greater than four data points. The slope of each linear regression plot was used to determine the rate of movement, and the average for all observed directed movements was calculated.
Half-spindle microtubules were measured by determining the distance from the spindle pole body to the outer edge of the half-spindle microtubules or tufts. Fluorescence intensity measurements on half-spindle microtubules were performed when the objective resolution (∼250–300 nm) limited the detection of half-spindle microtubule shortening. A 10 × 10 pixel region was placed over each half-spindle and the total integrated fluorescence intensity was measured. The background, determined by placing the same size region on a nonlabeled portion of the cell, was then subtracted from this value. This measurement was then standardized to the distance measurement of the last spindle pole body to the end of half-spindle measurement. Each decreasing intensity measurement, expressed as a decrease in distance from the spindle pole body, represented a loss in the total integrated fluorescence intensity for that particular half-spindle at the indicated time points.
All measurements using Z-series acquisitions were performed using the Metamorph Measure Pixel function. In-focus data points were measured using Cartesian coordinates. This allowed for manually calculated three-dimensional reconstructions to determine the localization of chromosome spot(s) or centromere spots, relative to the mitotic spindle. All collected data was exported into Microsoft Excel for analysis of rates and/or localization.
Image Presentation
Presentation images were prepared as previously described (Shaw et al. 1997; Maddox et al. 2000). Note that all analyses were performed using a manually calculated three-dimensional reconstruction with Cartesian coordinates. Kymographs were used to project all data points for an entire collected sequence of preanaphase movements at fast rate intervals. A region 5–8 pixels wide was drawn through the long axis of the mitotic spindle, and the brightest pixel was recorded on a single line. This was repeated for each time point and was displayed along the x axis to show the entire time course.
Results
Separation and Dynamic Movements of Centromeres and Surrounding DNA in Preanaphase Spindles
To test how sister centromeres and centromere proximal chromosome arms respond to forces generated by kinetochores attached to the mitotic spindle, we used fluorescent probes for centromeres, chromosome arms, and spindle pole bodies (Fig. 1). In live large budded cells before anaphase onset, centromeres labeled with Cse4–GFP were often observed as two clusters or groups separated on average by 0.58 ± 0.36 μm (minimum = 0 μm, maximum = 1.84 μm; n = 63) (Fig. 1 A; Table ). In contrast, average separation of the ∼1.1-kb centromere proximal spot was 0.26 ± 0.32 μm (min = 0 μm, max = 1.48 μm; n = 95) (Fig. 1 B; Table ). This is ∼0.32 μm less separated than the average separation of the Cse4–GFP centromere marker (Fig. 1A and Fig. B; Table ). Chromosome arms with lac operator repeats integrated more distal to the centromere were on average separated by 0.07 ± 0.19 μm (min = 0 μm, max = 0.85 μm; n = 94) at loci ∼12.7-kb CEN11 and were observed as a single unseparated spot when integrated ∼23 kb from CEN3 (n = 101) (Fig. 1C and Fig. D; Table ). The centromere separation and outward stretch of the arms near the centromeric region shown in Fig. 1 E is consistent with previous reports that centromeres are on average separated before anaphase onset by poleward forces (Goshima and Yanagida 2000; He et al. 2000; Tanaka et al. 2000).
Figure 1.
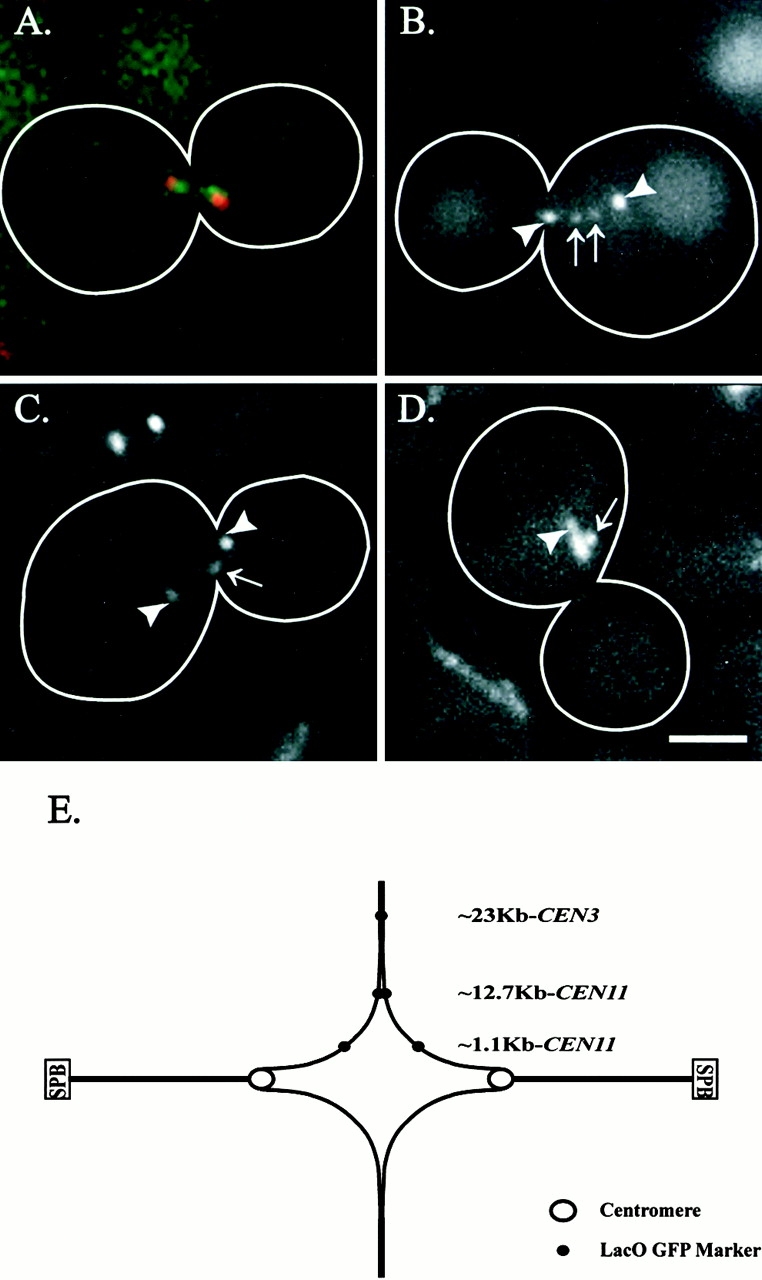
Preanaphase separation and positioning of sister centromeres. Centromeres displayed the greatest average separation with progressively decreasing separation of chromosome markers as they were placed distal from the centromere. (A) Pan-specific centromere marker for all centromeres, Cse4–GFP (green) with Spc29–CFP (red) marked spindle pole bodies. Centromeres clustered into discrete groups that were separated toward the spindle pole bodies. (B) LacO GFP marker placed ∼1.1 kb from CEN11 (arrows) with Spc72–GFP-labeled spindle pole bodies (arrowheads). (C) LacO GFP marker placed ∼12.7 kb from CEN11 (arrows) with Spc72–GFP-labeled spindle pole bodies (arrowheads). (D) LacO GFP marker placed ∼23 kb from CEN3 (arrows) with Tub3–GFP-labeled spindle (arrowhead). (E) Schematic representing the above average preanaphase separations (Table ). SPB, spindle pole body. Bar, 2 μm.
