Abstract
In mammalian cells, most membrane proteins are inserted cotranslationally into the ER membrane at sites termed translocons. Although each translocon forms an aqueous pore, the permeability barrier of the membrane is maintained during integration, even when the otherwise tight ribosome–translocon seal is opened to allow the cytoplasmic domain of a nascent protein to enter the cytosol. To identify the mechanism by which membrane integrity is preserved, nascent chain exposure to each side of the membrane was determined at different stages of integration by collisional quenching of a fluorescent probe in the nascent chain. Comparing integration intermediates prepared with intact, empty, or BiP-loaded microsomes revealed that the lumenal end of the translocon pore is closed by BiP in an ATP-dependent process before the opening of the cytoplasmic ribosome–translocon seal during integration. This BiP function is distinct from its previously identified role in closing ribosome-free, empty translocons because of the presence of the ribosome at the translocon and the nascent membrane protein that extends through the translocon pore and into the lumen during integration. Therefore, BiP is a key component in a sophisticated mechanism that selectively closes the lumenal end of some, but not all, translocons occupied by a nascent chain. By using collisional quenchers of different sizes, the large internal diameter of the ribosome-bound aqueous translocon pore was found to contract when BiP was required to seal the pore during integration. Therefore, closure of the pore involves substantial conformational changes in the translocon that are coupled to a complex sequence of structural rearrangements on both sides of the ER membrane involving the ribosome and BiP.
Keywords: BiP; protein integration; endoplasmic reticulum; translocon; fluorescence spectroscopy
Introduction
Nearly all membrane proteins in a mammalian cell are inserted into the membrane of the ER in a cotranslational manner at sites called translocons (Johnson and van Waes, 1999). The translocon is formed by ER membrane proteins that create an aqueous pore, and it also functions to translocate secretory proteins across the ER membrane. Translocation is the default pathway, and translocation of the nascent chain proceeds to completion unless a transmembrane (TM)* sequence in the nascent chain causes translocon function to convert to integration (Liao et al., 1997).
An important and intriguing mechanistic question is how TM segment insertion and the proper localization of cytoplasmic and lumenal domains can be achieved during cotranslational membrane protein integration without compromising the permeability barrier across the ER membrane. This barrier must be maintained because calcium ions are stored in the ER and the unregulated release of these potent second messengers would have severe metabolic consequences for the cell. During cotranslational translocation, the aqueous translocon pore is sealed at its cytoplasmic end by the tight binding of the ribosome to the translocon (Crowley et al., 1994). However, this mechanism is clearly not sufficient for membrane protein integration because the ribosome–translocon seal must be broken to allow newly synthesized cytoplasmic domains of the nascent membrane protein to move into the cytoplasm during the cotranslational integration process.
Liao et al. (1997) examined the integration of a signal-cleaved, single-spanning membrane protein, and found that the ribosome–translocon structure is tightly regulated during integration to ensure that the ER membrane remains sealed to ion flow. They discovered that after the TM sequence had been synthesized and was still inside the ribosome, the translocon pore was sealed from the lumenal side, and that shortly thereafter the cytoplasmic ribosome–translocon seal was opened. Presumably, this sequence of events constitutes a safety mechanism by ensuring that the lumenal end of the pore is closed before the cytoplasmic end is opened. These experiments also demonstrated that pore closure was mediated by a very long signal transduction pathway that extends from far inside the ribosome to the other side of the ER membrane (Liao et al., 1997).
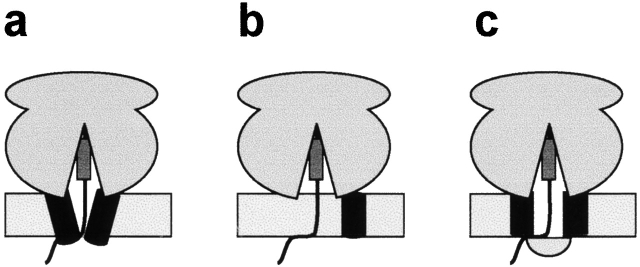
By what mechanism is the lumenal end of the translocon pore closed or gated during membrane protein integration? One possibility is that a conformational change in the translocon narrows the pore and thereby prevents ion flow (Fig. 1 a). Because the translocon has been shown to undergo structural changes that greatly alter pore diameter (Hamman et al., 1997, 1998), this is a likely possibility. Alternatively, the translocon may disassemble so that the pore disappears (Fig. 1 b). We consider this an unlikely scenario because the translocon pore remains intact even when no ribosome is bound to the translocon (Hamman et al., 1998). A third possibility is that a soluble lumenal protein effects, directly or indirectly, closure of the pore (Fig. 1 c). Consistent with such a scenario, BiP, a lumenal Hsp70, is responsible for closing the translocon pore before ribosome binding and for a short time after the ribosome is targeted to the translocon (Crowley et al., 1994; Hamman et al., 1998). However, a translocon at rest and a translocon engaged in integration differ structurally because the latter is bound to a ribosome and also has a nascent chain extending through the translocon (Liao et al., 1997). If some lumenal protein closes the pore during integration as depicted in Fig. 1c, the presence of the nascent chain makes the closure mechanism substantially more complicated than that of BiP closing a ribosome-free translocon pore that lacks a nascent chain. An even greater complication is the fact that not all pores occupied by a nascent chain will be closed: those pores translocating secretory proteins will not be closed (Crowley et al., 1994), and only some pores occupied by a nascent membrane protein will be closed at any given time (Liao et al., 1997). Thus, irrespective of the mechanism of pore closure, a sophisticated regulatory mechanism must have evolved for identifying which pores containing nascent chains need to be closed. In this study, a fluorescence quenching approach was used to examine these important and fundamental issues and to distinguish between the above three possibilities.
Figure 1.
Three possible mechanisms for sealing the lumenal side of the aqueous translocon pore during protein integration. (a) A conformational change in the translocon prevents ion flow through the pore from the lumenal end. (b) Disassembly of the translocon machinery maintains the permeability barrier of the membrane by eliminating the pore. (c) A soluble lumenal protein mediates, either directly or indirectly, the closure of the lumenal side of the aqueous translocon pore.
Results
Experimental approach
Fluorescent integration intermediates.
To directly monitor the exposure of a nascent membrane protein to each side of the ER membrane during integration, a fluorescent probe is incorporated into the nascent chain of an integration intermediate. A homogeneous population of integration intermediates is generated by in vitro translation of mRNAs that are truncated in the coding region (Liao et al., 1997). The ribosome halts when it reaches the end of such a truncated mRNA, but the nascent chain and peptidyl-tRNA remain bound to the ribosome because there is no stop codon. Therefore, the length of the nascent chain in an intermediate is dictated by the length of the truncated mRNA, and different stages in the process of integration can be examined simply by increasing the length of the truncated mRNA in a translation containing SRP and ER microsomes.
When in vitro translation is performed in the presence of a Lys-tRNA analogue with a fluorescent probe covalently attached to the lysine side chain, the fluorescent amino acid will be incorporated into the nascent chain in response to a lysine codon in the mRNA. Thus, a fluorescent probe can be incorporated into an integration intermediate at a defined location to serve as a probe of the local nascent chain environment during integration. For the experiments described here, we have employed Nɛ-(6-[7-nitrobenz-2-oxa-1,3-diazo-4-yl]aminohexanoyl)-Lys-tRNA (ɛNBD-Lys-tRNA) (Crowley et al., 1993).
