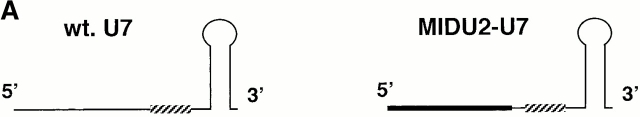Abstract
U2 small nuclear (sn)RNA contains a large number of posttranscriptionally modified nucleotides, including a 5′ trimethylated guanosine cap, 13 pseudouridines, and 10 2′-O-methylated residues. Using Xenopus oocytes, we demonstrated previously that at least some of these modified nucleotides are essential for biogenesis of a functional snRNP. Here we address the subcellular site of U2 internal modification. Upon injection into the cytoplasm of oocytes, G-capped U2 that is transported to the nucleus becomes modified, whereas A-capped U2 that remains in the cytoplasm is not modified. Furthermore, by injecting U2 RNA into isolated nuclei or enucleated oocytes, we observe that U2 internal modifications occur exclusively in the nucleus. Analysis of the intranuclear localization of fluorescently labeled RNAs shows that injected wild-type U2 becomes localized to nucleoli and Cajal bodies. Both internal modification and nucleolar localization of U2 are dependent on the Sm binding site. An Sm-mutant U2 is targeted only to Cajal bodies. The Sm binding site can be replaced by a nucleolar localization signal derived from small nucleolar RNAs (the box C/D motif), resulting in rescue of internal modification as well as nucleolar localization. Analysis of additional chimeric U2 RNAs reveals a correlation between internal modification and nucleolar localization. Together, our results suggest that U2 internal modification occurs within the nucleolus.
Keywords: snRNP, Sm binding site, modified nucleotide, snoRNA, localization
Introduction
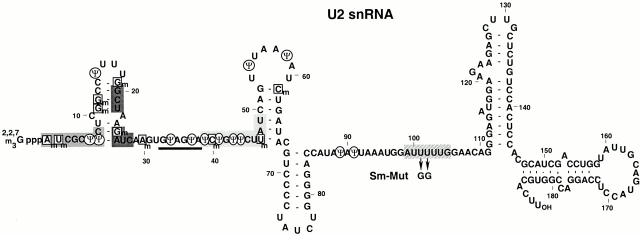
The spliceosomal small nuclear (sn)RNAs (U1, U2, U4, U5, and U6), which are essential for pre-mRNA splicing, are all posttranscriptionally modified (Reddy and Busch 1988; Massenet et al. 1998). Aside from 5′ cap trimethylation, numerous internal nucleotides are pseudouridylated or 2′-O-methylated. U2 is the most extensively modified of all the spliceosomal snRNAs. There are 10 2′-O-methylated residues and 13 pseudouridines in the Xenopus U2 snRNA (Fig. 1; Yu et al. 1998).
Figure 1.
Primary and secondary structure of Xenopus U2 snRNA. Internal modified nucleotides are highlighted by circles (pseudouridines) and squares (2′-O-methylated residues). The Sm binding site is indicated by a hatched gray box. Sequences known or predicted to be involved in intermolecular interactions are indicated: the thick line denotes bases interacting with the branch site of pre-mRNA; sequences boxed in dark gray, medium gray, and light gray are involved in U2-U6 interactions called Helix I, Helix II, and Helix III, respectively (Nilsen 1998). The arrows indicate mutations introduced into the Sm binding site (Sm-Mut).
The biogenesis of U2 and the other spliceosomal snRNPs has been extensively studied, principally using Xenopus oocytes (Mattaj 1988; Mattaj and Englmeier 1998). With the exception of U6, all major spliceosomal snRNAs are transcribed by RNA polymerase (Pol) II in response to specific promoter and termination signals (Dahlberg and Lund 1988). These Pol II transcripts are rapidly transported to the cytoplasm, where the RNAs are trimmed at their 3′ ends and a common set of proteins called the “core” Sm proteins are assembled onto the conserved Sm binding sequence present in each of these snRNAs (Mattaj and De Robertis 1985; Luhrmann 1988). Upon binding of the Sm proteins, the 5′-monomethylated guanosine cap becomes 2,2,7-trimethylated, an event that is sufficient for recognition by the protein snurportin 1, which facilitates the transport of these RNA-protein particles back to the nucleus (Huber et al. 1998). In addition to the common Sm proteins, each mature Sm snRNP contains specific proteins, the number of which varies between snRNPs and can be quite large (Luhrmann 1988; Behrens et al. 1993; Gottschalk et al. 1998, Gottschalk et al. 1999). Two distinct U2 snRNPs have been observed in nuclear extracts from HeLa cells (Behrens et al. 1993) and Xenopus oocytes (Yu et al. 1998), corresponding to 12S or 17S particles. The 12S particle contains all the core Sm proteins and two U2-specific proteins, whereas the 17S particle possesses at least nine additional loosely associated U2-specific proteins (Behrens et al. 1993). Seven of these nine proteins are the subunits of splicing factor 3 (SF3; Krämer 1996). Using microinjection into the Xenopus oocyte, we recently demonstrated that the modified nucleotides within the 5′ 27 nucleotides of U2 are essential for conversion of the 12S particle to the mature 17S particle (Yu et al. 1998).
Although it is clear that U2 and other Sm snRNAs undergo Sm protein binding and 5′ cap hypermethylation in the cytoplasm, it is not known where within the cell these Sm snRNAs are internally modified. It is established that the nucleolus harbors a large number of small nucleolar (sno)RNPs responsible for pseudouridylation and 2′-O-methylation of Pol I–transcribed ribosomal RNAs (Maxwell and Fournier 1995; Smith and Steitz 1997; Tollervey and Kiss 1997; Yu et al. 1999). Recent studies indicate that 2′-O-methylation of U6 snRNA, a Pol III transcript, is likewise mediated by snoRNPs (Tycowski et al. 1998; Ganot et al. 1999). Intranuclear localization studies indicate that the U6 snRNA travels through the nucleolus (Lange and Gerbi 2000; Narayanan, A., R.M. Terns, and M.P. Terns, unpublished data). The striking similarities in 2′-O-methylation between ribosomal RNAs (rRNAs) and the U6 snRNA raises the possibility that internal modification of Pol II–transcribed U2 and other Sm snRNAs also occurs in the nucleolus. In this study, we use the Xenopus oocyte microinjection system to show that internal modification of U2 (a product of Pol II transcription) occurs exclusively in the nucleus, and likely within the nucleolus.
Materials and Methods
Preparation of U2 Mutant and Chimeric RNAs
PCR was used to generate the Sm-mutant U2 construct, starting with a plasmid containing the wild-type U2 sequence (Yu et al. 1998). First, a 5′ and a 3′ Sm-mutated U2 fragment were produced using two pairs of DNA oligonucleotides. One pair (the 5′ primer [primer 1], 5′-TAATACGACTCACTATAG-3′, corresponding to the T7 promoter sequence located immediately upstream of the 5′ terminus of U2, and the 3′ primer [primer 2], 5′-CCTGTTCCAACCATCCATTTAATATATGG-3′, complementary to nucleotides 84–112 of Xenopus U2 except for two mismatches [underlined] in the second and third nucleotides of the Sm binding site) was used to amplify a 5′ half U2 fragment. The other pair (the 5′ primer [primer 3], 5′-TAAATGGATGGTTGGAACAGGGAG-3′, corresponding to nucleotides 92–115 of Xenopus U2, except for two changes in the second and the third nucleotides of the Sm binding site [underlined], and the 3′ primer [primer 4], 5′-GGGAAGTGCACCGGTCCTGGAGGT-3′, containing three G's 5′ to a 21–nucleotide sequence complementary to the 3′ end of Xenopus U2) was used to create a 3′ half U2 fragment. These two half DNA PCR products (containing an overlapping sequence, base pairs 92–112 with respect to the U2 sequence) were then mixed with primer 1 and primer 4 for a new round of PCR amplification, generating a mutant U2 construct with the second and the third U's of the Sm binding site changed to G's. The final PCR product was cloned into the SmaI site of pGEM-3Z vector (Promega) and sequenced. After linearization with SmaI, the plasmid was used as a template for GpppG-primed (for wild-type U2, ApppG was also used) transcription by T7 RNA polymerase.