Table 3.
Metaphase-like Alignment of Centromeres but Not Chromosome Arms
| Strain description | Foci and spot separation | Spindle length | % Population within medial 25% of spindle | n |
|---|---|---|---|---|
| μm | μm | |||
| CSE4-GFP (SPC29-CFP) | 0.58 ± 0.36 | 1.56 ± 0.41 | 83 | 63 |
| KBY2511 (∼1.1 kb CEN11) | 0.26 ± 0.32 | 1.97 ± 0.56 | 62 | 94 |
| KBY2512 (∼12.7 kb CEN11) | 0.07 ± 0.19 | 2.02 ± 0.64 | 47 | 95 |
| KBY2101 (∼23 kb CEN3) | 0.00 ± 0.00 | 1.19 ± 0.25 | 36 | 101 |
Centromere and chromosome spot localization along the length of G2/M spindles (spindle pole bodies labeled with Spc72–GFP, Spc29–CFP, or Tub3–GFP) indicate that centromeres persist within the medial 25% of the spindle. This is indicative of a metaphase-like conformation. Chromosome arms do not exhibit this phenotype as frequently. Optical Z-series were acquired for a population of cells. Using distance measurements with the Z-distance taken into account, the chromosome spot localization relative to the spindle was determined. For separated spots, the average distance relative to the spindle was used.
To measure the dynamic movements of centromeres in preanaphase cells, we recorded the ∼1.1-kb centromere proximal marker movements relative to GFP-labeled spindle pole bodies using rapid frame acquisition rates (∼1 frame/s) (Fig. 2). Fig. 2 shows that, in addition to dynamic movements between the separated chromosome spots, there were also distinctive oscillations of the centromere proximal spots along the mitotic spindle with Spc72–GFP-labeled spindle pole bodies. Two classes of centromere proximal spot movements are evident in Fig. 2: major oscillatory movements relative to the spindle pole body (Fig. 2 C, directed oscillation) and minor movements of <0.25 μm that may be due to diffusional motion. Analyses were repeated three times for each time-lapse sequence, and the mean was displayed, indicating that the smaller movements are not a product of tracking errors (Fig. 2 C). The centromere proximal markers separated ≤1.26 μm (Fig. 2 C, time point = 70.3 s) and reassociated with each other before reseparation (Fig. 2 C, time point = 199.0 s). There was no apparent strong coordination of the separated centromere spot oscillations relative to the spindle pole bodies, as determined by the independent movement of each spot relative to its sister (Fig. 2 C, directed oscillation). However, we did occasionally observe both separated spots to move toward a single pole in what appeared to be coordinated movements toward the spindle pole body (data not shown).
Figure 2.
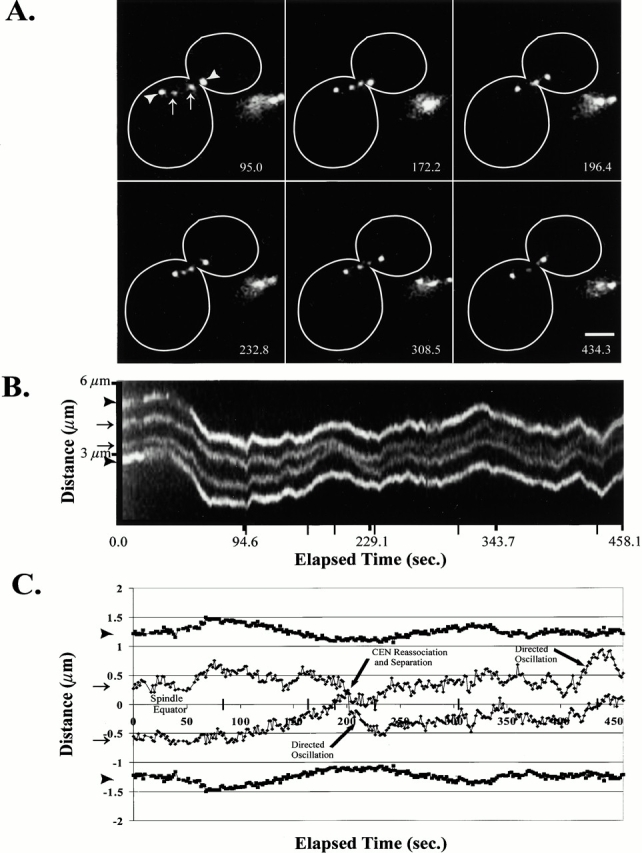
Centromere proximal spots exhibited dynamic separation and oscillations along the preanaphase mitotic spindle. The lacO marker was integrated ∼1.1 kb from CEN11 with Spc72–GFP-labeled spindle pole bodies. (A) Selected frames of an ∼7.5-min single-plane time-lapse. (B) A kymograph sequence of the entire time-lapse collected at ∼0.9-s intervals. Narrow tick marks indicate corresponding time points to the above selected frames. (C) Graphical plot of the above time course. In both A and B, arrowheads denote the spindle pole bodies, and arrows define the centromere proximal chromosome spots. Elapsed time provided in seconds. Bar, 2 μm.
To confirm the centromere oscillations observed using the ∼1.1-kb centromere proximal marker, we analyzed the movements of the pan-specific centromere marker Cse4–GFP. Fig. 3A and Fig. B, describes two distinct groups of centromeres at time point 145 s that separate into multiple spots, as observed with broader, less intense bands of fluorescence (Fig. 3 A, time point = 305 s, arrowheads), and then reassociate into two diffuse clusters or groups (Fig. 3 A, time point = 550 s). Transient Cse4–GFP movements were observed between the clustered groups of fluorescence (Fig. 3, arrowheads), indicating that centromeres exhibit dynamic movements relative to other centromeres in each clustered group. Furthermore, two-color imaging of Cse4–GFP with CFP-labeled spindle pole bodies shows that the centromeres move relative to the spindle pole bodies (data not shown).