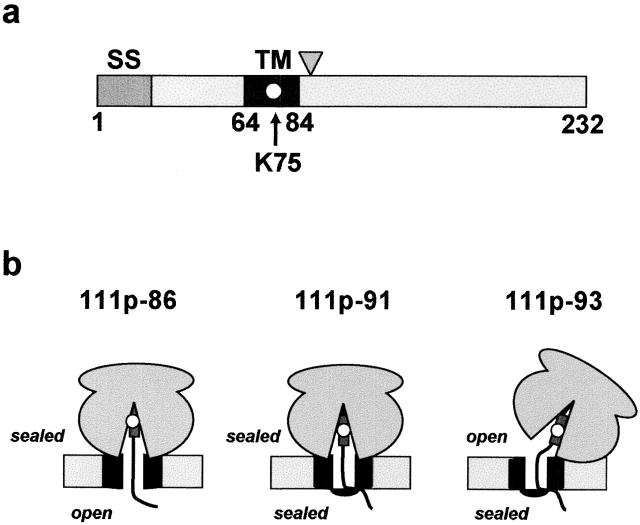
A chimeric single-spanning membrane protein designated 111p that has been well characterized previously (Liao et al., 1997) was used in this study (Fig. 2 a). The 111p coding sequence contains only a single lysine codon that positions a fluorescent probe in the center of the nascent chain TM sequence in each integration intermediate. At the lengths of nascent chain analyzed in this study, the TM sequence, and therefore the fluorescent probe, are located in the ribosomal nascent chain tunnel in each integration intermediate (Fig. 2 b).
Figure 2.
111p protein and integration intermediates. (a) The chimeric 111p single-spanning membrane protein was constructed using the TM sequence from vesicular stomatitus G protein (residues 65–84, dark gray) and lysine-free stretches of preprolactin and Bcl-2 (Liao et al., 1997). 111p contains only a single lysine codon at position 75 in the middle of the TM sequence (white circle). Note that the TM segment will still be nonpolar and uncharged when ɛNBD-Lys is incorporated at this location. The signal sequence (SS) is indicated in gray (residues 1–22), and the triangle indicates the approximate position of the truncations used in this study (residues 86, 91, and 93). (b) Accessibility of the fluorescent probe from either the cytoplasmic or lumenal sides of intact ER microsomes is indicated for each intermediate used in this study (Liao et al., 1997). The ribosome is shown bound to the translocon (black) at the ER membrane (light gray) in each case. The TM sequence is represented by a dark gray rectangle and the fluorescent probe by a white circle.
Fluorescence quenching.
Accessibility of the fluorescent probe in the nascent chain to either the cytoplasm or the lumen can be determined using hydrophilic collisional quenchers of NBD such as iodide ions. When a collisional quencher contacts an excited fluorescent dye, the excited state energy of the fluorophore is lost without the emission of a photon. The resulting decrease in fluorescence intensity is directly proportional to the number of collisions, and hence to the quencher concentration, as expressed in the Stern-Volmer equation: (Fo/F) – 1 = Ksv[Q], where Fo is the emission intensity in the absence of quencher, F is the emission intensity at quencher concentration [Q], and Ksv is the Stern-Volmer constant. Therefore, the extent of quenching is given by the Ksv, where higher Ksv values indicate increased quenching (Crowley et al., 1993, 1994).
When iodide ions are added to microsomes containing fully assembled integration intermediates, any nascent chain probes that are accessible to the aqueous cytosol will be collisionally quenched. To assess the accessibility of a nascent chain probe to the lumenal side of the ER membrane, iodide ions are introduced into the microsome interior using a pore-forming toxin to create large holes in the ER membrane (Crowley et al., 1994). Perfringolysin O (PFO) is a member of a family of cytolytic bacterial toxins that oligomerize to form holes ∼300 Å in diameter in cholesterol-containing membranes (Heuck et al., 2001). Thus, after PFO addition, quenchers can move freely into the lumenal compartment of the microsomes (Crowley et al., 1994; Hamman et al., 1997, 1998; Liao et al., 1997). Therefore, nascent chain probes exposed to either the cytoplasmic or the lumenal side of the ER are quenched by iodide ions in the presence of PFO.
Collisional quenching experiments can also be used to estimate the size of an aqueous pore that links two aqueous compartments. When a fluorescent probe is constrained to one compartment and quenchers are added to the other compartment, collisional quenching will be observed only if the hydrophilic quenchers are sufficiently small to pass through the aqueous pore connecting the two compartments. This was the approach used to determine that a ribosome-free translocon has a diameter of 9–15 Å (iodide ions, but not ADP molecules, pass through this pore) (Hamman et al., 1998). In contrast, a translocon engaged in translocation has a diameter of 40–60 Å (NAD+ molecules, but not a Fab fragment, pass through such a pore even though it also contains a nascent chain) (Hamman et al., 1997).
Microsomes with different lumenal contents.
To distinguish between the various mechanisms of sealing the translocon during integration (Fig. 1), microsomes were prepared that contained a full complement of soluble lumenal proteins, no soluble lumenal proteins, or a single purified lumenal protein. When salt-washed ER microsomes (KRMs) are treated with high pH buffer followed by dilution, their soluble lumenal contents are released (Bulleid and Freedman, 1988; Nicchitta and Blobel, 1993; Hamman et al., 1998). These lumen-extracted microsomes (XRMs) can be resealed without any loss in translocon function. Thus, integration intermediates prepared with XRMs can be used to determine whether a soluble lumenal protein is responsible for sealing the lumenal side of the translocon during integration (Fig. 1 c). This goal is addressed most directly by focusing on an integration intermediate that is normally sealed at both ends of the translocon pore so that the NBD-labeled nascent chain is inaccessible to quencher added from either side of the ER membrane (Liao et al., 1997). Such intermediates prepared with KRMs should exhibit no quenching by either cytoplasmic or lumenal iodide ions, whereas the same intermediates prepared with XRMs should be quenched if a soluble lumenal protein is involved in sealing the pore (Fig. 1 c). On the other hand, this XRM intermediate would remain inaccessible to quenchers if the lumenal end of the pore is closed by a mechanism independent of soluble lumenal proteins, such as a conformational change in the translocon pore (Fig. 1, a and b).
Empty XRMs can also be refilled with a purified protein(s) to form reconstituted microsomes (RRMs). To prepare RRMs, XRMs are first incubated with the purified protein in high pH buffer. When the pH is neutralized, the opened microsomes reseal and encapsulate the purified protein to yield sealed RRMs that contain the desired protein in their lumenal compartments. If a soluble lumenal protein is implicated in sealing the lumenal side of the translocon during integration, the identity of that protein can be determined by identifying which protein(s) encapsulated in RRMs are able to close the lumenal end of the pore in integration intermediates (Hamman et al., 1998).
A soluble protein is responsible for sealing the lumenal side of the translocon during integration
Liao et al. (1997) identified a stage in the integration process where the nascent chain was not accessible from either the cytoplasmic or the lumenal side of the ER membrane. When fluorescent integration intermediates containing an NBD-labeled, 91-residue 111p nascent chain (NBD-111p-91) and intact ER microsomes (KRM) were exposed to iodide ions, little collisional quenching of NBD fluorescence was observed (Ksv = 0.5 M−1) (Fig. 3 a; Table I). Because the Ksv is ∼2.5 M−1 when the same translation intermediate is not bound to the ER membrane (Liao et al., 1997), it is clear that the formation of the integration intermediate prevents iodide ion access to most nascent chain probes. As we have shown previously, residual quenching results from a combination of effects, each of which exposes a small number of NBD-labeled nascent chains to the cytoplasm. Some properly targeted integration intermediates dissociate from the translocon during the long spectroscopic measurements (Crowley et al., 1994), some nontargeted nascent chains adsorb to the outer microsomal membrane (this effect is worse with membrane proteins) (Hamman et al., 1997), and some ribosomal complexes are purified with the microsomes as part of a polysome (Hamman et al., 1997). Limited protease and ribonuclease treatment of the microsomes can eliminate nearly all of the NBD dyes exposed to (quenched by) cytoplasmic iodide ions and hence reduce the initial Ksv values to near 0 (Hamman et al., 1997). However, we have chosen here to avoid any risk of damaging the samples and focus instead on the difference, if any, in quenching between cytoplasmic and lumenal iodide ions (see below).