To generate U2-C/D motif chimeric RNAs, plasmids containing the wild-type or the Sm-mutant U2 sequence were used as templates for PCR. The 5′ primer, 5′-CCTAATACGACTCACTATAGGATATATATAATTTATGATCACGGTGATGAAATCGCTTCTCGGCC-TTTTG-3′, corresponded to the T7 promoter sequence, the 5′ 32 nucleotides of the yeast U14 snoRNA (including the terminal stem and box C sequences [Samarsky et al. 1998]), and the 5′ 19 nucleotides of U2. The 3′ primer, 5′-ATATATATAGTATACGATCACTCAGACAAGTGCACCGGTCCTGGAGG-3′, was complementary to the 3′ 27 nucleotides of yeast U14 (including the terminal stem and box D sequences) and the 3′ 20 nucleotides of Xenopus U2 RNA. After PCR, the DNA product was gel purified and used as a template for in vitro T7 RNA polymerase transcription.
To create the MID U2-U7 chimera, two oligodeoxynucleotides were used. The 5′ primer, 5′-CCTAATACGACTCACTATAGGCTAAGATCAAGTGTAGTATCTGTTCTCTATTTGTCTAGCAGG-3′, corresponded to the T7 promoter sequence, nucleotides 19–46 of Xenopus U2, and nucleotides 19–34 of Xenopus U7 snRNA. The 3′ primer, 5′-TGTGGCTCCTACAGAGTAAGAACCTGCTAGACAAATAG-3′, was complementary to the 3′ 38 nucleotides of Xenopus U7 snRNA. These two primers have a complementary sequence, nucleotides 19–34 with respect to the Xenopus U7 snRNA. After annealing, both strands were extended with Klenow DNA polymerase. The double-stranded DNA product served as a template for in vitro transcription of MID U2-U7 chimera by T7 RNA polymerase.
Anti-Sm (Y12) Immunoprecipitation
Immunoprecipitation with anti-Sm (Y12) antibodies was performed according to Lerner et al. 1981 and Yu et al. 1998. In brief, after injection of labeled U2 RNAs, nuclei were broken by sonication in Net-2 buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% NP-40). The nuclear suspension was clarified by centrifugation at 14,000 g for 5 min. The supernatant was mixed with Y-12 antibodies at 4°C for 2 h. After centrifugation, the pellet was washed three times with Net-2 buffer and digested with proteinase K. RNAs were recovered by phenol-chloroform-isoamylalcohol extraction and ethanol precipitation.
Xenopus Oocyte Microinjection and RNA Modification Assays
The procedures for microinjection and modification assays were essentially as described previously (Yu et al. 1998). In brief, various U2 RNAs (differentially capped wild-type U2, Sm-mutant U2, U2-C/D motif chimeras, and the U2-U7 chimera) uniformly labeled with α[32P]UTP (106 cpm/μl) were individually injected into Xenopus oocytes (32 nl for cytoplasmic injection, and 9 nl for nuclear injection [in most cases, 9 nl of U2 RNA constructs was directly injected into isolated nuclei under oil]). After 5 h at room temperature, RNAs were recovered from the nuclei or cytoplasm and assayed for modifications.
To assay pseudouridylation, RNAs were digested with nuclease P1 in 3 μl of 20 mM sodium acetate, pH 5.2, at 37°C for 1 h. The digested samples were analyzed on cellulose TLC PEI plates (Baker; Patton 1991). To determine the site specificity of pseudouridylation, endogenous U2 was first depleted from oocytes by cytoplasmic injection of an antisense U2 DNA oligonucleotide complementary to nucleotides 28–42 of U2 (Yu et al. 1998). Unlabeled U2 RNAs were then injected (∼1 ng/oocyte). Recovered RNAs were modified with 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate (CMC), treated with sodium bicarbonate buffer, pH 10.4, and analyzed by reverse transcription (Bakin and Ofengand 1993). Stops caused by pseudouridine-CMC modification were visualized on 6% sequencing gels. To analyze pseudouridylation of U2-C/D motif chimeras, a reverse transcription primer complementary to nucleotides 52–71 of U2 was used.
To assay 2′-O-methylation, labeled RNA preparations were site-specifically cleaved by RNase H directed by 2′-O-methyl RNA–DNA chimeras (5′-CmCmAmAmAmAmGmGmdCdCdGdAGmAmAmGmCmGmAmUm-3′, for position 11 of U2, and 5′-AmCmAmdCdTdTdGAmUmCmUmUmAmGmCm-CmAmAmAmAm-3′ for position 30 of U2). Cleaved and uncleaved U2 RNA constructs were analyzed on 6% denaturing gels (Yu et al. 1997, Yu et al. 1998).
Analysis of the Intranuclear Distribution of U2 snRNA and Variants
The procedure used to prepare Xenopus oocyte nuclear spreads and to determine the intranuclear distrution of injected fluorescein-labeled RNAs has been described (Narayanan et al. 1999). In brief, fluorescein-labeled RNAs were generated by in vitro transcription of linearized plasmid DNA in the presence of α[32P]GTP and fluorescein-12-UTP (1:1 ratio with UTP; Boehringer). The fluorescein-labeled RNAs (1 fmol each) were injected into the nuclei of stage V or stage VI oocytes. 32P-labeled U6 snRNA (retention control), U1 snRNA, and tRNA (export controls) were coinjected with each fluorescein-labeled test RNA to help make conclusions about the stability and nucleocytoplasmic distribution of wild-type U2 snRNA and U2 variants. The injected oocytes were incubated at 18°C for 5 h before nuclei were dissected and nuclear spreads were prepared. Images were obtained on a 63× magnification inverted fluorescence microscope (Axiovert S 100; ZEISS) equipped with differential interference contrast (DIC) optics (Thornwood) using a cooled charge-coupled device camera (Quantix-Photometrix) and IPLab Spectrum software (Signal Analytics). Localization analysis was performed using multiple independent sets of RNAs and oocytes, and at least four slides per time point. Representative data are shown (see Fig. 4). To determine the stability and nucleocytoplasmic distribution of the injected RNAs, RNAs present in the nuclear (N) and cytoplasmic (C) fractions were purified 5 h after injection (from the same set of oocytes analyzed by microscopy) and analyzed by 8% denaturing PAGE and autoradiography.
Figure 4.