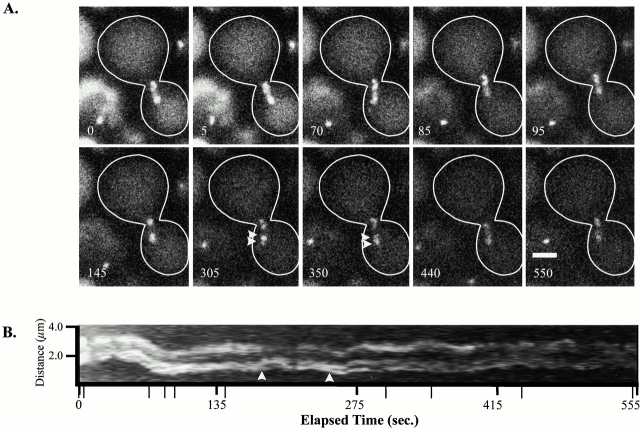
Figure 3.
Centromere dynamics of all chromosomes using the pan-specific centromere marker Cse4p fused to GFP (Cse4–GFP). Dynamic movements between the two clustered groups indicated a similar separation and dynamic movements to those observed for the ∼1.1-kb centromere proximal marker. Spindle pole bodies are not labeled. (A) Selected images of an ∼9-min single-plane time-lapse. (B) Kymograph of the entire time-lapse taken at 5-s intervals. Arrowheads in A and B indicate occurrences of GFP centromere fluorescence disjoining from the clustered group. Narrow tick marks indicate corresponding times to A. Elapsed time is in seconds. Bar, 2 μm.
Velocity Calculations of the Preanaphase ∼1.1-kb CEN Proximal Spot Oscillations
We have measured the rate of movement of centromere proximal spots relative to GFP-marked spindle pole bodies. Linear regression lines were drawn through directed chromosome movements (Fig. 2 C, directed movements; Materials and Methods), and the slope was used to determine the rate of centromere proximal movement. The mean rate for the ∼1.1-kb CEN11-directed spot movements relative to the spindle pole body was 1.51 ± 0.66 μm/min (n = 24). The average distance traveled during a directed movement was 0.42 ± 0.16 μm for 16.3 ± 9.6 s (n = 24). Arm spots labeled at both ∼12.7 and ∼23 kb from the centromere were also observed to undergo oscillatory movements; however, these movements did not persist in a single direction for as long a distance or time interval as observed for the centromere proximal marker (data not shown).
Centromeres but Not Chromosome Arms Exhibit a Metaphase Conformation
We considered a metaphase conformation to exist if the midpoint of sister chromatids aligned within a medial 25% of the mitotic spindle. To determine the average position of each chromosome marker along the spindle in a population of cells, we examined chromosome arm or centromere markers with GFP-labeled microtubules or spindle pole bodies. Z-series images of cell populations containing either Cse4–GFP, ∼1.1-kb CEN11, ∼12.7-kb CEN11, or ∼23-kb CEN3 chromosome markers were collected in three separate experiments. The localization of markers in each cell was measured by determining the midpoint of the separated Cse4–GFP sister clusters or the lacO–GFP spots relative to the spindle pole bodies.
The midpoint of separated Cse4–GFP foci were observed within the medial 25% of the spindle in 83% of the population of large budded cells (n = 63; Table ). The ∼1.1-kb CEN11 midpoint spot, ∼12.7-kb CEN11, and ∼23-kb CEN3 spots were found to be within the medial 25% of the spindle in 62, 47, and 36% of the populations, respectively (n = 94, 95, and 101, respectively; Table ). These results indicated that centromeres persist in a metaphase conformation, whereas the chromosome arms progressively decrease in spindle equator localization as the marker is placed further from the centromere.
Temporal Resolution of Anaphase A Centromere Movement and Microtubule Shortening to the Spindle Pole Bodies Relative to Anaphase B Spindle Pole Body Separation
To resolve the timing of anaphase A chromosome movement to the spindle pole body, we studied anaphase movements of GFP-marked chromosomes, centromeres, and spindle microtubules relative to anaphase B spindle elongation.
The ∼1.1-kb CEN11 lacO marker was integrated into strains containing spindle pole bodies marked with Nuf2–GFP or Spc72–GFP. Z-series time-lapses were acquired at 15–45 s. We observed the ∼1.1-kb CEN11 proximal spots to move toward the GFP-marked spindle pole bodies coincident with or within 3 min after anaphase onset, as defined by the start of anaphase B spindle elongation (Fig. 4A and Fig. B and Fig. 5). Fig. 5 B (arrow) shows the separation of the spindle pole bodies to begin at the 0.0-min time point. Movement of the CEN proximal spots toward the spindle pole body ensues at 1.0 min (Fig. 5 B, arrow). This delay results in a transient increase in the distance between the centromere proximal spot and its spindle pole body. The centromere proximal spot began anaphase A chromosome movement toward the spindle pole body at an average spindle length of 2.87 ± 0.51 μm (Table IV; n = 15). Regression lines were drawn through each plot of centromere proximal chromosome spot movement to the spindle pole body (Fig. 5 C). The average rate observed was 0.33 ± 0.16 μm/min (Fig. 5 C; Table IV; n = 23). The average distance moved was 0.98 ± 0.28 μm (Table III; n = 23).
Figure 4.