Figure 3.
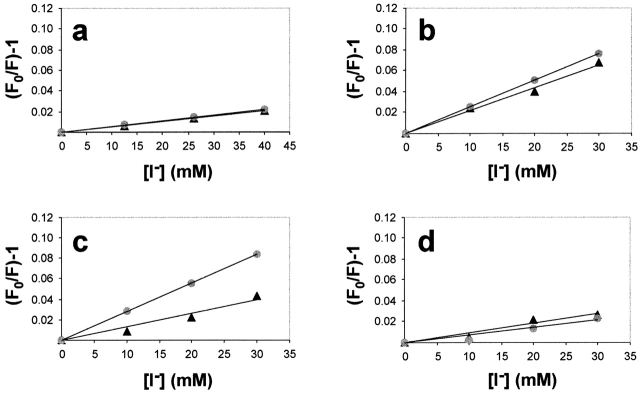
Iodide ion quenching of NBD-labeled 111p-91 integration intermediates prepared with various ER microsomes. Samples containing NBD-111p-91 integration intermediates prepared with (a) KRMs, (b) XRMs, (c) XRMs + αSec61α, or (d) RRMs (XRMs reconstituted with rBiP) were divided into four equal aliquots. Each sample then received the same total concentration of KCl and KI, but different amounts of KI as shown. Fo is the net fluorescence intensity in the absence of quencher, whereas F is the net fluorescence intensity at a given iodide ion concentration. Measurements were made in the absence (▴) or the presence (•) of pore-forming PFO toxin to introduce iodide ions into the lumen of the microsomes. The linear least-squares best-fit lines for data averaged from several independent experiments are shown. The results are also reported in Tables I and II.
Table I. Iodide ion quenching of NBD-labeled integration intermediates in intact and lumen-extracted ER microsomes.
| Intermediate components | Observed Ksv (M− 1) | |
|---|---|---|
| −PFO | +PFO | |
| 111p-86 • KRMa | 0.4 | 1.8 |
| 111p-86 • XRM | 1.7 | 1.8 |
| 111p-86 • XRM + αSec61αb | 0.9 | 1.7 |
| 111p-91 • KRMa | 0.5 | 0.5 |
| 111p-91 • XRM | 2.2 | 2.5 |
| 111p-91 • XRM + αSec61αb | 1.3 | 2.8 |
| 111p-93 • KRMa | 3.6 | 3.7 |
| 111p-93 • XRM | 3.0 | 2.8 |
The Ksv values shown are the average of 3–5 independent experiments except where indicated. The standard deviations for the Ksv values were: ±0.2 M−1 for KRM measurements; ±0.1–0.3 M−1 for XRM – PFO measurements; and ±0.1–0.4 M−1 for XRM + PFO measurements. Samples were prepared as described in Materials and methods.
Taken from Liao et al. (1997).
The Ksv values are the average of two independent experiments.
Because hydrophilic iodide ions do not pass through a phospholipid bilayer (Cranney et al., 1983; Crowley et al., 1994), nascent chain exposure to the lumenal side of the membrane can be evaluated upon introduction of iodide ions into the interior of the microsomes using PFO. When PFO was added to the above NBD-111p-91 integration intermediate sample, no increase in iodide ion quenching of NBD emission was observed (Fig. 3 a; Table I). The identical Ksv values before and after quenchers were introduced into the lumenal compartment show that the 111p-91 nascent chain and its fluorophore are not exposed to the lumenal side of the membrane. Thus, the lumenal end of the translocon pore is closed at this stage of the integration process.
The inaccessibility of the nascent chain from the cytoplasmic side is explained by the tight ribosome-translocon seal. However, in order to determine the mechanism of lumenal sealing at this stage of membrane protein integration, collisional quenching experiments were performed on integration intermediates prepared with XRM microsomes that lacked soluble lumenal proteins. Lumenal extraction removes the BiP that normally seals ribosome-free translocons (Hamman et al., 1998), and this allows cytoplasmic iodide ions to diffuse into the lumenal interiors of the XRMs, even in the absence of PFO, through the pores of those translocons that are not bound to ribosomes (∼50%) (Hamman et al., 1997). Such iodide ions would be expected to quench the probe in the NBD-111p-86 intermediate, as its pore is normally open to the ER lumen (Fig. 2 b). When NBD-111p-91 integration intermediates containing XRMs were treated with cytoplasmic iodide ion quenchers, maximal quenching was observed both in the presence and absence of PFO (Fig. 3 b; Table I). Because minimal quenching of NBD-111p-91 by iodide ions was observed in intermediates containing intact KRM microsomes both in the presence and absence of PFO (Fig. 3 a), lumenal extraction must compromise the seal in XRM-containing intermediates.
To determine whether the iodide ions were indeed entering the lumen via unoccupied translocon pores, we took advantage of previous work that had revealed that the binding of Sec61α-specific antibodies to ribosome-free translocons occludes the translocon so that iodide ion flow through the pore is severely inhibited (Hamman et al., 1998). Thus, addition of these affinity-purified antibodies to a sample before iodide ion addition would reduce the extent of quenching if iodide ions were accessing the nascent chains from the lumenal side of the membrane after passage through ribosome-free translocons. As shown by Fig. 3 c and the data in Table I, the addition of Sec61α-specific antibodies to samples dramatically reduced the observed Ksv values for both the NBD-111p-86 XRM and NBD-111p-91XRM intermediates in the absence of PFO. When PFO was added and iodide ions were allowed full access to the lumen, quenching of both intermediates reached the maximal level (Fig. 3 c; Table I). Therefore, iodide ions had to first pass through a ribosome-free translocon before being able to collisionally quench the nascent chain probes from the lumenal side.
The lumenal end of the pore in the 111p-91 integration intermediate is normally closed to the ER lumen (Fig. 2 b; Table I). Two of the possible mechanisms for pore closure, gating of the translocon pore by a conformational change or disassembly of the translocon (Fig. 1, a and b), would be independent of the soluble lumenal protein content of the microsomes. Therefore, if a conformational change in the translocon or its disassembly were responsible for sealing the lumenal side of the translocon during integration, we would have expected no difference in the iodide ion quenching for NBD-111p-91 between intact and lumen-extracted microsomes. However, when microsomes are devoid of soluble lumenal protein contents, maximal quenching of the fluorescent probe in NBD-111p-91 XRM intermediates was observed from the lumenal side of the translocon (Fig. 3, b and c; Table I). These results strongly indicate that a soluble lumenal protein is responsible for maintaining the permeability barrier at this stage of integration (Fig. 1 c).
BiP is responsible for maintaining the permeability barrier during cotranslational integration
Because a soluble lumenal protein appears to be required to seal the lumenal side of the translocon, the protein responsible for closing the pore can be identified by reconstituting purified lumenal proteins back into lumen-extracted microsomes and determining which single protein or combination of proteins restores the seal at the lumenal end of the pore of the NBD-111p-91 intermediate. A good candidate protein is the lumenal Hsp70, BiP, because Hamman et al. (1998) showed that BiP is required to seal the pore in a ribosome-free translocon. Although the translocon structure is likely to be substantially different during cotranslational integration because of the bound ribosome (Hamman et al., 1997) and the nascent chain threading through the translocon, it was reasonable to first test whether BiP was involved in maintaining the permeability barrier of the ER membrane during integration.