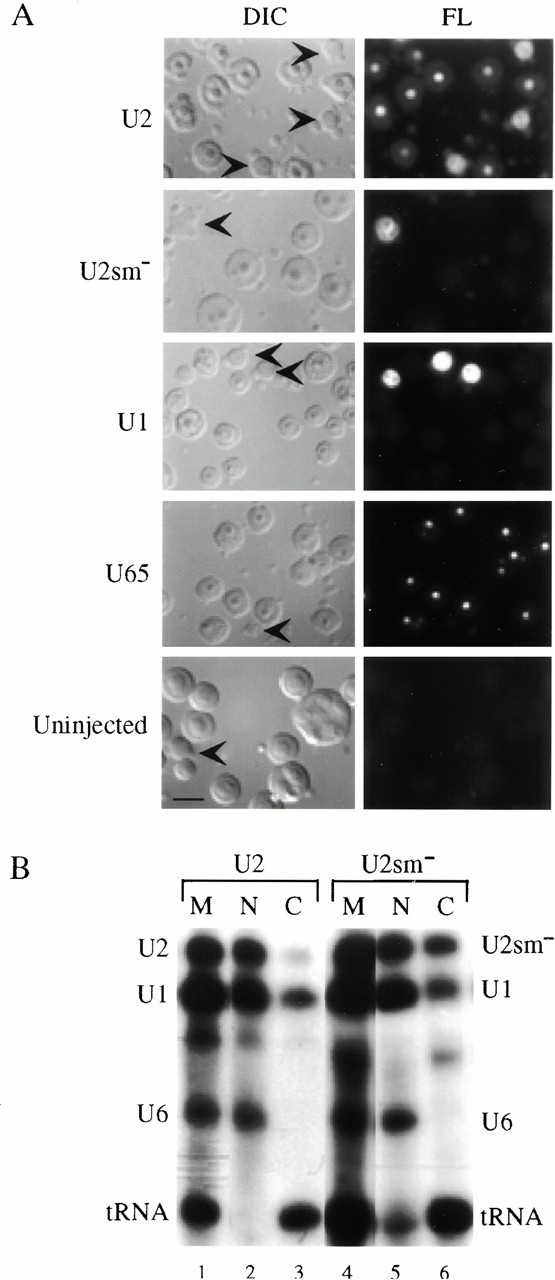
(A) Intranuclear localization of wild-type U2 and Sm-mutant U2 (U2sm−) snRNAs. 1 fmol of 32P- and fluorescently labeled, in vitro–transcribed U2, Sm-mutant U2, U1, or U65 was injected into Xenopus oocyte nuclei. Nuclear spreads were prepared 5 h later. U1 was included as a positive control for Cajal body localization and as a negative control for nucleolar localization (Narayanan et al. 1999). U65 snoRNA served as a positive control for nucleolar localization and as a negative control for Cajal body localization (Narayanan et al. 1999). A nuclear spread prepared from an uninjected oocyte was included to control for background fluorescence of the preparations. The nuclear spreads were analyzed by DIC and fluorescence (FL) microscopy. Each panel includes several nucleoli and a few Cajal bodies. Cajal bodies are indicated by arrowheads in the DIC panels. (B) Nucleocytoplasmic distribution of U2 and Sm-mutant U2 (U2sm−) snRNAs. Injected oocytes from the same batch analyzed above were dissected into nuclear and cytoplasmic fractions after 5 h. Samples were analyzed by denaturing PAGE and autoradiography to determine the stability and nucleocytoplasmic distribution of the injected RNAs. tRNA was used as a positive control for export and U6 served as a nuclear retention control (Narayanan et al. 1999; Speckmann et al. 1999). Nuclear (N) RNAs are in lanes 2 and 5, cytoplasmic (C) RNAs are in lanes 3 and 6, and marker (M) lanes 1 and 4 show RNAs before injection. Bar, 10 μm.
Results
U2 Modification Occurs Exclusively in the Nucleus
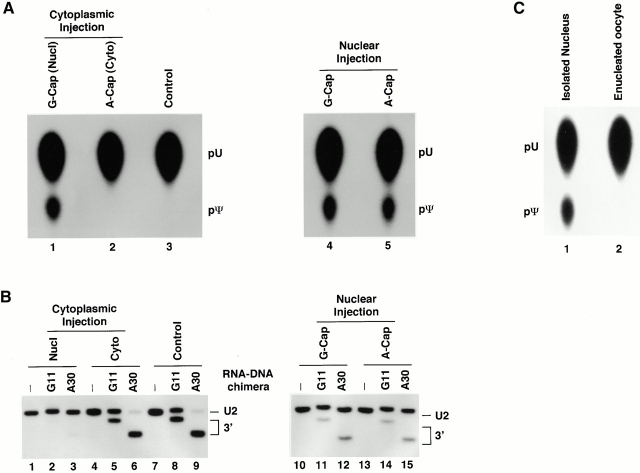
Given the fact that U2 snRNA cycles through the cytoplasm before returning to the nucleus, we first tested whether the internal modification (pseudouridylation and 2′-O-methylation) occurs in the nucleus and/or in the cytoplasm. In these experiments, we exploited the fact that after injection into the cytoplasm of Xenopus oocytes, GpppG-capped U2 is hypermethylated and rapidly transported into the nucleus, whereas ApppG-capped U2 is not trimethylated and remains in the cytoplasm (Fischer et al. 1991). 5 h after injection of differentially capped, uniformly α[32P]UTP-labeled U2 snRNA into the cytoplasm, the cytoplasm and the nucleus were manually separated. The RNA in each cellular compartment was assayed for pseudouridylation by complete nuclease P1 digestion followed by TLC analysis (Fig. 2 A) and for 2′-O-methylation by RNase H site-specific cleavage directed by 2′-O-methyl RNA–DNA chimeras (Fig. 2 B; Yu et al. 1997, Yu et al. 1998). The G-capped U2 recovered from nuclei was efficiently pseudouridylated (Fig. 2 A, lane 1) and 2′-O-methylated (Fig. 2 B, lanes 1–3). In contrast, the A-capped U2 recovered from the cytoplasm was not detectably modified (Fig. 2A and Fig. B, lanes 2 and 4–6, respectively). To ensure that the cap structure does not affect the ability to serve as a substrate for internal modification, we injected both A-capped and G-capped U2 RNA directly into nuclei and found that both were modified to a comparable extent (Fig. 2 A, compare lanes 4 and 5, and 2 B, compare lanes 10–12 and 13–15).
Figure 2.
U2 internal modifications occur exclusively in the nucleus. α[32P]UTP uniformly labeled G- or A-capped U2 was injected into the cytoplasm of oocytes (A, lanes 1 and 2; B, lanes 1–6) or directly into nuclei (A, lanes 4 and 5; B, lanes 10–15). After cytoplasmic injection, only G-capped U2 entered the nucleus; after nuclear injection, both G- and A-capped U2 RNAs were retained in the nucleus. RNAs recovered from nuclei (A, lanes 1, 4, and 5; B, lanes 1–3, 10–15) or from cytoplasm (A, lane 2; B, lanes 4–6) were assayed for pseudouridylation (A) and 2′-O-methylation (B). In the 2′-O-methylation assay, two different chimeras were used to test two positions (G11 and A30). The control is uninjected U2 RNA. (C) Oocyte nuclei were separated from the cytoplasm under oil. α[32P]UTP uniformly labeled U2 was then injected into the isolated nuclei (lane 1) or enucleated oocytes (lane 2). After a 5-h incubation, U2 RNAs were recovered and assayed for pseudouridylation. The positions of uridylate, pseudouridylate, uncleaved U2, and 3′ fragments generated by RNase H site-specific cleavage are indicated on the side of each gel. In these experiments, >80% of the expected level of pseudouridylation and 2′-O-methylation was observed.
To confirm these results, we physically separated oocyte nuclei from the cytoplasm under oil (Lund and Paine 1990) and then injected U2 snRNA containing either the A or G cap directly into the isolated nuclei or the enucleated oocytes. Again, only U2 RNA recovered from isolated nuclei, not from the enucleated oocytes, was pseudouridylated (Fig. 2 C, compare lanes 1 and 2) and 2′-O-methylated (data not shown). To ensure that the enucleated oocytes were functional, we conducted anti-Sm and anti–trimethyl guanosine cap immunoprecipitation experiments. Our data showed that both G- and A-capped U2 snRNAs injected into the cytoplasm were able to assemble into immunoprecipitable Sm snRNPs and that G-capped U2 snRNA became 5′ trimethylated (data not shown). We conclude that pseudouridylation and 2′-O-methylation occur exclusively in the nucleus and do not require prior 5′ cap trimethylation.