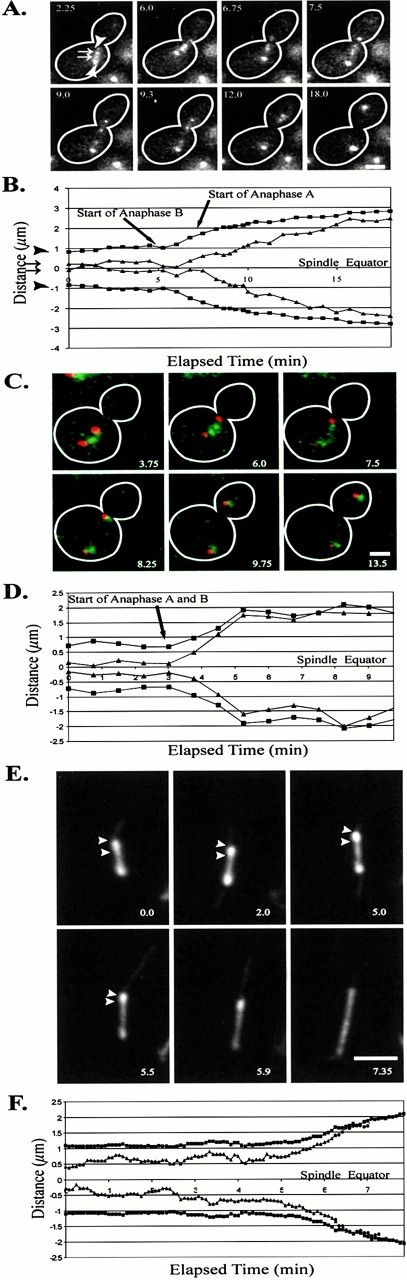
Anaphase A centromere movement to the spindle pole body and half-spindle shortening began coincident with or shortly after the onset of anaphase B spindle pole body separation. (A) Progression through anaphase was followed using the ∼1.1-kb CEN11 marker and Nuf2–GFP to label the spindle pole bodies. Selected frames from a Z-series time-lapse sequence taken at 45-s intervals. (B) Graphical plot of the above time-lapse (A), using measurements with the Z-distance taken into account. In A and B, arrowheads denote the spindle pole bodies, and arrows define centromere proximal chromosome spots. (C) Centromere movement to the spindle pole body was coincident with the onset of anaphase B spindle pole body separation, as determined using Cse4–GFP to label all centromeres. Spindle pole bodies were labeled using Spc29–CFP, allowing two-color imaging of anaphase onset. Centromere clusters underwent a synchronous separation at anaphase onset. Frames are from a Z-series time-lapse sequence taken at 45-s intervals. (D) Graphical plot of the above time-lapse (C), with the Z-distance taken into account. (E) Half-spindle microtubule or shortening to the spindle pole body began near the onset of anaphase B spindle pole body separation. Using GFP–Tub1, we show that, upon elongation of the spindle, the fluorescent tufts emanating from each spindle pole body, presumed to be kinetochore microtubules, shortened toward the spindle pole bodies. Plots also indicate that there were dynamic movements in the lengths of half-spindle microtubules emanating from the spindle pole body. When the half-spindle had shortened to within the resolution limit of the light microscope (∼250–300 nm), integrated fluorescent intensities, standardized to the last distance measurement (6.26 min), were plotted to show that fluorescence intensities decreased, indicating that microtubule polymer further decreased from the optical resolution limited tufts (▴). Images were from a single plane acquisition time-lapse sequence at 5-s intervals. Arrows indicate the spindle pole bodies and plus ends of half-spindles. (F) Graphical plot of the above time-lapse showing plots of each spindle pole body (▪) relative to the ends of each fluorescent tuft (▴; 0–6.3 min). At 6.26–7.26 min, the fluorescent intensity of each tuft was standardized to the last distance measurement at 6.26 min and plotted relative to their respective spindle pole body (•, 6.26–7.26 min). Elapsed time for A–E is in minutes. Bars, 2 μm.
Figure 5.
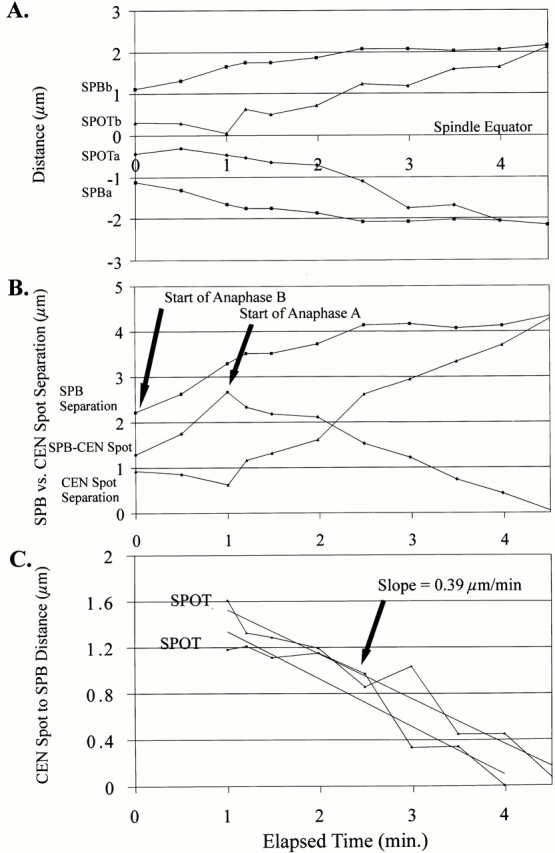
Centromere proximal (∼1.1 kb) marker movement to the spindle pole body during anaphase A. Graphical plot of a single representative early anaphase onset (15 observed) using Nuf2–GFP to mark the spindle pole bodies and the ∼1.1-kb CEN11 chromosome marker. Centromere movement to the spindle pole body (anaphase A) began at or shortly after the onset of anaphase B spindle pole body separation. (A) Plot of anaphase onset. (B) Plot showing the separation of spindle pole bodies (▪) and CEN spots (▴) and the distance between the spindle pole body and the CEN proximal spot (•). The onset of anaphase A was defined by the decrease in the distance between the spindle pole body and the ∼1.1-kb spots, indicating that chromosomes were moving toward the spindle pole bodies. (C) Plot of each individual CEN proximal spot to spindle pole bodies with linear regression lines representing the general rate of anaphase A movement to the spindle pole body. The average slope was 0.33 ± 0.16 μm/min (Table ).
Cse4–GFP and Spc29–CFP facilitated the visualization of all centromeres relative to the spindle pole bodies. We found the centromere movement to the spindle pole body did not begin until anaphase B spindle pole body separation (Fig. 4C and Fig. D). Fig. 4 D describes the coincident incidence of anaphase A relative to anaphase B. At 3 min, spindle pole body separation and centromere movement to the spindle pole body began (Fig. 4 D, arrow). We did not observe an increase in the distance between the spindle pole bodies and the Cse4–GFP clusters, as observed for the CEN11 proximal marker (Fig. 5 B; data not shown). The delay observed between the start of anaphase B and anaphase A observed for the CEN11 proximal marker (Fig. 5 B) may be due to the timing of release of sister chromatid cohesion from the centromere proximal region.
EM and fluorescent analysis of the mitotic spindle shows that two populations of microtubules emanate from the spindle pole bodies: 4–8 interpolar microtubules between the spindle pole bodies and 16 presumptive kinetochore microtubules, creating a half-spindle from each spindle pole body (Winey et al. 1995; O'Toole et al. 1999; Maddox et al. 2000). Using GFP–Tub1 to analyze the behavior of the presumptive kinetochore microtubules, we measured shortening of the mitotic half-spindle as the length of the interpolar spindle increased during anaphase B. Fig. 4 E (arrows) shows images of fluorescent half-spindles or tufts emanating from the spindle pole body as they shortened after anaphase B onset. The half-spindle distance was measured until their length decreased to the resolution limit of the light microscope, ∼250–300 nm (Fig. 4 F, solid line; to 6.26 min). Fluorescence intensity measurements, standardized to the 6.26-min time point, were then determined for the remaining time points measured. The fluorescence intensity decreased, indicating a further loss in microtubule polymer within the resolution-limited half-spindles (Fig. 4 F, dashed line). In addition to half-spindle shortening at the onset of anaphase B, Fig. 4 F shows several dynamic changes in the length of the half-spindles relative to their spindle pole bodies that may reflect either dynamic movements of clustered centromeres or slight changes in the focal plane. Also, note that the half-spindle emanating from each spindle pole body only decreases in length after spindle pole body separation, indicating that anaphase A microtubule shortening to the spindle pole body does not begin before anaphase B has initiated (Fig. 4 F).
