Abstract
Although the budding yeast centromere is extremely short (125 bp) compared to those of other eukaryotes, the kinetochore that assembles on this DNA displays a rich molecular complexity. Here, we describe recent advances in our understanding of kinetochore function in budding yeast and present a model describing the attachment that is formed between spindle microtubules and centromeric DNA. This analysis may provide general principles for kinetochore function and regulation.
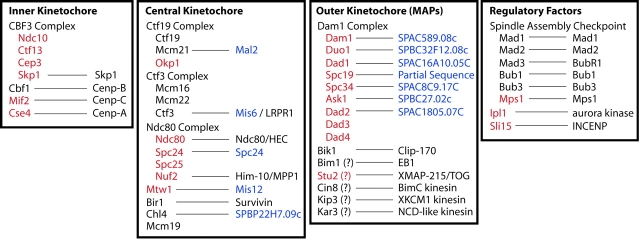
Chromosome segregation during mitosis requires a physical connection between spindle microtubules and chromosomes. This attachment occurs at proteinaceous structures called kinetochores that assemble on centromeric DNA. Studies in the budding yeast Saccharomyces cerevisiae have been particularly revealing for understanding kinetochore function. Recently, the development of sensitive assays for detecting kinetochore association has allowed the identification of >30 yeast kinetochore proteins, many of which are conserved among eukaryotes (Fig. 1). In this review, we discuss the current molecular understanding of how the budding yeast kinetochore assembles on centromeric DNA, the nature of the higher order kinetochore structure, the mechanism by which the kinetochore attaches to spindle microtubules, and how this kinetochore–microtubule attachment is regulated.
Figure 1.
Budding yeast kinetochore proteins and their homologues. Classification of budding yeast kinetochore proteins based on their function and interactions within the kinetochore. Essential genes are shown in red, and nonessential genes are shown in black. When applicable, the metazoan homologue of each protein is listed. For proteins with no identifiable metazoan homologue, the S. pombe homologue is listed in blue. In cases where kinetochore function has not been definitively established, that protein is indicated with a question mark.
The complexity of the yeast kinetochore makes the prospect of achieving a complete molecular understanding of its function and regulation a daunting prospect. Fortunately, this task is somewhat simplified by the fact that subsets of kinetochore proteins interact physically in discrete complexes or function together in signaling modules. To further simplify our discussion of the yeast kinetochore, here we propose a classification of individual kinetochore proteins and these complexes according to whether they function at the interface with centromeric DNA (inner kinetochore proteins), at the interface with spindle microtubules (outer kinetochore proteins), or at the interface between the inner and outer kinetochore proteins (central kinetochore proteins).
The inner kinetochore
Correct chromosome segregation requires that one and only one kinetochore assembles on each chromosome. To achieve this, a subset of kinetochore proteins functions to specifically recognize and bind to centromeric DNA. In budding yeast, centromeric DNA is 125 bp long and is conserved among the different chromosomes (Fitzgerald-Hayes et al., 1982). In contrast, metazoan centromeric DNA can be megabases in length and does not contain easily identifiable DNA consensus sequences (for review see Choo, 1997). Despite the differences between yeast and metazoan centromeric DNA, the kinetochores that assemble on this DNA in both cases are organized around centromeric nucleosomes that contain specialized histone H3-like proteins (yeast Cse4p or its metazoan homologue CENP-A [Meluh et al., 1998]).
Since Cse4p/CENP-A–containing nucleosomes are found only at centromeres, there must be a mechanism to target these nucleosomes specifically to centromeric DNA. Yeast have solved this problem in part through the activities of additional DNA-binding kinetochore proteins. The yeast centromeric nucleosome binds to an 80-bp sequence (termed CDEII) that spans the middle of the centromere. The DNA sequences on either side of CDEII (termed CDEI and CDEIII) also serve as binding sites for distinct proteins. The most important of these is the CBF3 complex (Ndc10p, Cep3p, Ctf13p, and Skp1p), which binds to CDEIII (Lechner and Carbon, 1991). In the absence of CBF3, kinetochore function is abolished in vivo and in vitro (Goh and Kilmartin, 1993; Sorger et al., 1994), and the association of all known kinetochore proteins with the centromere, including Cse4p (Ortiz et al., 1999), is disrupted. In contrast, the association of CBF3 with centromere DNA in vivo does not require Cse4p (Measday et al., 2002). Therefore, the specific binding of CBF3 to CDEIII helps define the position of the yeast kinetochore.