The Sm Binding Site of U2 snRNA Is Required for Internal Modification
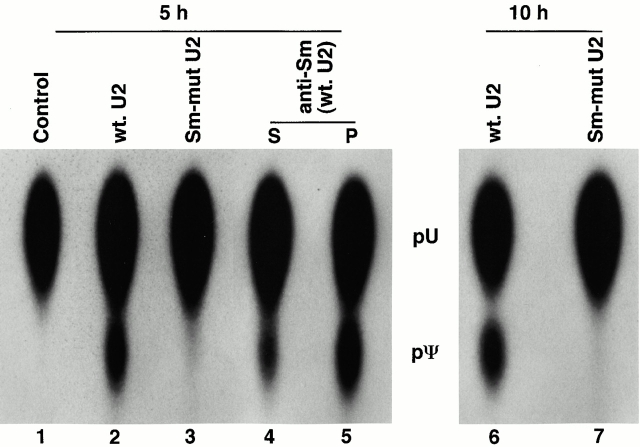
The Sm binding site is critical for U2 snRNP biogenesis (Luhrmann 1988; Mattaj 1988) and is located in the 3′ half of the RNA, where essentially no modified nucleotides are present. We mutated this element by changing the second and the third uridines in the Sm binding site to guanosines (Fig. 1). 5 h after injection of RNA that was uniformly labeled with α[32P]UTP into isolated nuclei, total RNAs were recovered and modification assays were performed. Although the wild-type U2 was pseudouridylated (Fig. 3, lane 2) and 2′-O-methylated (data not shown), no modified nucleotides were detected in the Sm-mutant RNA (Fig. 3, lane 3 and data not shown), even after a prolonged (10-h) incubation (Fig. 3, lane 7), suggesting that the Sm site is required for internal modification. The Sm site is important as a site for binding of Sm proteins, which normally occurs in the cytoplasm; however, assembly of the Sm RNP can occur in the nucleus under some conditions, apparently via exchange of proteins with endogenous snRNPs (Terns and Dahlberg 1994; Terns et al. 1995). Assembly of the injected wild-type U2 into particles in our experiments was confirmed by anti-Sm immunoprecipitation: ∼50% of injected RNAs were immunoprecipitated (Fig. 3, compare lanes 4 and 5). The U2 RNA bound by Sm proteins was pseudouridylated (Fig. 3, lane 5), whereas only a trace amount of pseudouridylate was detected in the supernatant (unbound) fraction (Fig. 3, lane 4), presumably because of incomplete immunoprecipitation. We conclude that the Sm binding site, and likely the binding of Sm proteins, is required for U2 internal modification.
Figure 3.
Binding of Sm proteins is required for U2 internal modification. [α32P]UTP uniformly labeled wild-type (lanes 2, 4, 5, and 6) or Sm-mutant U2 (lanes 3 and 7; see Fig. 1) was injected into isolated nuclei under oil. After 5 h (lanes 1–5) or 10 h (lanes 6 and 7) at room temperature, RNAs were recovered by phenol-chloroform-isoamylalcohol extraction and ethanol precipitation and subjected to the pseudouridylation assay (lanes 1–3, 6, and 7). For the wild-type U2, ∼40% (lane 2, 5 h) or 50% (lane 6, 10 h) of the expected level of modification was observed. Alternatively, wild-type U2 was recovered by anti-Sm (Y-12 antibody) immunoprecipitation after 5 h at room temperature and then subjected to the pseudouridylation assay (lanes 4 and 5). More than 85% of the expected level of modification was observed in immunoprecipitated RNA (lane 5). S, supernatant; P, pellet. The control is an uninjected U2 RNA.
Wild-Type U2, but Not Sm-mutant U2, Localizes to the Nucleolus
The Sm binding site could contribute to U2 internal modification in several ways. For instance, mutation of the Sm binding site could directly or indirectly impair recognition of the snRNA by the modifying machinery. Alternatively, the nucleus contains several subcompartments (e.g., nucleoplasm, nucleoli, Cajal bodies, gems, speckles, and snurposomes) and the binding of Sm core proteins could target the U2 snRNA to nuclear subcompartments where internal modification occurs. Alteration of the Sm binding site resulting in mislocalization would thus affect modification.
We analyzed the intranuclear distribution of fluorescently labeled RNAs to pinpoint the location of the wild-type and the Sm-mutant U2 within the nucleus. Fluorescently and radio-labeled U2 RNAs, along with radiolabeled control RNAs (U1 snRNA, U6 snRNA, and tRNA), were injected into oocyte nuclei and the intranuclear distribution of U2 was analyzed by fluorescence microscopy of nuclear spreads prepared 5 h after injection (Fig. 4 A). We also extracted the RNAs from the nucleus and cytoplasm and analyzed their nucleocytoplasmic distribution and stability by gel electrophoresis (Fig. 4 B). The wild-type U2 snRNA was localized to nucleoli and Cajal bodies (Fig. 4 A, U2). The U2 nucleolar signal was less than that observed for U65 snoRNA, but it was significantly greater than the level observed for U1 snRNA and the background fluorescence of uninjected oocytes (Fig. 4 A, compare U2, U1, U65, and uninjected). In contrast, the Sm-mutant U2 RNA was observed only in Cajal bodies; no signal was detected in nucleoli (Fig. 4 A, U2sm−). Control RNAs (U1, U6, and tRNA) behaved as expected (Fig. 4 B). The correlation between nucleolar localization and efficient modification suggests that U2 internal modifications take place in the nucleolus.
Nucleolar Targeting of Sm-mutant U2 Restores Internal Modification
The observation that a double point mutation in the Sm binding site resulted both in a loss of nucleolar localization and internal modification of U2 RNA (Sm-mutant U2) prompted us to test whether targeting of this RNA to nucleoli can restore internal modification. Therefore, we introduced a well-characterized nucleolar localization signal, the snoRNA Box C/D motif (containing boxes C and D and the terminal stem; Samarsky et al. 1998; Narayanan et al. 1999; Speckmann et al. 1999), into Sm-mutant U2 as well as into wild-type U2 RNA. Specifically, the 5′-terminal sequence of U14 snoRNA, including box C and one strand of the terminal stem, was fused to the 5′ terminus of U2, while the 3′-terminal sequence of U14, including box D and the other strand of the terminal stem, was linked to the 3′ terminus of U2 (Fig. 5 A). In addition, the 5′ half (nucleotides 1–100) and the 3′ half (101–188) of U2 were each inserted into the same box C/D motif (Fig. 5 A). These chimeric RNAs were injected into isolated nuclei, incubated for 5 h, and assayed for modification.
Figure 5.