Thus, lumen-extracted microsomes (Bulleid and Freedman, 1988; Nicchitta and Blobel, 1993; Hamman et al., 1998) were reconstituted with purified recombinant hamster BiP protein (rBiP) (Gaut and Hendershot, 1993) to yield RRMs. Integration intermediates prepared with these rBiP-containing RRMs were then analyzed to determine the exposure of the fluorescent nascent chain to the cytoplasm and the ER lumen. As expected, the purified rBiP protein mediated the closure of ribosome-free translocon pores, as evidenced by the inability of cytoplasmic iodide ions to enter the lumenal compartment of the RRM microsomes and quench the NBD-111p-86 nascent chain probe in the absence of PFO (Table II). This rBiP-dependent protection of the nascent chain from access to cytoplasmic iodide ions was effected from the lumenal side of the ER membrane, and was not a nonspecific effect because the nascent chain in the NBD-111p-93 RRM intermediate was maximally quenched by cytoplasmic iodide ions in the absence of PFO (Table II). Because the same RRMs were used to prepare the NBD-111p-86, NBD-111p-91, and NBD-111p-93 intermediates, the NBD-111p-93 results serve as an important internal control for this spectroscopic approach, and also show, as expected (Liao et al., 1997), that the NBD-111p-93 nascent chain is exposed to the cytosol during integration (Fig. 2 b).
Table II. Iodide ion quenching of NBD-labeled integration intermediates in ER microsomes reconstituted with rBIP.
| Intermediate components | Observed Ksv (M− 1) | |
|---|---|---|
| −PFO | +PFO | |
| 111p-86 • RRM | 0.4 | 2.0 |
| 111p-86 • RRM + αBiP | 1.8 | 2.0 |
| 111p-86 • RRM + AMPPNP | 2.0 | 2.0 |
| 111p-91 • RRM | 0.9 | 0.8 |
| 111p-91 • RRM + αBiP | 2.4 | 2.4 |
| 111p-91 • RRM + AMPPNP | 2.5 | 2.6 |
| 111p-93 • RRM | 2.6 | 3.1 |
The Ksv values shown are the average of 3–4 independent experiments. The standard deviations were ±0.2 M−1 for RRM – PFO measurements, and between 0.2 and 0.4 M−1 for all other measurements. Samples were prepared as described in Materials and methods.
When iodide ions were introduced into the lumenal compartment of RRMs using PFO, NBD-111p-86 was quenched as in the KRM-containing intermediates, thereby showing that the translocon is open on the lumenal side at this stage of integration (Table II). However, with NBD-111p-91 RRM intermediates, no additional quenching was observed after PFO was added (Fig. 3 d; Table II). Therefore, the seal at the lumenal end of the translocon in this integration intermediate is restored upon reconstitution of the microsomes with purified rBiP protein. The sealing by rBiP is specific to the 111p-91 nascent chain intermediate, and does not result from indiscriminate sealing of translocon pores because the 111p-86 intermediate is not sealed under the same conditions. Therefore, the 111p-86 and 111p-91 data constitute another important internal control by demonstrating that translocon closure is not due to a nonspecific effect such as BiP binding to partially unfolded translocon proteins. These data also demonstrate that the ribosome–translocon seal in these integration intermediates is not disrupted by the extraction or reconstitution procedures.
Thus, rBiP reconstituted into RRMs is able to seal the lumenal side of the translocon pore during cotranslational integration. The rBiP-reconstituted microsomes behave exactly as the KRMs in the iodide ion collisional quenching experiments, demonstrating that BiP is the only soluble lumenal protein required to restore the seal (compare Table I, KRM, and Table II, RRM).
Several control experiments were performed to demonstrate that the closure of the lumenal side of the translocon pore during integration is due specifically to the rBiP protein. When affinity-purified antibodies against the BiP protein are added to an rBiP sample before reconstitution into the microsomes, maximum quenching by iodide ions is observed in the absence and presence of PFO for the NBD-111p-86 and NBD-111p-91 intermediates (Table II). The extents of quenching are equivalent to those obtained with lumen-extracted microsomes that contain no soluble lumenal proteins (compare Table I, XRM, and Table II, RRM + αBiP). Thus, the binding of the BiP-specific IgG molecules to the rBiP molecules prevented the latter from sealing the ER translocons. These results strongly indicate that the BiP protein is solely responsible for gating the translocons actively engaged in integration.
The BiP protein is an Hsp70 family member with a highly conserved NH2-terminal ATPase domain (Bukau and Horwich, 1998; Gething, 1999). To determine whether closure of the translocon pore during integration requires ATP hydrolysis, AMPPNP, a nonhydrolyzable ATP analogue, was reconstituted into RRMs in place of ATP. When rBiP protein was preincubated with an excess of AMPPNP and then reconstituted into RRMs, the rBiP protein was no longer able to close the lumenal side of the translocon in the 111p-91 integration intermediate (Table II). In agreement with previous results (Hamman et al., 1998), rBiP treated with AMPPNP was also unable to seal the ribosome-free translocon pores (Table II, 111p-86, and 111p-91, -PFO). This ATP dependence of translocon closure is consistent with BiP involvement, and also reveals the mechanistic involvement of ATP hydrolysis in the BiP-mediated closure of the lumenal end of the translocon pore during cotranslational integration.
The translocon pore contracts during integration
We previously showed that the diameter of the aqueous translocon pore was 40–60 Å during translocation (Hamman et al., 1997), but was dramatically reduced to 9–15 Å after translation terminated and the ribosome dissociated from the translocon (Hamman et al., 1998). Is the size of the pore altered during integration? As noted earlier, a direct method for estimating the minimum diameter of an aqueous pore is to determine whether a hydrophilic collisional quencher of known dimensions is able to diffuse passively through the pore. In the following experiments, the fluorescent probe was located inside the aqueous nascent chain tunnel of a ribosome that forms a tight seal with the translocon, whereas quenchers were introduced into the aqueous lumenal compartment of the ER microsomes. Quenching was then observed only if the quencher moieties were small enough to pass through the pore in the translocon, the only aqueous pathway into the ribosomal tunnel.
Large quenchers such as nicotinamide adenine dinucleotide (NAD+), with anhydrous dimensions of ∼11 × 12 × 20 Å (Bell and Eisenberg, 1996; Li et al., 1996) and a single net negative charge, are able to traverse the translocon pore of a translocation intermediate containing a preprolactin (pPL) nascent chain (Table III; Hamman et al., 1997). Once introduced into the ER lumen (Table III, +PFO), the NAD+ is able to pass through the translocon and far into the nascent chain tunnel to quench the probes in the pPL nascent chain (Ksv = 1.8 M−1). Thus, during secretory protein translocation at the ER membrane, the translocon pore is large enough to accommodate and freely pass both the nascent chain and large hydrated NAD+ molecules (Hamman et al., 1997). However, during membrane protein integration, different quenching results are obtained that indicate a structural change at the translocon.
Table III.
NAD + quenching of NBD-labeled translocation and integration intermediates in lumen-extracted ER microsomes
| Intermediate components | Observed Ksv (M− 1) | |
|---|---|---|
| −PFO | +PFO | |
| pPL-sK-78 • XRM | 0.4 | 1.8 |
| 111p-86 • XRM | 0.0 | 0.1 |
| 111p-91 • XRM | 0.5 | 0.5 |
| 111p-93 • XRM | 1.8 | 1.7 |
The Ksv values shown are the average of 2–3 independent experiments. The standard deviations were between 0.1 and 0.4 M−1 for all measurements. Samples were prepared as described in Materials and methods. The modified pPL construct, pPL-sK-78, is described in Crowley et al. (1994).