Synchrony of Chromosome Separation at Anaphase Onset
Analysis of the anaphase movements of centromeres labeled with Cse4–GFP indicated that the initiation of anaphase A movement toward the spindle pole body was synchronous. Distinct groups of fluorescence, presumed to be a cluster of the 16 sister pairs, were observed to undergo anaphase A movements toward the spindle pole body with very few centromeres moving distinct from each discrete cluster of centromeres (Fig. 4 C). This is consistent with our observations that ∼1.1-kb centromere proximal spots on two different chromosomes in the same cell separated synchronously during anaphase onset (data not shown).
Chromatin Stretching Occurs between the Centromere and the Centromere Proximal Spot upon Anaphase B Spindle Pole Body Separation
Upon anaphase B spindle elongation, we often observed (12/13 measured) the distance between the spindle pole bodies and their respective centromere proximal spot to increase before undergoing anaphase A shortening to the spindle pole body (Fig. 5 B, 0.0–1.0 min). This length increase was due to either the lengthening of the microtubules attached to chromosomes, stretching of the kinetochore, and/or stretching of the chromatin between the centromere and the ∼1.1-kb CEN11 marker. The distance between the Cse4–GFP cluster and the spindle pole body did not increase upon spindle pole body separation, nor did the half-spindle length measured, using GFP–Tub1 (Fig. 4, C–F; data not shown). This indicates that the increased distance between the spindle pole body and the ∼1.1–kb CEN11 spot is likely due to further stretch in the chromatin between the centromere and the lacO marker.
Rapid Poleward Recoil of Chromosomal DNA upon Anaphase B Spindle Pole Body Separation
The above analyses indicate that chromatin surrounding the centromere is stretched and therefore predict that release of cohesions should result in poleward chromatin recoil. To test this prediction, lacO markers, labeling chromosomes at ∼1.1 or ∼23 kb from their respective centromeres, were imaged using single plane and <1-s acquisition intervals without spindle pole body markers to determine their behavior and rate of separation upon anaphase onset. Chromosome separation rates similar to those observed for biphasic spindle elongation rates previously characterized (Kahana et al. 1995; Yeh et al. 1995; Straight et al. 1998) would be expected if the chromosomes passively followed the separating spindle pole bodies. Anaphase A movements also contribute to the rate of early chromatid separation (Fig. 4 and Fig. 5). Therefore, the rate of spot separation would be expected to be the sum of spindle pole body separation in early anaphase (∼1 μm/min) (Kahana et al. 1995; Yeh et al. 1995; Straight et al. 1998) and anaphase A rates for both spots moving to their respective spindle pole bodies (1.66 μm/min; Table ).
Table 4.
Analysis of Anaphase A Movements to the SPB
| Description | Rate of movement to SPB | Spot separation | Spot to SPB distance | Anaphase A displacement | Duration | n | Spindle length | n |
|---|---|---|---|---|---|---|---|---|
| μm/min | μm | μm | μm | min | μm | |||
| KBY2511_∼1.1 kb CEN11 spot to SPB movement | 0.33 ± 0.16 | 1.03 ± 0.63 | 1.13 ± 0.23 | 0.98 ± 0.28 | 3.33 ± 1.85 | 23 | 2.87 ± 0.51 | 15 |
Centromere proximal spot (∼1.1 kb) movements to the spindle pole body (SPB) during anaphase A were recorded by acquiring optical Z-series images at 15-s intervals. Sister centromere movements to the spindle pole body were plotted (Fig. 5 C), and linear regression lines were drawn through each persistent movement to the spindle pole body.
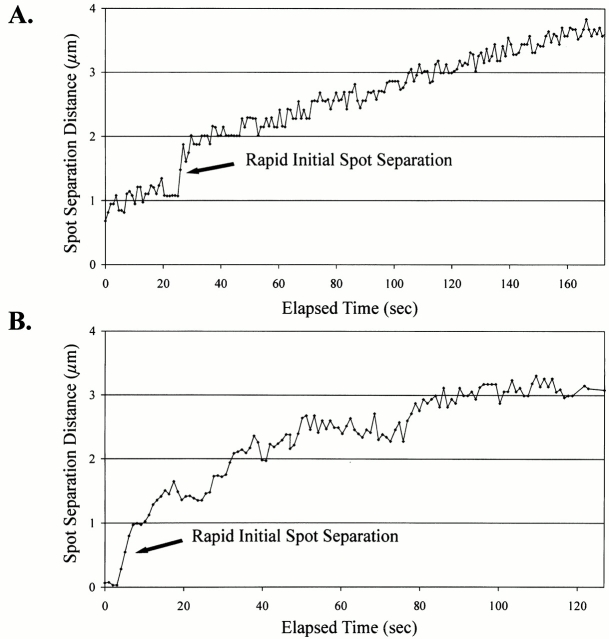
The centromere proximal spot was generally observed to separate at rates slightly greater than those observed for fast anaphase B spindle pole body separation. This is consistent with anaphase A and B movements being responsible for the rates of sister centromere separation (1.21 ± 0.46 μm/min; Table ). However, infrequently (3/16 observed; Table ), the initial rate of separation was observed to be much greater than that expected for the separation of sister chromatids (see above). Fig. 6 A shows initial separation of the ∼1.1-kb centromere proximal spots with a short rapid separation (arrow). These rates were 2.73–9.67 μm/min (average = 5.22 ± 3.87 μm/min; Table , initial separation) for a distance of 1.16 ± 0.22 μm (Table ). Although there was some evidence for chromatin recoil at the centromere proximal marker, it is not a frequent event with a significant contribution to chromosome poleward movement.
Table 5.