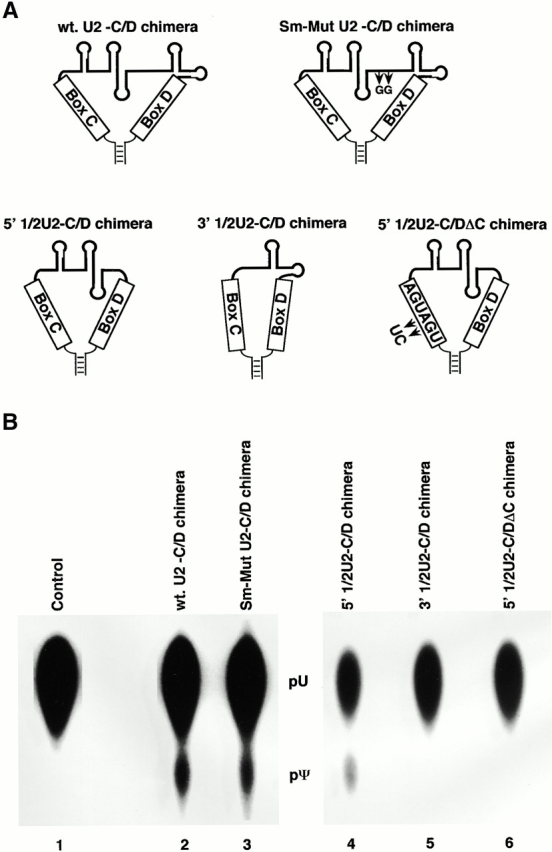
Introduction of a snoRNA nucleolar localization signal restores internal modification of Sm-mutant U2. (A) Structures of the wild-type U2-C/D motif chimera, the Sm-mutant U2-C/D motif chimera, the 5′ 1/2U2-C/D motif chimera, the 3′ 1/2U2-C/D motif chimera, and the 5′ 1/2U2-C/DΔC motif chimera, each containing a nucleolar localization signal derived from U14 snoRNA (box C, box D, and a short terminal stem), are shown schematically. The two arrows in the Sm-mutant U2-C/D chimera depict the double point mutation in the Sm binding site. The two arrows in 5′ 1/2U2-C/DΔC indicate the sequence alterations in box C. (B) [α32P]UTP uniformly labeled wild-type U2-C/D motif chimera (lane 2), Sm-mutant U2-C/D motif chimera (lane 3), 5′ 1/2U2-C/D motif chimera (lane 4), 3′ 1/2U2-C/D motif chimera (lane 5), or 5′ 1/2U2-C/DΔC motif chimera (lane 6) was injected into isolated nuclei under oil. 5 h later, RNAs were recovered and assayed for modification. 40–50% of the calculated level of modification was observed in lanes 2–4. The positions of uridylate and pseudouridylate are indicated.
Remarkably, the Sm-mutant U2-C/D motif chimera not only became modified (Fig. 5 B, lane 3), but was modified to a level equivalent to that of the wild-type U2-C/D motif chimera (Fig. 5 B, lane 2). As expected, since all the internally modified nucleotides are located exclusively in the 5′ half of U2, the 5′ half U2-C/D motif chimera was efficiently modified (lane 4). No modification was detected in the 3′ half U2-C/D motif chimera (lane 5). Modification was dependent on the C/D motif, as changes in the box elements completely abolished modification (lane 6, and data not shown). The accuracy of the restored modifications was confirmed by RNase H site-specific cleavage directed by 2′-O-methyl RNA–DNA chimeras (for 2′-methylation; Yu et al. 1997, Yu et al. 1998) and by CMC modification followed by reverse transcription (for pseudouridylation, see Materials and Methods; Bakin and Ofengand 1993; data not shown).
Visual evidence of nucleolar localization conferred on these chimeric RNAs by the box C/D motif was obtained by fluorescence microscopy using nuclear spreads prepared 5 h after injection of fluorescently labeled chimeric RNAs into oocyte nuclei. Fig. 6 shows that introduction of the C/D motif indeed resulted in targeting of the Sm-mutant U2-C/D motif chimera and the 5′ half U2-C/D motif chimera to nucleoli (U2sm−-CD and 5′ half U2-CD). The nucleolar localization of these chimeric RNAs was dependent on the presence of the C/D motif, as point mutations within the motif resulted in loss of localization to nucleoli but not Cajal bodies (Fig. 6, Fig. 5′ half U2-CDΔC). Introduction of the C/D motif into wild-type U2 did not alter its nucleolar targeting (compare Fig. 6, U2-CD and Fig. 4 A, U2).
Figure 6.
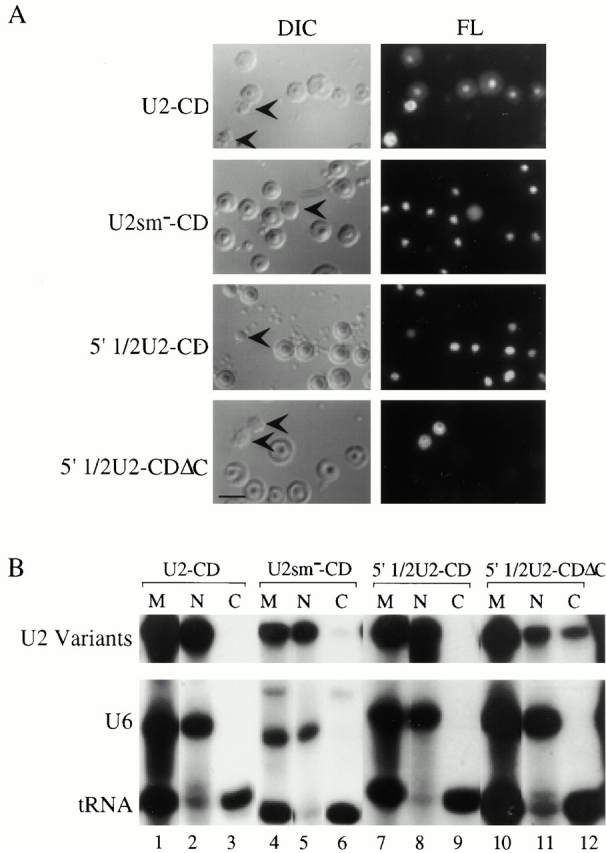
(A) Intranuclear localization of U2-C/D motif chimeras. 1 fmol of fluorescein-labeled wild-type U2-C/D motif chimera (U2+CD), Sm-mutant U2-C/D motif chimera (U2sm−-CD), 5′ 1/2U2-C/D motif chimera (5′ 1/2U2-CD), or 5′ 1/2U2-C/DΔC motif chimera (5′ 1/2U2+CDΔC) was injected into Xenopus oocyte nuclei, and nuclear spreads were prepared 5 h later. DIC and fluorescence (FL) panels are shown for each field. The arrowheads in the DIC panels indicate Cajal bodies. (B) Nucleocytoplasmic distribution of the U2-C/D motif chimeras. The distribution of the injected RNAs within the nuclear and cytoplasmic oocyte compartments was determined as described in the legend to Fig. 4 B. Bar, 10 μm.