The yeast inner kinetochore also contains two additional DNA-binding proteins. CDEI serves as a binding site for a homodimer of Cbf1p (Mellor et al., 1990). Although Cbf1p is not essential for kinetochore function, it induces the bending of DNA (Niedenthal et al., 1993) and may therefore contribute to the higher order structure of the kinetochore. Cbf1p has structural similarity and limited sequence identity to CENP-B, which binds to metazoan centromeric DNA and also induces DNA bending (Tanaka et al., 2001). Physical and genetic evidence suggests that Mif2p, a protein with similarity to metazoan CENP-C, also binds to centromeric DNA near Cbf1p (Meluh and Koshland, 1995, 1997). However, despite the presence of DNA-binding motifs (Meluh and Koshland, 1995) Mif2p has not been shown to bind directly to this DNA sequence in vitro.
The central kinetochore
Based on identified physical interactions, it appears that inner kinetochore proteins do not associate directly with microtubules or the microtubule-binding components of the outer kinetochore. Therefore, we propose that “central kinetochore” proteins mediate the linkage between the inner and outer kinetochore proteins. One important central kinetochore component appears to be the Ctf19 complex (Ctf19p, Mcm21p, and Okp1p), which binds to each of the inner kinetochore components described above (Ortiz et al., 1999). By virtue of its two-hybrid interactions, the Ctf19 complex also appears well positioned to link together the Ctf3 complex (Ctf3p, Mcm16p, and Mcm22p [Measday et al., 2002]) and the Ndc80 complex (Ndc80p, Spc24p, Spc25p, and Nuf2p [Janke et al., 2001; Wigge and Kilmartin, 2001]). The Ndc80 complex is especially important for kinetochore function, since mutants in this complex are completely defective for chromosome segregation, similar to CBF3 mutants.
There are also a variety of kinetochore proteins which are less defined in terms of their physical interactions. Since these proteins have not been shown to bind to either microtubules or centromeric DNA, we have tentatively classified them here as central kinetochore proteins. These include Mtw1p, which is essential for kinetochore function with mtw1-1 mutants, showing highly abnormal DNA segregation (Goshima and Yanagida, 2000). Other kinetochore proteins, including Slk19p (Zeng et al., 1999), Mcm19p (Ghosh et al., 2001), Chl4p (Roy et al., 1997), and Bir1p (Yoon and Carbon, 1999), are not essential for viability and therefore might play redundant or nonessential roles in kinetochore function or structural integrity. Although initial studies have indicated that Bir1p, a homologue of the metazoan kinetochore passenger protein Survivin, shows genetic and two-hybrid interactions with the CBF3 complex (Yoon and Carbon, 1999), future work will be required to establish the specific roles that these nonessential proteins play at the kinetochore.
The outer kinetochore
The most critical function of a kinetochore is to connect chromosomes to microtubules. However, until recently it was unclear how yeast kinetochores attach to spindle microtubules. In metazoans, microtubule-associated motors, including CENP-E, dynein, and XKCM1, play roles in mediating kinetochore–microtubule attachments and in subsequent chromosome movements during congression and anaphase A (for review see Heald, 2000). Yeast lack CENP-E, and yeast dynein appears restricted to cytoplasmic microtubules. However, yeast Kip3p may be an XKCM1 homologue (Severin et al., 2001), suggesting that it may function at kinetochores. Cin8p, a BimC-related kinesin, associates with kinetochores in vivo (He et al., 2001), and in vitro assays have suggested that the motor protein Kar3p plays a role in kinetochore function (Middleton and Carbon, 1994). Although deletion of CIN8, KAR3, or KIP3 individually does not dramatically affect chromosome segregation, cin8Δ kip3Δ and kip3Δ kar3Δ double mutants are inviable (Miller et al., 1998), possibly reflecting redundant roles for these motors at the kinetochore.