In the absence of BiP, iodide ions (anhydrous diameter ∼4 Å) are able to move freely through the translocon pore of a NBD-111p-91 XRM integration intermediate despite the presence of the 111p-91 nascent chain in the translocon pore and ribosomal tunnel (Fig. 3 b; Table I). However, when the larger NAD+ molecules were used as quenchers with the same integration intermediates, no NAD+ was able to move from the lumenal compartment into the nascent chain tunnel. This was shown by the absence of any increase in quenching when PFO was added to introduce NAD+ into the lumenal compartments of the microsomes (Table III). The inability of NAD+ to quench the 111p-91 probe from the lumenal side of the ER membrane is not due to the inability of NAD+ to move into the ribosomal tunnel, because NAD+ molecules are able to efficiently quench the nascent chain probes inside the ribosome both in the pPL translocation intermediate and also in the NBD-111p-93 XRM intermediate (Table III). In the latter case, NAD+ is able to enter and move into the nascent chain tunnel from the cytosol rather than through the translocon pore because the ribosome–translocon seal has been broken (Fig. 2 b). This result also reveals that when the ribosome–translocon seal is broken during integration to allow the cytoplasmic domain of the nascent chain to enter the cytoplasm, the opening between the ribosome and the membrane is sufficiently large to freely pass NAD+ molecules.
Thus, NAD+ is unable to quench the probe in the 111p-91 XRM intermediate because the quencher cannot pass through the translocon pore into the ribosome. Because NAD+ can move through the translocon pore when it is opened to the lumen during translocation (Hamman et al., 1997; Table III), it is clear that the translocon contracts during integration. However, the pore is not completely closed in the absence of BiP, as shown by the quenching of the probe in the NBD-111p-91 XRM intermediate by iodide ions (Table I). Because the pore contracts in the 111p-91 XRM intermediates even though BiP is absent, it is possible that this structural change is part of the signal transduction mechanism (Liao et al., 1997) by which changes inside the ribosome and at the ribosome–translocon interface are transmitted to the other side of the membrane to signal the need to initiate BiP-mediated closure of the pore. In any event, these results demonstrate once again the dynamic nature of translocon structure (Johnson and van Waes, 1999).
Discussion
A critical factor in mammalian cell viability is its ability to maintain and regulate the permeability barrier of the ER membrane. In particular, Ca2+ ion flow must be controlled during cotranslational protein translocation and integration. We have shown in this study that BiP is required to seal the lumenal end of the aqueous translocon pore during the cotranslational integration of a nascent membrane protein, and hence to maintain the permeability barrier of the ER membrane during this complex process. BiP alone is sufficient to effect the closure of the translocon pore in the absence of all other soluble lumenal proteins, and ATP hydrolysis is required to seal the pore.
This discovery identifies a new functional role for the BiP protein. BiP is best known for its activity as a resident chaperone of the ER lumen, where BiP assists protein folding and assembly by ATP-dependent cycles of binding to hydrophobic stretches in polypeptides (Gething and Sambrook, 1992; Simons et al., 1995; Gething, 1999). However, BiP is clearly a multifunctional protein because it is also required for posttranslational translocation in yeast (Nguyen et al., 1991; Sanders et al., 1992; Brodsky and Schekman, 1993; Panzner et al., 1995; Brodsky, 1996; Matlack et al., 1999; Pilon and Schekman, 1999). Furthermore, although BiP was not required to detect cotranslational translocation in a purified, reconstituted system (Görlich and Rapoport, 1993), other studies have shown that BiP is required for cotranslational translocation in yeast (Brodsky et al., 1995; Young et al., 2001) and possibly in mammals (Nicchitta and Blobel, 1993). BiP also plays a role in retrotranslocation, and is upregulated as a result of the unfolded protein response (Chapman et al., 1998; Brodsky and McCracken, 1999; Gething, 1999; Plemper and Wolf, 1999; Römisch, 1999; Johnson and Haigh, 2000; Patil and Walter, 2001). In addition, BiP seals the aqueous pore of a ribosome-free ER translocon in mammalian cells (Hamman et al., 1998).
This last function of BiP, closing ribosome-free translocon pores, is structurally and mechanistically distinct from its role in closing translocon pores that are actively engaged in integrating nascent membrane proteins into the membrane bilayer. Ribosome-free translocons have aqueous pores with a small diameter (9–15 Å), and there is no nascent chain in the pore when BiP effects pore closure (Hamman et al., 1998). In contrast, we have shown that the nascent chain extends through the translocon and into the ER lumen when the pore is closed during integration (Liao et al., 1997). Therefore, the presence of the nascent chain in the translocon and of the ribosome at the translocon creates a structural problem in terms of how to seal the pore, and also in terms of when to seal the pore. For example, it is easy to imagine that an empty translocon pore would automatically and immediately be recognized by BiP as a substrate for closure. Furthermore, BiP-mediated closure of such a pore could be elicited either by BiP binding directly to the lumenal end of the pore and plugging it (compare Fig. 1 c) and/or by binding elsewhere to a translocon protein and thereby initiating a conformational change that closes the pore (compare Fig. 1 a).
However, the mechanism by which BiP seals a translocon pore during integration is distinguished by the presence of the nascent chain. Despite the complication caused by the nascent chain, BiP could still mediate closure either directly by plugging the pore with the nascent chain in it (Fig. 1 c) and/or indirectly by initiating a conformational change that closes down upon the nascent chain (Fig. 1 a). In fact, pore closure during integration appears to involve a combination of the mechanisms depicted in Fig. 1, a and c. The detection of the TM sequence in the nascent chain inside the ribosome (Liao et al., 1997) causes the translocon pore to contract from its large NAD+-permeable diameter to a diameter that is too small to pass NAD+ (Table III). But this conformational change does not completely close the pore, as iodide ions can still move through the pore (Table I). BiP and ATP hydrolysis are then required to seal the pore (Table II). However, it remains to be seen exactly how the pore is closed with the nascent chain present and exactly where the nascent chain is located in the sealed translocon.
The timing of BiP-mediated pore closure is also more complicated when a nascent chain occupies a translocon pore. Only some of the translocons bound to ribosomes are synthesizing membrane proteins, and of those, only a fraction require sealing by BiP at any given time to allow nascent chain exposure to the cytoplasm. Therefore, recognition of a translocon engaged in integration and distinguishing it from one engaged in translocation is much more difficult for BiP than is recognition of a nascent chain-free translocon. A sophisticated mechanism must recognize which of the translocons engaged in integration require pore closure to allow a cytoplasmic domain of a nascent membrane protein to enter the cytosol. One possible mechanism is the ribosome-mediated contraction of the translocon pore, as suggested in the last section of the Results and discussed further below.
Whatever mechanism triggers pore closure, it is conceivable that BiP transiently and repeatedly binds to nascent chain-containing translocons to identify those in need of closure. However, it seems more likely that BiP remains bound to a translocon throughout translation so that it can either close or open the pore as necessary during the synthesis and integration of a nascent membrane protein. A few proteins have been identified that could serve to localize BiP at the ER membrane. One mammalian ER membrane protein, MTJ1, has been shown to bind and activate BiP (Chevalier et al., 2000). In yeast, the BiP homologue, Kar2p, is bound to the ER membrane surface via Sec63p (Lyman and Schekman, 1995; Corsi and Schekman, 1997; Misselwitz et al., 1999). Because Young et al. (2001) found that Sec63p is required for SRP-dependent translocation in vivo in yeast, it seems likely that Sec63p is also involved in binding and localizing BiP at the translocon during cotranslational translocation and integration. A mammalian homologue of Sec63p was recently discovered and found to be associated with the translocon (Skowronek et al., 1999; Meyer et al., 2000; Tyedmers et al., 2000), but the mammalian Sec63p has not yet been shown to bind BiP.