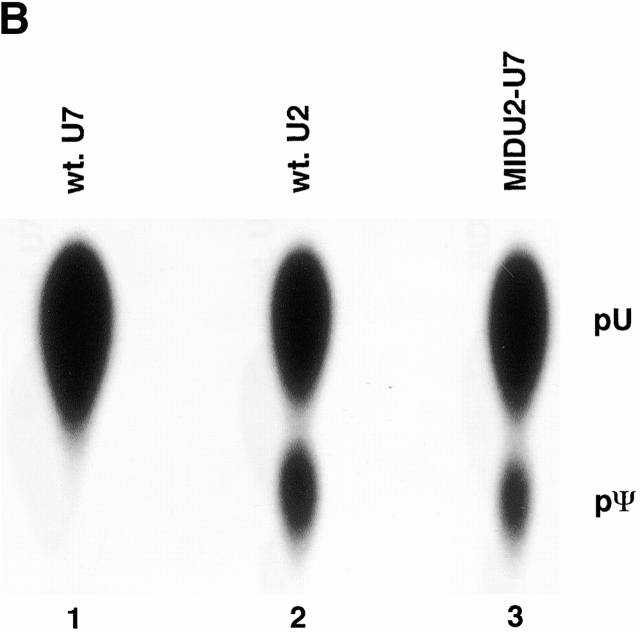
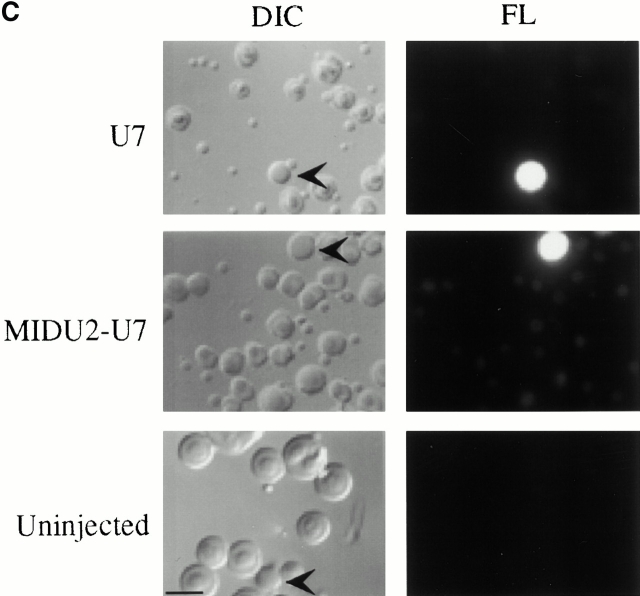
Although our observations suggest that the nucleolus is the site of U2 internal modification, U2 constructs that become efficiently modified are detected in both nucleoli and Cajal bodies. We attempted to assess any potential role of Cajal bodies in modification by fusing a 5′ fragment (nucleotides 19–46) of U2 RNA to a 3′ half sequence of U7 snRNA, which contains a distinct Sm binding site known to target U7 snRNAs to Cajal bodies (Wu et al. 1996). Surprisingly, this U2-U7 chimera became modified (Fig. 7 A, lane 3). As expected, the RNA localized to Cajal bodies (Fig. 7 B, MIDU2-U7). However, we also detected nucleolar signals that were weak but distinctly greater than background, suggesting that these U2-U7 chimeric constructs may transiently cycle through nucleoli for modification (see Discussion).
Figure 7.
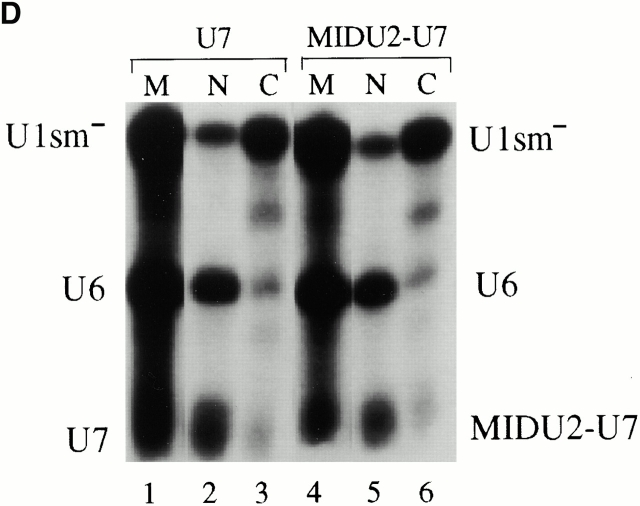
(A) Structures of Xenopus wild-type U7 snRNA and MIDU2-U7 chimera are shown schematically. The U7 Sm binding site is indicated by a hatched box. Thin lines represent the U7 RNA chain. In the MID U2-U7 chimera, the first 18 nucleotides of U7 are substituted with a Xenopus U2 sequence (nucleotides 19–46) indicated by a thicker line. (B) The U7 Sm binding site also supports U2 internal modification. [α32P]UTP uniformly labeled wild-type U7 (lane 1), U2-U7 chimera (MID U2-U7; lane 2), or wild-type U2 (lane 3) was injected into isolated nuclei under oil. 5 h later, RNAs were recovered and assayed for modification. The positions of uridylate and pseudouridylate are indicated. (C) Intranuclear localization of U7 and the U2-U7 chimera. 32P- and fluorescently labeled wild-type U7 and MID U2-U7 chimera were synthesized by in vitro transcription and 1 fmol of each RNA was microinjected into the nuclei of Xenopus oocytes. Nuclei were isolated 5 h later and nuclear spreads were prepared. Spreads were also prepared from uninjected oocytes as controls. DIC and fluorescence (FL) images are shown for each field. The arrowheads in the DIC panels point to Cajal bodies. Prominent Cajal body labeling was observed for both U7 and the MID U2-U7 chimera. Weak but above background nucleolar signal was also detected for the MID U2-U7 chimera. (D) The nucleocytoplasmic distribution of U7 and the U2-U7 chimera RNAs was determined as described in the legend to Fig. 4 B. Bar, 10 μm.
Discussion
Using the Xenopus oocyte microinjection system, we have demonstrated that U2 internal modification is a nuclear event. A combination of mutant analyses and fluorescence microscopy further showed that efficiently modified U2 RNA variants are localized to nucleoli (and Cajal bodies), but that RNAs that are not localized to nucleoli are not modified. Strikingly, a modification-deficient Sm-mutant U2 RNA can be converted to a modification proficient state by addition of an exogenous nucleolar localization signal (the box C/D motif). Our findings suggest that U2 internal modifications occur in the nucleolus.
Previous studies suggested that U2 internal modification might not occur in the nucleolus. Ganot et al. 1999 reported that a fragment of U2 was not modified when fused to an rRNA minigene and transcribed by RNA Pol I in the nucleolus. Although the studies were done using a different cell system (mouse versus Xenopus oocytes), two additional points should be considered. First, it is possible that the short U2 sequence (22 nucleotides), fused to a large rRNA sequence, formed a structure that prevented U2 from being accessed by the modifying machinery. Second, modification of U2 may require prior exposure to other nuclear environments, which might be forfeited when the RNA is synthesized in the nucleolus. In our experiments RNAs were injected into nuclei of oocytes, which may allow trafficking of the RNA in the nucleus and association with relevant nuclear factors or structures. In other work it was shown that pseudouridylation of position 44 of yeast U2 snRNA can be catalyzed by a yeast tRNA pseudouridylase, Pus1p (Massenet et al. 1999), which resides in the nucleoplasm (Simos et al. 1996). Several other known yeast pseudouridylases, including Cbf5p (NAP57), were tested and found to be negative (Massenet et al. 1999), suggesting that pseudouridylation of this site is not mediated by snoRNPs in the nucleolus in yeast. It is possible that internal modification of yeast Sm snRNAs is different from that of vertebrate Sm snRNAs and that modification of the other residues of yeast Sm snRNAs is catalyzed by snoRNPs in the nucleolus.
Do Cajal Bodies as Well as Nucleoli Harbor Machinery for Internal Modification of U2?