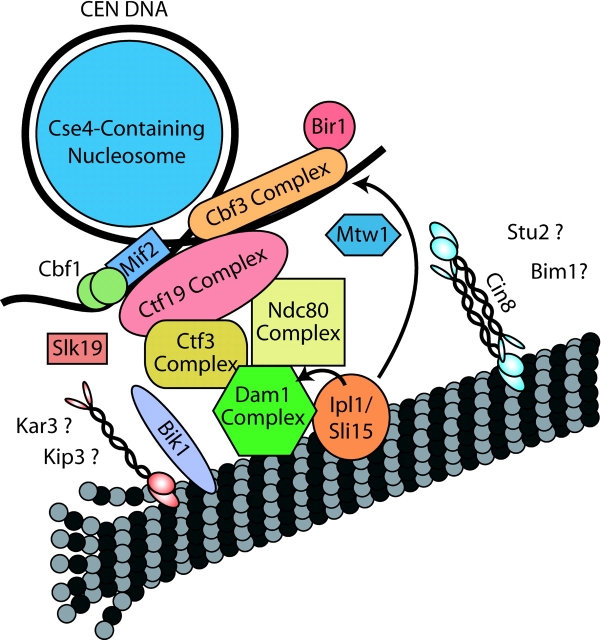
Nonmotor microtubule-associated proteins (MAPs)* also appear to play roles in mediating kinetochore–spindle attachments. The first kinetochore-associated MAP to be identified was Dam1p, a component of the Dam1p complex (Dam1p, Duo1p, Dad1p, Spc19p, Spc34p, Dad2p, Ask1p, Dad3p, and Dad4p [Cheeseman et al., 2001a; Janke et al., 2002; Li et al., 2002; unpublished data]). This complex localizes to kinetochores in an Ndc10- and Ndc80-dependent manner (Enquist-Newman et al., 2001; Jones et al., 2001; Janke et al., 2002) and is essential for chromosome segregation. Phenotypic analyses of dam1 mutants showed that their spindles have monopolar attachments to paired sister chromatids (Cheeseman et al., 2001b; He et al., 2001), possibly reflecting an inability to form new kinetochore–microtubule attachments or a role in chromosome biorientation (Janke et al., 2002). The Dam1p complex interacts physically with central kinetochore proteins of both the Ctf3 and Ndc80 complexes (Fig. 2) (Cheeseman et al., 2001a; Measday et al., 2002).
Figure 2.
Schematic diagram of the kinetochore. This model is based on the organization of the DNA-binding proteins (Espelin et al., 1997; Meluh and Koshland, 1997; Meluh et al., 1998) and the known physical interactions of the different kinetochore proteins (Cheeseman et al., 2001a).
Electron microscope studies of vertebrate kinetochores revealed that microtubule plus ends make end-on attachments with the kinetochore. This observation suggests a role for plus-end–binding MAPs in the microtubule kinetochore linkage. Although there is no direct evidence for such an end-on attachment in yeast, Bik1p, a member of the Clip-170 family of plus-end tracking MAPs, does localize to kinetochores independent of its microtubule-binding activity (He et al., 2001; Lin et al., 2001). bik1 mutants deleted for the kinetochore interaction domain have normal microtubule dynamics. However, in polyploid cells, highly sensitive to disrupted kinetochore function because of numerous spindle connections, these bik1 mutants show defects in chromosome segregation (Lin et al., 2001). Bim1p, a plus-end–binding MAP of the EB1 family, may also play a role in kinetochore function. In fact, bim1Δ mutants show genetic interactions with genes encoding a variety of kinetochore components (Tong et al., 2001).
Additional studies have also implicated Stu2p, a homologue of the microtubule stabilizing protein XMAP215, in kinetochore function (He et al., 2001). A similar role was suggested independently for the Schizosaccharomyces pombe Stu2p homologues, Dis1 and Mtc1 (Nakaseko et al., 2001). However, as with other MAPs that play multiple roles in spindle function the significance of these results is unclear. Although stu2 mutants show altered chromosome dynamics (He et al., 2001), Stu2p plays a key role in modulating microtubule dynamics (Severin et al., 2001). Therefore, altered chromosome dynamics may reflect microtubule defects instead of a role for Stu2p in mediating kinetochore–microtubule interactions. Nevertheless, it appears that multiple MAPs play roles at the kinetochore.