Chromosome Spot Separation Rates at Anaphase A Start
| Fast initial separation | Rapid phase | Slow phase | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Description | Spot separation rate | Time interval measured | Distance measured | n | Spot separation rate | Time interval measured | Distance measured | n | Spot separation rate | Time interval measured | Distance measured | n |
| μm/min | s | μm | μm/min | min | μm | μm/min | min | μm | ||||
| KBY2501 (∼1.1-kb CEN11) | 5.22 ± 3.87 | 17.5 ± 11.3 | 1.16 ± 0.22 | 3 | 1.21 ± 0.46 | 2.26 ± 1.36 | 2.54 ± 0.96 | 13 | 0.27 ± 0.20 | 9.55 ± 11.18 | 1.04 ± 0.41 | 3 |
| KBY2001 (∼23-kb CEN3) | 10.48 ± 6.38 | 8.6 ± 4.2 | 1.26 ± 0.28 | 7 | 0.99 ± 0.67 | 3.13 ± 2.73 | 2.79 ± 0.53 | 12 | 0.19 ± 0.05 | 10.5 ± 3.12 | 5.24 ± 0.73 | 4 |
The rate of spot separation at anaphase onset for chromosome markers at ∼1.1 and 23 kb from the centromere. All (7/7) ∼23-kb spot separations exhibited a fast initial separation followed by rapid and slow velocity phases, as characterized for anaphase B spindle pole separation rates. The ∼1.1-kb CEN11 marker also exhibited fast spot separations; however, these separations were slower and were observed in only 3 of 16 observed events.
Figure 6.
Upon anaphase onset chromosome spots exhibit rapid separation from their sister chromosome GFP spots at rates much greater than anaphase A and B movements. (A) Representative plot of sister chromatid spot separation for the ∼1.1-kb CEN11 marker upon anaphase onset. (B) Representative plot of sister chromatid separation for the ∼23-kb CEN3 marker upon anaphase onset. Sequences of rapid spot separation were acquired at single-plane, short acquisition intervals (>1 frame/s).
In time-lapse sequences for the ∼23-kb CEN3 chromosome arm marker, all observations (7/7 observed; Table ) of the initial separation of sister chromatids showed a rapid initial separation of sister chromosome arm markers at an average separation rate of 10.48 ± 6.38 μm/min (n = 7; Table ) for an average distance of 1.26 ± 0.28 μm (Fig. 6 B, arrow; Table ) . Although there was a large variability in the rates of separation, 5.12–21.77 μm/min, the ∼23-kb CEN3 lacO chromosome arm spot was observed to consistently separate at rapid rates. Poleward recoil of the ∼23-kb lacO spot indicates that stretch of the chromatin ∼23 kb from the centromere makes a significant contribution to the kinetics of chromosome arm movement.
Discussion
We describe a dynamic process for which mitotic chromosomes in budding yeast maintain a bipolar attachment. Tension across the centromere results in separation of the sister centromeres and proximal chromatin before the onset of anaphase. Centromere-specific proteins labeled with GFP, such as Cse4p (Cse4–GFP), show that the separation between sister centromeres spans a large part of the spindle and the midpoint of the separated centromere clusters preferentially localizes to the spindle equator in a metaphase-like conformation (Fig. 1 E and 7). This preanaphase separation of sister centromeres is likely important for the maintenance of bipolar chromosome attachment and the functional processes of properly segregating the duplicated genome. Similar stretching in centromere chromatin has been observed in human tissue culture cells (Shelby et al. 1996) and in the diatoms (Pickett-Heaps and Tippit 1978; Tippit et al. 1980). The kinetochore microtubule attachment in meiotic grasshopper spermatocytes is stabilized with tension (Nicklas and Koch 1969), and the direction of chromosome motility is also sensitive to tension (Nicklas 1977; Inoue and Salmon 1995; Skibbens et al. 1995). Additionally, tension in higher eukaryotes has roles in regulating the cell cycle progression from metaphase to anaphase (McIntosh 1991; Nicklas 1997; Rieder and Salmon 1998).
Our observation of clustered centromeres based on Cse4–GFP localization allowed us to corroborate in vivo the mitotic spindle structure previously proposed based on EM studies (Peterson and Ris 1976; Winey et al. 1995; O'Toole et al. 1999). The spindle structure based on EM work shows kinetochore microtubules to be localized in a parallel structure contradictory to the splayed mitotic structure seen in higher eukaryotes. Centromere-linked lacO spots integrated at various loci along chromosome arms were clearly off the spindle axis; however, this is likely because these markers do not represent the actual localization of the centromeres. Therefore, localization of clustered centromeres between the spindle pole bodies using Cse4–GFP leads us to postulate that the kinetochore microtubule arrays are organized in parallel arrays consistent with EM data. This view of the budding yeast spindle organization is not consistent with the splayed mitotic spindle proposed by He et al. 2000.
The large difference in the average separation of the ∼1.1-kb CEN11 chromosome marker and Cse4–GFP foci indicates that the ∼1.1 kb of DNA between these markers is highly elongated by an average distance of 0.16 or 0.15 μm/kb of DNA (Fig. 1; Table ) before the onset of anaphase. Naked B form DNA is 0.34 μm/kb, and a sevenfold nucleosomal compaction predicts that 1 kb of mitotic chromatin covers a distance of 0.05 μm (Finch et al. 1977). Our results predict that the level of DNA compaction at the centromere and surrounding chromatin in large budded cells is threefold less than nucleosomal DNA. Upon anaphase onset, this stretching of DNA is further amplified due to large forces generated by the elongating central spindle. Functionally, this centromeric DNA conformation may be indicative of an active structure that can be dynamically stretched and is responsible for proper centromere function. Importantly, our lacO sequence is 1.7 kb in length, and it has been shown that lacO sequences can be stretched by forces generated from the mitotic spindle (He et al. 2000). Therefore, the measured spot for the ∼1.1-kb CEN11 lacO marker may represent a distance greater than the ∼1.1-kb (≳2.8 kb) distance from the centromere, because portions of the lacO array are stretched toward the spindle pole bodies.
Centromeres undergo oscillatory movements relative to each other and to the spindle pole bodies, as shown for vertebrate kinetochores in tissue cells (Skibbens et al. 1993; Rieder and Salmon 1994, Rieder and Salmon 1998). The microtubule- and kinetochore-dependent separation of sister centromeres in S. cerevisiae (Goshima and Yanagida 2000; He et al. 2000; Tanaka et al. 2000) before anaphase onset indicates that the functions involved at the interface between the microtubule and kinetochore are important for chromosome dynamics in mitosis. Fluorescence recovery after photobleaching studies of the mitotic half-spindle also show that microtubule dynamics are evident before the onset of anaphase (Maddox et al. 2000). We show that these half-spindle microtubules shorten upon anaphase B spindle pole body separation, as would be expected for kinetochore microtubule shortening during anaphase A (Fig. 4 E and 7). Interestingly, the rate of chromosome movement we observed before the onset of anaphase (see Results) is similar to the in vivo growth and shortening rates of wild-type astral microtubule plus ends (Tirnauer et al. 1999). Capped microtubule minus ends shown by EM studies in yeast provide evidence that microtubule minus ends may be stable, whereas the flared microtubule plus ends may be dynamic (O'Toole et al. 1999). These results lead us to further postulate that the minus end microtubule dynamics of kinetochore microtubules or treadmilling may have little involvement in chromosome movements, whereas plus end dynamics at the kinetochore are important in budding yeast (Maddox et al. 2000).