An intriguing mechanistic feature of the integration process for a signal-cleaved membrane protein is that BiP-mediated closure of the translocon pore occurs while the TM sequence is located far inside the ribosome (Liao et al., 1997). Thus, there is not only a means to identify a TM sequence in a nascent chain inside the ribosome, but also a mechanism for transmitting this information through the ribosome and then across the ER membrane to elicit closure of the pore. Interestingly, an elongated protein adjacent to the nascent chain tunnel in the large ribosomal subunit of the 50S subunit of the bacterial ribosome may be the conduit for such signal transduction through the ribosome (Nissen et al., 2000), though the extent of structural homology between the prokaryotic and eukaryotic ribosomes has yet to be established in this region. As for transmittance of a signal across the ER membrane to the lumenal side of the translocon, several transmembrane ER proteins found at the translocon could interact with the pertinent ribosomal component and carry out this function. In particular, both Sec61α and Sec63p are attractive candidates if, as is thought, they bind to the ribosome and/or BiP, respectively, in mammals. Although the other components of this very long, membrane-spanning signal transduction pathway are not yet known, we have shown here that BiP is essential for this communication pathway. Thus, BiP is the first component of this important signal transduction mechanism to be identified, a discovery that further increases the number of functional roles attributed to BiP.
Images of translocation intermediates reconstructed from cryo-EM data show a gap between the ribosome and Sec61 (Ménétret et al., 2000; Beckmann et al., 2001). Although the resolution of the EM technique is not sufficient to detect all of the polypeptide present (e.g., the nascent chain itself is not visible), the apparent gap has led to speculation that the ribosome does not form a seal or permeability barrier upon binding to the translocon. Instead, Ca2+ efflux from the ER lumen is presumed to be prevented because the pore is too small to permit ion transport, though no experimental measurements of pore size in a membrane with an intact translocon were reported (Ménétret et al., 2000; Beckmann et al., 2001). Because the translocation intermediates (Ménétret et al., 2000) and the purified Sec61 proteins (Beckmann et al., 2001) were solubilized in detergent before analysis in these EM studies, the putative translocation intermediates lack a membrane and some of the translocon-associated proteins (e.g., TRAM, oligosaccharyltransferase, and signal peptidase). In contrast to the EM studies, the fluorescence experiments were done with intact membranes and complete translocation intermediates, and these studies revealed an ion-impermeable ribosome-translocon junction and a large (40–60 Å in diameter) translocon pore (Crowley et al., 1994; Hamman et al., 1997).
Thus, the simplest explanation for the apparent contradiction between the EM and fluorescence experiments is that the tight ribosome–translocon junction and the large pore of the translocation intermediates cannot be maintained or formed in the absence of some translocon components and/or in the absence of the membrane. This would explain why no significant difference was observed between the ribosome-Sec61 images obtained in samples containing translation intermediates with nascent chains and those containing nontranslating ribosomes (Beckmann et al., 2001). Consistent with this possibility, the small (<20 Å) diameter of the translocon in EM reconstructions is similar to that of a ribosome-free translocon in an intact ER membrane determined by fluorescence (Hamman et al., 1998). Hence, translocation intermediates in the Ménétret et al. (2000) study may have been unable to withstand detergent treatment, even in the presence of the nascent chain, and the translocon reverted to its ribosome-free conformation when the dissolution of the membrane and the loss of translocon components caused the ribosome to disengage and break the tight junction or seal between the ribosome and the translocon. Similarly, a purified ribosome–nascent chain complex in the Beckmann et al. (2001) study may not be able to bind to a detergent-purified Sec61 complex in the same way as it would to a fully assembled translocon in an intact membrane.
However, the ribosomes and translocons in the EM studies clearly do interact in a meaningful way because the ribosomal exit site is localized close to the Sec61 complex in all of the EM images. In fact, the ribosome and the Sec61 complex are aligned by four specific connections that orient them, but leave a gap between them (Beckmann et al., 2001). These interactions are both strong and membrane independent because they survive EM preparation procedures (Beckmann et al., 1997, 2001; Ménétret et al., 2000). Indeed, these same interactions may be responsible for keeping the ribosome near the translocon when the ribosomal seal is broken to allow nascent chain movement into the cytosol during integration (Fig. 4 d). However, because no gap larger than 9 Å (the diameter of a hydrated iodide ion) between the cytoplasm and the ribosomal nascent chain tunnel is detected in the fluorescence experiments with intact membranes and translocons, we conclude that the gap observed in reconstructed EM images of translocation intermediates does not accurately depict all of the ribosome interactions with a complete translocon in an intact membrane.
Figure 4.
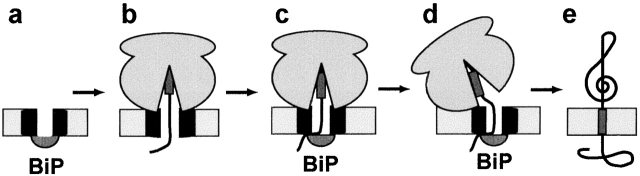
Mechanism for maintaining the permeability barrier of the ER membrane during cotranslational membrane protein integration. (a) Prior to integration, the ribosome-free translocon is sealed on the lumenal side by BiP. (b) After SRP-dependent targeting of a ribosome-nascent chain complex to the translocon and translation to yield a nascent chain longer than 70 amino acids (Crowley et al., 1994), the ribosome–translocon seal is intact and the lumenal end of the pore is open (e.g., 111p-86). (c) After the TM sequence has been synthesized and is still near the peptidyltransferase center far inside the ribosome (Liao et al., 1997), the lumenal end of the translocon pore is closed by the action of BiP (111p-88, 111p-91). Although BiP is shown here physically plugging the pore, BiP may effect closure indirectly by binding to another protein(s) that physically closes the pore. At this point, the ribosome–translocon seal is still intact. (d) The ribosome–translocon seal is then broken, whereas the BiP-dependent seal at the other end of the pore remains intact (111p-93). Although the ribosome is depicted here as rotating relative to the translocon, the nature and magnitude of this structural change is not yet known. (e) After termination of translation, the TM sequence is integrated into the ER membrane.
Maintenance of the permeability barrier during cotranslational integration of a signal-cleaved single-spanning membrane protein at the ER membrane therefore includes, at a minimum, the structural states of the ribosome–translocon complex shown in Fig. 4. Prior to integration, the small aqueous pore of the ribosome-free translocon (9–15 Å diameter) is sealed on the lumenal side by BiP either directly (as shown in Fig. 4 a) or indirectly (Hamman et al., 1998). After SRP-dependent targeting to the ER membrane, a ribosome synthesizing a nascent protein binds tightly to the cytoplasmic side of the translocon to seal the large (40–60 Å) ribosome-bound pore that is open at its lumenal end after the nascent chain reaches a length of ∼70 residues (Fig. 4 b) (Crowley et al., 1994; Hamman et al., 1997). The appearance of a complete TM sequence in the nascent chain inside the ribosome elicits translocon pore contraction and the closure of the lumenal end of the pore while the cytoplasmic end remains closed (Fig. 4 c) (Liao et al., 1997). We have indicated in Fig. 4, c and d, that BiP blocks the pore directly, but BiP-mediated lumenal closure may be accomplished indirectly. The ribosome–translocon seal is then broken to allow a cytoplasmic domain of the nascent chain to enter the cytosol, whereas the BiP-dependent lumenal seal remains closed (Fig. 4 d). After translation of the nascent chain has been completed, the new membrane protein is released from the translocon and inserted into the ER bilayer (Do et al., 1996). Therefore, BiP plays a critical role in maintaining the permeability barrier of the ER membrane during cotranslational membrane protein integration. It will now be interesting to determine which domains and residues of BiP are involved in this function, and whether they are the same as those involved in its other functions.