Regulation at the kinetochore
To ensure that the kinetochore proteins described above function properly, the conserved mitotic checkpoint (Fig. 1) monitors the formation of bipolar kinetochore–microtubule attachments and, in the event of an error, arrests a cell in metaphase (for review see Amon, 1999). In budding yeast, the primary signal for this checkpoint is the absence of an attachment between the centromere and the spindle, implicating kinetochore proteins as the source of this signal. In fact, mutants in several kinetochore proteins, including Ndc10p, Ctf13p, Cep3p, Spc24p, and Spc25p, are defective for the mitotic checkpoint (Gardner et al., 2001; Janke et al., 2001), indicating either that an intact kinetochore is required for checkpoint function or that these proteins signal to the mitotic checkpoint.
In addition to sensing attachment defects, the yeast mitotic checkpoint also appears to monitor tension on the kinetochore. Bipolar attachments exert sufficient force on the kinetochore to pull centromeric regions apart before anaphase (Goshima and Yanagida, 2000; He et al., 2000). Interestingly, a lack of tension can temporarily activate the yeast mitotic checkpoint even when kinetochore–microtubule connections are intact (Stern and Murray, 2001). Although the classically defined checkpoint components are necessary for this tension checkpoint, it additionally requires Ipl1p (Biggins and Murray, 2001), an aurora protein kinase.
It is less clear how yeast kinetochores change through the cell cycle. Centromeres are positioned near the spindle poles throughout the cell cycle (Jin et al., 2000). This localization requires microtubules and functional kinetochores, suggesting that active attachments between the spindle and the kinetochore exist during the majority of the cell cycle. However, changes in kinetochore function may occur during events such as the assembly of a new kinetochore, during spindle assembly to facilitate the formation of bipolar attachments, and during anaphase A when kinetochores move to the spindle poles.
Several proteins have been identified that may regulate these and other aspects of kinetochore function. The best characterized of these regulatory factors is the Ipl1p protein kinase, which is essential for chromosome segregation. Ipl1p can associate with kinetochores (Biggins and Murray, 2001; Kang et al., 2001) and directly with microtubules (Kang et al., 2001). Therefore, it is well positioned to regulate kinetochore–microtubule attachments. Ipl1p associates closely in vivo with Sli15p, an INCENP homologue that plays a role in activating Ipl1p's kinase activity and possibly in substrate recognition (Kang et al., 2001). Both ipl1 and sli15 mutants show high frequencies of chromosome missegregation and monopolar attachments of paired sister chromatids to the spindle (Biggins et al., 1999; Kim et al., 1999; He et al., 2001). Ipl1p inhibits kinetochore–microtubule attachments in vitro by phosphorylating the CBF3 subunit Ndc10p (Biggins et al., 1999). In addition, Ipl1p phosphorylates Dam1p in vivo and in vitro (Kang et al., 2001), consistent with the indistinguishable chromosome missegregation phenotypes observed for mutants of each protein. Recently, it was shown that kinetochores are unable to detach from the spindle pole in ipl1 mutants, suggesting that a primary function for Ipl1p is promoting the turnover of these attachments to facilitate sister chromatid biorientation (Tanaka et al., 2002).
A working model
The work described here has led to the most detailed molecular picture of an intact kinetochore in any organism. Figs. 1 and 2 are an attempt to synthesize what is currently known about the protein composition of the budding yeast kinetochore and how these proteins associate with each other, centromeric DNA, and the spindle. Although it is possible that some yeast kinetochore proteins remain unidentified, the diagram shown in Fig. 2 provides a useful working model for understanding kinetochore function and should allow predictions to be made on the order of assembly of proteins at the kinetochore. However, many key questions regarding kinetochore function remain to be answered. Future work must address issues such as the specific functions of individual kinetochore proteins, how kinetochores move to the poles in anaphase A, how kinetochore assembly and function are regulated, and how unattached kinetochores activate the mitotic checkpoint. With a large list of kinetochore proteins in hand and with knowledge of the specific interactions and biological importance of many kinetochore components, the answers to these questions should now be accessible.
Acknowledgments
We thank K. Kozminski, S. Gadde, R. Heald, and J. Sharp for critical reading of the article. We apologize to those researchers whose work we were unable to cite due to space constraints.
Our work is supported by a grant from the National Institute of General Medical Sciences to G. Barnes (GM-47842) and a National Science Foundation Graduate Research fellowship to I.M. Cheeseman.