U7 snRNA is targeted to Cajal bodies by its Sm site (Wu et al. 1996), but is not detected in nucleoli (Fig. 7 C; Wu et al. 1996). We were surprised to find that when attached to the U7 Sm binding site, a 5′ fragment of U2 was efficiently modified (Fig. 7a and Fig. b). The U2-U7 chimera was targeted to Cajal bodies, but distinct nucleolar signals were also observed (Fig. 7 C). We favor the explanation that the U2-U7 chimeras are modified during a transient association with nucleoli. The work of several labs suggests that many types of RNAs progress through nucleoli and Cajal bodies during their maturation, even if they ultimately come to reside in other cellular subcompartments (Pederson 1998; for review see Olson et al. 2000). For instance, box C/D snoRNAs, including U3, U8, and U14, transiently associate with Cajal bodies before localization to nucleoli (Narayanan et al. 1999; Speckmann et al. 1999). On the other hand, U6 snRNA transiently localizes to nucleoli, (Lange and Gerbi 2000; Lukowiak, A. Narayanan, R.M. Terns, and M.P. Terns, submittedmanuscript for publication) presumably for modification mediated by box C/D snoRNAs (Tycowski et al. 1998; Ganot et al. 1999). Fibrillarin and NAP57, which are the presumed 2′-O-methylase and pseudouridine synthase components of the box C/D and box H/ACA snoRNPs, respectively, have been reported to be present in both nucleoli and Cajal bodies (Raska et al. 1991; Meier and Blobel 1994; Gall et al. 1999). The Lamond group has also shown that mammalian spliceosomal snRNPs accumulate in Cajal bodies, which, after treatment with a phosphatase inhibitor, coalesce with nucleoli (Lyon et al. 1997; Sleeman et al. 1998). The accumulation of spliceosomal snRNPs in Cajal bodies and nucleoli is a transient event that occurs before localization to speckles (Sleeman and Lamond 1999), suggesting that Cajal bodies and/or nucleoli may be the site(s) for spliceosomal snRNA modification (Bohmann et al. 1995a,Bohmann et al. 1995b; Sleeman and Lamond 1999). Our data indicate that internal modification of U2 snRNA occurs in the nucleolus, but does not exclude a role for Cajal bodies. Further analyses are needed to establish whether U2 internal modification also takes place within Cajal bodies.
Role of the Sm Binding Site in Intranuclear Trafficking
We have shown that the Sm binding site is required not only for U2 internal modification but also for nucleolar localization. A double point mutation in the U2 Sm binding site results in a modification defective phenotype and disruption of nucleolar targeting, even though a significant fraction of the RNA remained in nuclei (including Cajal bodies) at the time analyzed (Fig. 4). This observation suggests a role for the Sm binding site in nucleolar targeting.
Interestingly however, two other Sm snRNAs, U1 and U7, were detected in Cajal bodies but not nucleoli (Fig. 4 and Fig. 7). Why do these two Sm RNAs have different intranuclear localization profiles? If U2 is internally modified in the nucleolus, where are U1 and U7 modified? One explanation is that U1 and U7 may pass through nucleoli too quickly to generate detectable signals. The different nucleolar transit times of the RNAs could correlate with the number of modifications acquired by the various RNAs. U2 is modified at 23 internal sites (by pseudouridylation and 2′-O-methylation; Massenet et al. 1998; Reddy and Busch 1988) and is detected in the nucleolus (Fig. 4), whereas U1 is modified at only five sites (Reddy and Busch 1988; Massenet et al. 1998) and is not detected there (Fig. 4). Similarly, U7 snRNA contains no pseudouridylation (Fig. 7 B, lane 1) (it is unknown whether U7 is 2′-O-methylated) and is not detected in the nucleolus. Interestingly, we also detect a transient nucleolar localization of U6 snRNA (Lukowiak, A. Narayanan, R.M. Terns, and M.P. Terns, manuscript submitted for publication), which contains 13 internal modifications (Reddy and Busch 1988). Finally, addition of a 5′ fragment (nucleotides 19–46) of U2, which lacks an Sm site but contains 10 sites of modification, to U7, which is not detected in nucleoli, results in detectable levels of nucleolar localization (Fig. 7; U2-U7). Alternatively, differences in nucleolar transit time may be a result of functional differences between the Sm binding sites. Comparing sites from various species, we note a striking difference in the sequences for U1 and U7 versus U2: AUUUGUG for U1 (AUUUCUG in frog), AUUUGUC for U7, and AUUUUUG for U2. Perhaps a difference in the fifth position of the Sm site mediates association with distinct factors that affect nucleolar trafficking rates. Interestingly, the Sm binding sites of U1 and U5 (AUUUUUUG, more similar to U2 than to U1 and U7 Sm binding sites) are not functionally exchangeable in protein binding (Jarmolowski and Mattaj 1993). Even between U1 and U7, the Sm binding sites are not exchangeable, since replacement of the U7 Sm site with the U1 Sm site generates a nonfunctional U7 snRNP (Stefanovic et al. 1995). Further studies are required to understand the detailed differences between the functions of various Sm binding sites. Our work indicates that the Sm site targets U2 to nucleoli where it is modified. Other snRNAs may transit through nucleoli with different kinetics.
At What Stage in U2 Biogenesis Does Modification Occur?
Although our results demonstrate that U2 internal modification occurs in the nucleus rather than the cytoplasm, we still have no definitive answer to the question of whether modifications are introduced before export to the cytoplasm or after reentry into the nucleus. For three reasons, we consider the latter more likely. First, it is well established that the Sm snRNAs are rapidly transported to the cytoplasm after synthesis in the nucleus (Yang et al. 1992; Terns et al. 1993; Jarmolowski et al. 1994). By contrast, U2 internal modification is relatively slow, in our hands requiring more than 6 h to complete (Yu et al. 1998; data not shown). Second, we have shown previously that internal modifications directly contribute to the conversion of the 12S U2 particle into the functional 17S particle, which possesses at least nine extra proteins (Behrens et al. 1993). This suggests that the internal modifications are introduced at the stage between 12S and 17S snRNP assembly, i.e., after U2 transport back to the nucleus. Third, we have demonstrated here that an intact Sm site, and presumably the binding of Sm proteins, is required for U2 internal modification (Fig. 3). Since assembly with Sm proteins normally occurs in the cytoplasm (Mattaj 1988), internal modification could occur only after reentry into the nucleus.
Is Internal Modification of Sm snRNAs Guided by snoRNAs in the Nucleolus?
Our evidence that U2 internal modification occurs in the nucleolus strongly supports the notion that the nucleolus harbors the modifying machinery responsible for pseudouridylation and 2′-O-methylation of several different types of RNAs. These include ribosomal RNAs (Pol I transcripts; Cavaille et al. 1996; Kiss-Laszlo et al. 1996, Kiss-Laszlo et al. 1998; Ganot et al. 1997; Ni et al. 1997; Bortolin et al. 1999), U6 snRNA (a Pol III transcript; Tycowski et al. 1998; Ganot et al. 1999; Lange and Gerbi 2000), and U2 and perhaps other Sm snRNAs (Pol II transcripts; this study). Modification of eukaryotic rRNAs and U6 snRNA is catalyzed by snoRNPs, where the snoRNA component serves as a guide to direct modification at a specific site (Cavaille et al. 1996; Kiss-Laszlo et al. 1996, Kiss-Laszlo et al. 1998; Ganot et al. 1997, Ganot et al. 1999; Ni et al. 1997; Tycowski et al. 1998; Bortolin et al. 1999). Are the Pol II–transcribed Sm snRNAs (U2 and others) modified by the same mechanism? Since all of these RNAs share a common modification subcompartment, the nucleolus, we predict the existence of similar snoRNPs responsible for internal modification (2′-O-methylation and pseudouridylation) of Pol II–transcribed Sm snRNAs.
Acknowledgments
The work described in this article was supported by grant GM26154 (to J.A. Steitz) and grant GM54682 (to M.P. Terns) from the National Institutes of Health, and by a Start Up Fund from the University of Rochester (to Y.-T. Yu). J.A. Steitz is an investigator of the Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper: CMC, 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate; DIC, differential interference contrast; Pol, polymerase; rRNA, ribosomal RNA; sn, small nuclear; sno, small nucleolar.