Materials and methods
Plasmids, mRNA, and protein purification
Construction of plasmid pJL111 encoding the 111p protein was described previously (Do et al., 1996). Plasmid pSPJL111 was created by transferring the KpnI–BssHII (Klenow-blunted) DNA fragment of pJL111 into pGEM4Z using the KpnI and HindIII (Klenow-blunted) sites (unpublished data). To produce nascent chains of defined lengths, plasmids were cleaved with restriction endonucleases within the coding region and transcribed in vitro using SP6 RNA polymerase as before (Liao et al., 1997). Transcription of EcoRI-, SpeI-, and SalI-cut pSPJL111 yielded truncated mRNAs coding for the first 86, 91, and 93 amino acids of the 111p protein.
Plasmid pQEwtBiP, a gift of Dr. Linda Hendershot (St. Jude's Children's Research Hospital, Memphis, TN), directs the IPTG-inducible expression of mature hamster BiP protein with an amino-terminal 6×-histidine tag. Recombinant hamster BiP protein was overexpressed and purified from Escherichia coli as described previously (Gaut and Hendershot, 1993).
Integration intermediates
Integration intermediates containing 111p nascent chains of different lengths were prepared as before (Liao et al., 1997). Translations were performed in vitro as described previously (Crowley et al., 1993, 1994) using wheat germ extract, either ɛNBD-Lys-tRNALys or unmodified Lys-tRNALys (Crowley et al., 1993), canine SRP, and ∼100 equivalents of canine KRMs, XRMs, or RRMs prepared as described below. After translation, KOAc, pH 7.5, was added to a final concentration of 0.5 M to newly synthesized integration intermediates, and they were incubated on ice for 10 min. Integration intermediates were then purified by gel filtration chromatography using a Sepharose CL−2B column (0.7 cm i.d. × 50 cm) equilibrated and run in buffer A (50 mM Hepes, pH 7.5, 40 mM KOAc, pH 7.5, 5 mM MgCl2). Prior to sample loading, a 2-ml preload of high-salt buffer A (50 mM Hepes, pH 7.5, 0.5 M KOAc, pH 7.5, 5 mM MgCl2) was run into the column to allow microsome separation from salt-sensitive NBD-containing adsorbed material before the intermediates moved into buffer A in the column. Where indicated, affinity-purified antibodies against Sec61α were incubated with XRM-containing intermediates for 20 min on ice following their purification by gel filtration and before fluorescence measurements. After fluorescence measurements, the samples were analyzed biochemically to assess the extent of ɛNBD-Lys incorporation and membrane association of ribosome–nascent chain complexes (Crowley et al., 1993).
Extraction of soluble lumenal proteins
Extraction of soluble lumenal proteins was performed by high pH treatment as described (Nicchitta and Blobel, 1993; Hamman et al., 1998). A 100-μl volume of buffer B (500 mM Hepes/500 mM CAPS titrated to pH 9.5) was added to 100 equivalents of canine rough microsomes in 100 μl of buffer C (50 mM Hepes, pH 7.5, 1 mM DTT, 200 mM sucrose), and this mixture was diluted to 1 ml with water. After a 20-min incubation on ice, the XRMs were collected by sedimentation through a 0.5-M sucrose cushion in buffer A and resealed by resuspension in 100 μl buffer C at 4°C.
Reconstitution
Reconstitution was performed essentially as described in Bulleid and Freedman (1988), a procedure similar to that used in our lab previously (Hamman et al., 1998). XRMs were prepared as described above, except that the microsomes were collected by sedimentation through a high pH cushion (0.5 M sucrose in 0.1 × buffer B). Microsomes were reconstituted by resuspension in ∼250 μl of a solution containing 10–20 μM of purified hamster BiP (Gaut and Hendershot, 1993) and 200 mM sucrose in buffer B at pH 9.5. The microsomes were incubated on ice for 5 min, at which time ATP was added to a final concentration of 5 mM and the pH was then immediately reduced to 7 by the addition of 50 μl of 1.0 M Hepes, pH 6.8. After a further 5-min incubation on ice, the microsomes were diluted to 1 ml with buffer C and collected by sedimentation (Beckman TLA 100.2 rotor, 60,000 rpm, 20 min, 4°C) through a 200-μl cushion of 0.5 M sucrose in buffer A. The microsomes were then resuspended in 100 μl buffer C to yield sealed microsomes reconstituted with rBiP (RRMs). For AMPPNP experiments, the purified BiP protein was preincubated with a 600-fold molar excess of AMPPNP for ∼60 min to replace any bound nucleotide (Wei and Hendershot, 1995) and no ATP was added before resealing. Where indicated, affinity-purified antibodies against rBiP (Hendershot et al., 1996) were incubated with purified rBiP protein at neutral pH on ice for 10–20 min before reconstitution as above.
Fluorescence spectroscopy
Collisional quenching experiments were performed on an SLM-8100 spectrofluorimeter in Buffer A at 4°C as described previously (Crowley et al., 1994; Hamman et al., 1997). Fluorescent (ɛNBD-Lys-containing) and nonfluorescent (Lys-containing) samples were prepared and analyzed in parallel in order to subtract scatter and background signals from the measured intensities of the NBD-containing samples. To introduce large holes into the ER microsomes, purified PFO protein was added to samples at a final concentration of ∼100 nM and allowed to react for 30 min at room temperature before further fluorescence measurements were made.
Acknowledgments
We are grateful to Yiwei Miao and Yuanlong Shao for superb technical assistance, to Dr. Linda Hendershot for the rBiP plasmid, BiP antibodies, and valuable advice, and to current and former members of the Johnson lab for helpful discussions.
This work was supported by an American Cancer Society Postdoctoral Fellowship (PF-00–151–01-CSM to N.G. Haigh), a National Institutes of Health grant (GM26494 to A.E. Johnson), and the Robert A. Welch Foundation (A.E. Johnson).
Footnotes
Abbreviations used in this paper: KRM, salt-washed ER microsome; NAD+, nicotinamide adenine dinucleotide; NBD, 7-nitrobenz-2-oxa-1,3-diazole; PFO, perfringolysin O; pPL, preprolactin; RRM, reconstituted microsome; TM, transmembrane; XRM, lumen-extracted microsome.