The duration and frequency of yeast sister centromere movements relative to each other is important to the mechanism for how a chromosome maintains a bipolar attachment to the mitotic spindle and may also be important for why sister centromeres separate and reassociate before the onset of anaphase. Fig. 2 provides an example of sister centromere rejoining and separation. During these sequences, we observed centromere proximal spots to reassociate for only 0.4%, or 19.5 s, of the entire imaging time (25.5 min), beginning with separated sister centromeres. These reassociations were observed in only two of eight time-lapse sequences, indicating that once centromeres have separated, they infrequently reassociate. Consistent with He et al. 2000, we did not observe the centromere proximal spots to remain separated for longer than 8 min before rejoining, however our low sample time may have been limiting to these analyses. The rejoining of sister centromere proximal spots may be due to unstable attachments of sister kinetochores to kinetochore microtubules, thereby collapsing the separated lacO spots. However, because we do not observe rapid inward recoil of separated spots, it is more likely that the rejoining of centromere proximal spots is due to directional instability of kinetochore microtubules (Skibbens et al. 1993). This behavior may be analogous to the tension-dependent regulatory mechanism in higher eukaryotes, where coordinated movements between sister kinetochores regulates the direction of chromosome movement essential for congression to a metaphase plate (Skibbens et al. 1993, Skibbens et al. 1995).
In large budded cells, centromeres, and not the chromosome arms, adopt a metaphase-like conformation. The Cse4–GFP localization (Table ) persists within the medial 25% of the mitotic spindle, whereas chromosome arms do not. This centromere configuration is similar to tissue cell kinetochores that are separated and aligned at the spindle equator during metaphase (Skibbens et al. 1993, Skibbens et al. 1995; Khodjakov and Rieder 1996; Waters et al. 1996, Waters et al. 1998). Our metaphase observation is consistent with Peterson and Ris 1976 original EM observation that there is a metaphase plate that separates into two “anaphase plates” upon anaphase onset. A single metaphase plate, rather than two separated clusters surrounding the spindle equator, may have been observed in their studies because chromatin proximal to the centromere was likely detectable in a single thick EM image plane between the separated centromeres. More recent EM studies show a population of shorter kinetochore microtubules found to be of approximately the same length, consistent with a metaphase alignment of chromosomes (O'Toole et al. 1999). Chromosome congression to a metaphase-like conformation is believed to have evolved with the spindle assembly checkpoint (Nicklas and Arana 1992). Although some organisms do not contain a metaphase plate (Kubai 1975), this conformation may have significant importance to the physical processes of segregating DNA in an organized and regulated manner.
The greatest physical evidence for tension-dependent chromatin stretching and elastic recoil during mitosis is apparent upon anaphase onset, when chromatin snaps toward the separating spindle pole body at rates much greater than exhibited by centromeres during anaphase A and B movements. A previous report indicated that a marker ∼23 kb from CEN3 separated by 1.8 μm in the 26-s interval observed, predicting a rapid separation rate of 3.6 μm/min (Straight et al. 1997). Using rapid image acquisition, we were able to obtain velocity measurements of these fast chromosome separation rates. Upon anaphase onset, spindle elongation and centromere movement to the spindle pole body began coincidentally and was followed by further stretching of the chromatin linking sister centromeres (Fig. 5 B and 7). This increase in distance from the spindle pole body is not evident in the GFP–Tub1 or Cse4–GFP analyses, indicating that the stretching occurs between the centromere and the chromosome markers. This stretching is presumably before the release of cohesion, since chromosome arms have not segregated (Fig. 7). Loss of cohesion creates a release of tension in the chromatin stretched between sister centromeres and causes chromosome arms to undergo a rapid elastic recoil toward the spindle pole body (Fig. 7). Because markers closer to the centromere segregate earlier than telomere proximal markers (Straight et al. 1997), the simplest model for the elastic recoil is a “zippering” effect that results in a progressive separation and recoil of the DNA toward the spindle pole bodies (Fig. 7). Interestingly, the rate of spindle elongation (labeled with Spc29–GFP, GFP–Tub1, Spc72–GFP, or Nuf2–GFP) did not exhibit rapid transient movements distinguishable from the normal rates of spindle elongation. This indicates that the polar forces involved in maintaining the proper spindle length during anaphase B spindle pole body separation are much greater than the forces provided by stretched chromosomes. The ability for stretched DNA to recoil in vivo is analogous to a histone-dependent folding mechanism where stretched nucleosomal DNA is placed under tension and then undergoes a reversible repacking (Cui and Bustamante 2000; Poirier et al. 2000).
Figure 7.
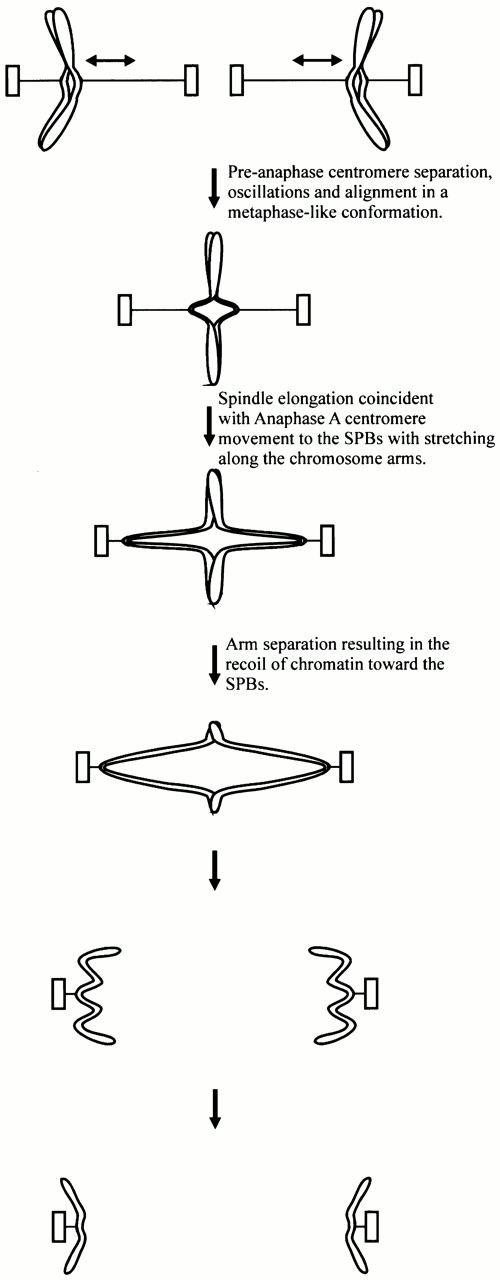
Model of chromosome oscillations, alignment, and segregation.