References
- Bakin A., Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase centeranalysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- Behrens S.-E., Tyc K., Kastner B., Reichelt J., Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol. Cell. Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K., Ferreira J.A., Lamond A.I. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus J. Cell Biol. 131 1995. 817 831a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K., Ferreira J., Santama N., Weis K., Lamond A.I. Molecular analysis of the coiled body J. Cell Sci. Suppl. 19 1995. 107 113b [DOI] [PubMed] [Google Scholar]
- Bortolin M.L., Ganot P., Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:457–469. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J., Nicoloso M., Bachellerie J.-P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- Dahlberg J.E., Lund E. The genes and transcription of the major small nuclear RNAs. In: Birnstiel M.L., editor. Small Nuclear Ribonucleoprotein Particles. Springer-Verlag; Heidelberg: 1988. pp. 38–70. [Google Scholar]
- Fischer U., Darzynkiewicz E., Tahara S.M., Dathan N.A., Luhrmann R., Mattaj I.W. Diversity in the signals required for nuclear accumulation of U snRNPs and variety in the pathways of nuclear transport. J. Cell Biol. 1991;113:705–714. doi: 10.1083/jcb.113.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J.G., Bellini M., Wu Z., Murphy C. Assembly of the nuclear transcription and processing machineryCajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P., Bortolin M.L., Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- Ganot P., Jady B.E., Bortolin M.L., Darzacq X., Kiss T. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol. 1999;19:6906–6917. doi: 10.1128/mcb.19.10.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A., Tang J., Puig O., Salgado J., Neubauer G., Colot H.V., Mann M., Seraphin B., Rosbash M., Luhrmann R., Fabrizio P. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A., Neubauer G., Banroques J., Mann M., Luhrmann R., Fabrizio P. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:4535–4548. doi: 10.1093/emboj/18.16.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J., Cronshagen U., Kadokura M., Marshallsay C., Wada T., Sekine M., Luhrmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Mattaj I.W. The determinants for Sm protein binding to Xenopus U1 and U5 snRNAs are complex and non-identical. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:223–232. doi: 10.1002/j.1460-2075.1993.tb05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens W., Izaurralde E., Mattaj I. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo Z., Henry Y., Bachellerie J.-P., Caizergues-Ferrer M., Kiss T. Site-specific ribose methylation of preribosomal RNAa novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo Z., Henry Y., Kiss T. Sequence and structure elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Lange T.S., Gerbi S.A. Transient nucleolar localization of U6 small nuclear RNA in Xenopus laevis oocytes. Mol. Biol. Cell. 2000;11:2419–2428. doi: 10.1091/mbc.11.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M.R., Boyle J.A., Hardin J.A., Steitz J.A. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. 1981;211:400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- Luhrmann R. snRNP proteins. In: Birnstiel M.L., editor. Small Nuclear Ribonucleoprotein Particles. Springer-Verlag; Heidelberg: 1988. pp. 71–99. [Google Scholar]
- Lund E., Paine P. Nonaqueous isolation of transcriptionally active nuclei from Xenopus oocytes. Methods Enzymol. 1990;181:36–41. doi: 10.1016/0076-6879(90)81110-g. [DOI] [PubMed] [Google Scholar]
- Lyon C.E., Bohmann K., Sleeman J., Lamond A.I. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp. Cell Res. 1997;230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- Massenet S., Mougin A., Branlant C. Posttranscriptional modifications in the U small nuclear RNAs. In: Grosjean H., Benne R., editors. The Modification and Editing of RNA. ASM Press; Washington, DC: 1998. pp. 201–228. [Google Scholar]
- Massenet S., Motorin Y., Lafontaine D.L.J., Hurt E.C., Grosjean H., Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol. Cell. Biol. 1999;19:2142–2154. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. UsnRNP assembly and transport. In: Birnstiel M.L., editor. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer-Verlag; Heidelberg: 1988. pp. 100–114. [Google Scholar]
- Mattaj I.W., De Robertis E.M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985;40:111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W., Englmeier L. Nucleocytoplasmic transportthe soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Maxwell E., Fournier M. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Meier U., Blobel G. NAP57, a mammalian nucleolar protein with a putative homologue in yeast and bacteria. J. Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A., Speckmann W., Terns R., Terns M.P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Tien A.L., Fournier M.J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. RNA/RNA interactions in nuclear pre-mRNA splicing. In: Simons R., Grunberg-Manago M., editors. RNA Structure and Function. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1998. pp. 279–307. [Google Scholar]
- Olson M.O., Dundr M., Szebeni A. The nucleolusan old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- Patton J.R. Pseudouridine modification of U5 RNA in ribonucleoprotein particles assembled in vitro. Mol. Cell. Biol. 1991;11:5998–6006. doi: 10.1128/mcb.11.12.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I., Andrade L.E., Ochs R.L., Chan E.K., Chang C.M., Roos G., Tan E.M. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Reddy R., Busch H. Small nuclear RNAsRNA sequences, structure, and modifications. In: Birnstiel M.L., editor. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer-Verlag; Heidelberg: 1988. pp. 1–37. [Google Scholar]
- Samarsky D.A., Fournier M.J., Singer R.H., Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G., Tekotte H., Grosjean H., Segref A., Sharma K., Tollervey D., Hurt E.C. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E., Lamond A.I. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 1999;9:1065–1074. doi: 10.1016/s0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- Sleeman J.E., Lyon C.E., Platani M., Kreivi J.-P., Lamond A.I. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp. Cell Res. 1998;243:290–304. doi: 10.1006/excr.1998.4135. [DOI] [PubMed] [Google Scholar]
- Smith C.M., Steitz J.A. Sno storm in the nucleolusnew roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Speckmann W., Narayanan A., Terns R., Terns M.P. Nuclear retention elements of U3 small nucleolar RNA. Mol. Cell. Biol. 1999;19:8412–8421. doi: 10.1128/mcb.19.12.8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic B., Hackl W., Lührmann R., Schümperli D. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 1995;23:3141–3151. doi: 10.1093/nar/23.16.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M.P., Dahlberg J.E. Retention and 5′ cap trimethylation of U3 snRNA in the nucleus. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- Terns M.P., Dahlberg J.E., Lund E. Multiple cis-acting signals for export of pre-U1 snRNA from the nucleus. Genes Dev. 1993;7:1898–1908. doi: 10.1101/gad.7.10.1898. [DOI] [PubMed] [Google Scholar]
- Terns M.P., Grimm C., Lund E., Dahlberg J.E. A common maturation pathway for small nucleolar RNAs. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Kiss T. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Tycowski K.T., You Z.-H., Graham P.J., Steitz J.A. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Murphy C., Gall J.G. The Sm binding site targets U7 snRNA to coiled bodies (spheres) of amphibian oocytes. RNA. 1996;2:811–823. [PMC free article] [PubMed] [Google Scholar]
- Yang H., Moss M.L., Lund E., Dahlberg J.E. Nuclear processing of the 3′-terminal nucleotides of pre-U1 RNA in Xenopus laevis oocytes. Mol. Cell. Biol. 1992;12:1553–1560. doi: 10.1128/mcb.12.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.-T., Shu M.-D., Steitz J.A. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- Yu Y.-T., Shu M.-D., Steitz J.A. U2 modifications are required for snRNP assembly and pre-mRNA splicing. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.-T., Scharl E.C., Smith C.M., Steitz J.A. The growing world of small nuclear ribonucleoprotein particles. In: Gesteland R., Cech T., Atkins J., editors. The RNA World. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1999. pp. 487–524. [Google Scholar]