References
- Beckmann, R., D. Bubeck, R. Grassucci, P. Penczek, A. Verschoor, G. Blobel, and J. Frank. 1997. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 278:2123–2126. [DOI] [PubMed] [Google Scholar]
- Beckmann, R., C.M.T. Spahn, N. Eswar, J. Helmers, P.A. Penczek, A. Sali, J. Frank, and G. Blobel. 2001. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 107:361–372. [DOI] [PubMed] [Google Scholar]
- Bell, C.E., and D. Eisenberg. 1996. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry. 35:1137–1149. [DOI] [PubMed] [Google Scholar]
- Brodsky, J.L. 1996. Post-translational protein translocation: not all hsc70s are created equal. Trends Biochem. Sci. 21:122–126. [PubMed] [Google Scholar]
- Brodsky, J.L., and A.A. McCracken. 1999. ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol. 10:507–513. [DOI] [PubMed] [Google Scholar]
- Brodsky, J.L., and R. Schekman. 1993. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 123:1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J.L., J. Goeckeler, and R. Schekman. 1995. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 92:9643–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau, B., and A.L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell. 92:351–366. [DOI] [PubMed] [Google Scholar]
- Bulleid, N.J., and R.B. Freedman. 1988. Defective co-translational formation of disulphide bonds in protein disulphide-isomerase-deficient microsomes. Nature. 335:649–651. [DOI] [PubMed] [Google Scholar]
- Chapman, R., C. Sidrauski, and P. Walter. 1998. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu. Rev. Cell Dev. Biol. 14:459–485. [DOI] [PubMed] [Google Scholar]
- Chevalier, M., H. Rhee, E.C. Elguindi, and S.Y. Blond. 2000. Interaction of murine BiP/GRP78 with the DnaJ homologue MTJ1. J. Biol. Chem. 275:19620–19627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi, A.K., and R. Schekman. 1997. The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J. Cell Biol. 137:1483–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranney, M., R.B. Cundall, G.R. Jones, J.T. Richards, and E.W. Thomas. 1983. Fluorescence lifetime and quenching studies on some interesting diphenylhexatriene membrane probes. Biochim. Biophys. Acta. 735:418–425. [Google Scholar]
- Crowley, K.S., G.D. Reinhart, and A.E. Johnson. 1993. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 73:1101–1115. [DOI] [PubMed] [Google Scholar]
- Crowley, K.S., S. Liao, V.E. Worrell, G.D. Reinhart, and A.E. Johnson. 1994. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 78:461–471. [DOI] [PubMed] [Google Scholar]
- Do, H., D. Falcone, J. Lin, D.W. Andrews, and A.E. Johnson. 1996. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell 85:369–378. [DOI] [PubMed] [Google Scholar]
- Gaut, J.R., and L.M. Hendershot. 1993. Mutations within the nucleotide binding site of immunoglobulin-binding protein inhibit ATPase activity and interfere with release of immunoglobulin heavy chain. J. Biol. Chem. 268:7248–7255. [PubMed] [Google Scholar]
- Gething, M.J. 1999. Role and regulation of the ER chaperone BiP. Semin. Cell Dev. Biol. 10:465–472. [DOI] [PubMed] [Google Scholar]
- Gething, M.J., and J. Sambrook. 1992. Protein folding in the cell. Nature. 355:33–45. [DOI] [PubMed] [Google Scholar]
- Görlich, D., and T.A. Rapoport. 1993. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 75:615–630. [DOI] [PubMed] [Google Scholar]
- Hamman, B.D., J.-C. Chen, E.E. Johnson, and A.E. Johnson. 1997. The aqueous pore through the translocon has a diameter of 40-60 Å during cotranslational protein translocation at the ER membrane. Cell. 89:535–544. [DOI] [PubMed] [Google Scholar]
- Hamman, B.D., L.M. Hendershot, and A.E. Johnson. 1998. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 92:747–758. [DOI] [PubMed] [Google Scholar]
- Hendershot, L.M., J. Wei, J.R. Gaut, J. Melnick, S. Aviel, and Y. Argon. 1996. Inhibition of immunoglobulin folding and secretion by dominant negative BiP ATPase mutants. Proc. Natl. Acad. Sci. USA. 93:5269–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuck, A.P., R.K. Tweten, and A.E. Johnson. 2001. β-Barrel pore forming toxins: Intriguing dimorphic proteins. Biochemistry. 40:9065–9073. [DOI] [PubMed] [Google Scholar]
- Johnson, A.E., and M.A. van Waes. 1999. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15:799–842. [DOI] [PubMed] [Google Scholar]
- Johnson, A.E., and N.G. Haigh. 2000. The ER translocon and retrotranslocation: is the shift into reverse manual or automatic. Cell. 102:709–712. [DOI] [PubMed] [Google Scholar]
- Li, M., F. Dyda, I. Benhar, I. Pastan, and D.R. Davies. 1996. Crystal structure of the catalytic domain of Pseudomonas exotoxin A complexed with a nicotinamide adenine dinucleotide analog: implications for the activation process and for ADP ribosylation. Proc. Natl. Acad. Sci. USA. 93:6902–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, S., J. Lin, H. Do, and A.E. Johnson. 1997. Both lumenal and cytosolic gating of the aqueous ER translocon pore is regulated from inside the ribosome during membrane protein integration. Cell 90:31–41. [DOI] [PubMed] [Google Scholar]
- Lyman, S.K., and R. Schekman. 1995. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J. Cell Biol. 131:1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack, K.E.S., B. Misselwitz, K. Plath, and T.A. Rapoport. 1999. BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell. 97:553–564. [DOI] [PubMed] [Google Scholar]
- Ménétret, J.-F., A. Neuhof, D.G. Morgan, K. Plath, M. Radermacher, T.A. Rapoport, and C.W. Akey. 2000. The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell. 6:1219–1232. [DOI] [PubMed] [Google Scholar]
- Meyer, H.-A., H. Grau, R. Kraft, S. Kostka, S. Prehn, K.-U. Kalies, and E. Hartmann. 2000. Mammalian Sec61 is associated with Sec62 and Sec63. J. Biol. Chem. 275:14550–14557. [DOI] [PubMed] [Google Scholar]
- Misselwitz, B., O. Staeck, K.E.S. Matlack, and T.A. Rapoport. 1999. Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J. Biol. Chem. 274:20110–20115. [DOI] [PubMed] [Google Scholar]
- Nguyen, T.H., D.T.S. Law, and D.B. Williams. 1991. Binding protein BiP is required for translocation of secretory proteins into the endoplasmic reticulum in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 88:1565–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta, C.V., and G. Blobel. 1993. Lumenal proteins of the mammalian endoplasmic reticulum are required to complete protein translocation. Cell. 73:989–998. [DOI] [PubMed] [Google Scholar]
- Nissen, P., J. Hansen, N. Ban, P.B. Moore, and T.A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science. 289:920–930. [DOI] [PubMed] [Google Scholar]
- Panzner, S., L. Dreier, E. Hartmann, S. Kostka, and T.A. Rapoport. 1995. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 81:561–570. [DOI] [PubMed] [Google Scholar]
- Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349–355. [DOI] [PubMed] [Google Scholar]
- Pilon, M., and R. Schekman. 1999. Protein translocation: how Hsp70 pulls it off. Cell. 97:679–682. [DOI] [PubMed] [Google Scholar]
- Plemper, R.K., and D.H. Wolf. 1999. Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem. Sci. 24:266–270. [DOI] [PubMed] [Google Scholar]
- Römisch, K. 1999. Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J. Cell Sci. 112:4185–4191. [DOI] [PubMed] [Google Scholar]
- Sanders, S.L., K.M. Whitfield, J.P. Vogel, M.D. Rose, and R.W. Schekman. 1992. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell. 69:353–365. [DOI] [PubMed] [Google Scholar]
- Simons, J.F., S. Ferro-Novick, M.D. Rose, and A. Helenius. 1995. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J. Cell Biol. 130:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronek, M.K., M. Rotter, and I.G. Haas. 1999. Molecular characterization of a novel mammalian DnaJ-like Sec63p homolog. Biol. Chem. 380:1133–1138. [DOI] [PubMed] [Google Scholar]
- Tyedmers, J., M. Lerner, C. Bies, J. Dudek, M.H. Skowronek, I.G. Haas, N. Heim, W. Nastainczyk, J. Volkmer, and R. Zimmermann. 2000. Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc. Natl. Acad. Sci. USA. 97:7214–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J., and L.M. Hendershot. 1995. Characterization of the nucleotide binding properities and ATPase activity of recombinant hamster BiP purified from bacteria. J. Biol. Chem. 270:26670–26676. [DOI] [PubMed] [Google Scholar]
- Young, B.P., R.A. Craven, P.J. Reid, M. Willer, and C.J. Stirling. 2001. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 20:262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]