Centromere-specific and spindle pole body fluorescent probes used in combination with sensitive high temporal resolution imaging techniques allowed us to characterize the structural and dynamic features of chromosome movements during metaphase and anaphase in S. cerevisiae. The major features of these movements are similar to mitotic chromosome movements in mammalian tissue cells making budding yeast a useful tool to study the fundamental molecular mechanisms that produce and regulate chromosome motility and segregation.
Acknowledgments
We thank E. Yeh, D. Thrower, and M. Karthikayen for stimulating discussions. We thank E. Yeh, D. Thrower, D. Beach, L. Topper, and K. Shannon for helpful and critical comments on the manuscript. We thank the Cell Division Group at the Marine Biological Laboratory, Woods Hole, MA, for helpful discussion and laboratory space.
This work was supported by National Institutes of Health grant GM 32238 to K.S. Bloom and GM 24364 to E.D. Salmon.
Footnotes
Abbreviations used in this paper: CFP, cyan fluorescent protein; Cse4p, centromere-specific histone; GFP, green fluorescent protein; lacI, lac repressor; YFP, yellow fluorescent protein.
References
- Chen X., Yin H., Huffaker T. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J. Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Baker R.E., Keith K.C., Harris K., Stoler S., Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher order structure. Proc. Natl. Acad. Sci. USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J.T., Lutter L.C., Rhodes D., Brown R.S., Rushton B., Levitt M., Klug A. Structure of nucleosomal core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Goshima G., Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P.K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Inoue S., Salmon E.D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortiz J., Lechner J., Shevchenko A., Shevchenko A., Magiera M.M., Schramm C., Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO (Eur. Mol. Biol. Organ.). 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana J.A., Schnapp B.J., Silver P.A. Kinetics of spindle pole body separation in budding yeast. Proc. Natl. Acad. Sci. USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C.L. Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J. Cell Biol. 1996;135:315–327. doi: 10.1083/jcb.135.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubai, D.F. 1975. Mitosis and fungal phylogeny. Nuclear Division in the Fungi. I.B. Heath, editor. 177–229.
- Maddox P., Bloom K., Salmon E.D. Polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae . Nat. Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T., Ginkel L.M., Hunter A.W., Wordeman L. The kinetochore of higher eucaryotesa molecular view. Int. Rev. Cyt. 2000;194:67–131. doi: 10.1016/s0074-7696(08)62395-5. [DOI] [PubMed] [Google Scholar]
- McIntosh J.R. Structural and mechanical control of mitotic progression. Cold Spring Harbor Symp. Quant. Biol. 1991;56:613–619. doi: 10.1101/sqb.1991.056.01.070. [DOI] [PubMed] [Google Scholar]
- Meluh P.B., Yang P., Glowczewski L., Koshland D., Smith M.M. cse4p is a component of the core centromere of Saccharomyces cerevisiae . Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Nicklas, R.B. 1977. Chromosome movements: facts and hypotheses. Mitosis Facts and Questions. M. Little, N. Paweletz, C. Petzelt, H. Ponstingl, D. Schroeter, and H.-P. Zimmerman, editors. 150–155.
- Nicklas R.B. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Nicklas R.B., Arana P. Evolution and the meaning of metaphase. J. Cell Sci. 1992;102:681–690. doi: 10.1242/jcs.102.4.681. [DOI] [PubMed] [Google Scholar]
- Nicklas R.B., Koch C.A. Chromosome micromanipulation III. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol. 1969;43:40–50. doi: 10.1083/jcb.43.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne M.A., Schlenstedt G., Jinks T., Silver P.A. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J. Cell Biol. 1994;125:853–866. doi: 10.1083/jcb.125.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole E.T., Winey M. The spindle cycle in budding yeast. Nat. Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- O'Toole E.T., Winey M., McIntosh J.R. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae . Mol. Biol. Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J.B., Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae . J. Cell Sci. 1976;22:219–242. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J.D., Tippit D.H. The diatom spindle in perspective. Cell. 1978;14:455–467. doi: 10.1016/0092-8674(78)90232-5. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J.D., Tippit D.H., Porter K.R. Rethinking mitosis. Cell. 1982;29:729–744. doi: 10.1016/0092-8674(82)90435-4. [DOI] [PubMed] [Google Scholar]
- Pidoux A.L., Allshire R.C. Centromeresgetting a grip of chromosomes. Curr. Opin. Cell Biol. 2000;12:308–319. doi: 10.1016/s0955-0674(00)00094-6. [DOI] [PubMed] [Google Scholar]
- Poirier M., Eroglu S., Chatenay D., Marko J.F. Reversible and irreversible unfolding of mitotic newt chromosomes by applied force. Mol. Biol. Cell. 2000;11:269–276. doi: 10.1091/mbc.11.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S.L., Yeh E., Bloom K., Salmon E.D. Imaging green fluorescent protein fusion proteins in Saccharomyces cerevisiae . Curr. Biol. 1997;7:701–704. doi: 10.1016/s0960-9822(06)00299-5. [DOI] [PubMed] [Google Scholar]
- Shelby R.D., Hahn K.M., Sullivan K.F. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V., Skeen V.P., Salmon E.D. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cellsa push–pull mechanism. J. Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V., Rieder C.L., Salmon E.D. Kinetochore motility after severing between sister centromeres using laser microsurgeryevidence that kinetochore directional instability and position is regulated by tension. J. Cell Sci. 1995;108:2537–2548. doi: 10.1242/jcs.108.7.2537. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Belmont A.S., Robinett C.C., Murray A.W. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr. Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Marshall W.F., Murray A.W. Mitosis in living budding yeastanaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Sedat J.W., Murray A.W. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Fuchs J., Loidl J., Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Tippit D.H., Pickett-Heaps J.D., Leslie R. Cell division in two large pennate diatoms Hantzschia and Nitzschia IIIa new proposal for kinetochore function during prometaphase. J. Cell Biol. 1980;86:402–416. doi: 10.1083/jcb.86.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J.S., O'Toole E.O., Berrueta L., Bierer B.E., Pellman D. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Waters J., Chen R., Murray A., Salmon E.D. Localization of Mad2 to kinetochores depends on microtubule attachement, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J.C., Mitchison T.J., Rieder C.L., Salmon E.D. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell. 1996;7:1547–1558. doi: 10.1091/mbc.7.10.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Kilmartin J.V. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Mamay C.L., O'Toole E.T., Mastronarde D.N., Giddings T.H., Jr., McDonald K.L., McIntosh J.R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Skibbens R.V., Cheng J.W., Salmon E.D., Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae . J. Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]