Footnotes
Abbreviation used in this paper: MAP, microtubule-associated protein.
References
- Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69–75. [DOI] [PubMed] [Google Scholar]
- Biggins, S., and A.W. Murray. 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15:3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., F.F. Severin, N. Bhalla, I. Sassoon, A.A. Hyman, and A.W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., C. Brew, M. Wolyniak, A. Desai, S. Anderson, N. Muster, J.R. Yates, T.C. Huffaker, D.G. Drubin, and G. Barnes. 2001. a. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., M. Enquist-Newman, T. Müller-Reichert, D.G. Drubin, and G. Barnes. 2001. b. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 152:197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, K.H.A. 1997. The Centromere. Oxford University Press, Oxford, UK. 304 pp.
- Enquist-Newman, M., I.M. Cheeseman, D. Van Goor, D.G. Drubin, P. Meluh, and G. Barnes. 2001. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell. 12:2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin, C.W., K.B. Kaplan, and P.K. Sorger. 1997. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J. Cell Biol. 139:1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes, M., L. Clarke, and J. Carbon. 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 29:235–244. [DOI] [PubMed] [Google Scholar]
- Gardner, R.D., A. Poddar, C. Yellman, P.A. Tavormina, M.C. Monteagudo, and D.J. Burke. 2001. The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics. 157:1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S.K., A. Poddar, S. Hajra, K. Sanyal, and P. Sinha. 2001. The IML3/MCM19 gene of Saccharomyces cerevisiae is required for a kinetochore-related process during chromosome segregation. Mol. Genet. Genomics. 265:249–257. [DOI] [PubMed] [Google Scholar]
- Goh, P.-Y., and J.V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 100:619–633. [DOI] [PubMed] [Google Scholar]
- He, X., S. Asthana, and P.K. Sorger. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 101:763–775. [DOI] [PubMed] [Google Scholar]
- He, X., D.R. Rines, C.W. Espelin, and P.K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 106:195–206. [DOI] [PubMed] [Google Scholar]
- Heald, R. 2000. Motor function in the mitotic spindle. Cell. 102:399–402. [DOI] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, J. Lechner, A. Shevchenko, M.M. Magiera, C. Schramm, and E. Schiebel. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, T.U. Tanaka, J. Lechner, and E. Schiebel. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q.-w., J. Fuchs, and J. Loidl. 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113:1903–1912. [DOI] [PubMed] [Google Scholar]
- Jones, M.H., X. He, T.H. Giddings, and M. Winey. 2001. Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc. Natl. Acad. Sci. USA. 98:13675–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.-s., I.M. Cheeseman, G. Kallstrom, S. Velmurugan, G. Barnes, and C.S.M. Chan. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H., J.S. Kang, and C.S. Chan. 1999. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145:1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, J., and J. Carbon. 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 64:717–725. [DOI] [PubMed] [Google Scholar]
- Li, Y., J. Bachant, A.A. Alcasabas, Y. Wang, J. Qin, and S.J. Elledge. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H., P. de Carvalho, D. Kho, C.-Y. Tai, P. Pierre, G.R. Fink, and D. Pellman. 2001. Polyploids require Bik1 for kinetochore-microtubule attachment. J. Cell Biol. 155:1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday, V., D.W. Hailey, I. Pot, S. Givan, K.M. Hyland, G. Cagney, S. Fields, T.N. Davis, and P. Hieter. 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor, J., W. Jiang, M. Funk, J. Rathjen, C.A. Barnes, T. Hinz, J.H. Hegemann, and P. Philippsen. 1990. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 9:4017–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and D. Koshland. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 6:793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., P. Yang, L. Glowczewski, D. Koshland, and M.M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 94:607–613. [DOI] [PubMed] [Google Scholar]
- Middleton, K., and J. Carbon. 1994. KAR3-encoded kinesin is a minus-end-directed motor that functions with centromere binding proteins (CBF3) on an in vitro yeast kinetochore. Proc. Natl. Acad. Sci. USA. 91:7212–7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R.K., K.K. Heller, L. Frisen, D.L. Wallack, D. Loayza, A.E. Gammie, and M.D. Rose. 1998. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell. 9:2051–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko, Y., G. Goshima, J. Morishita, and M. Yanagida. 2001. M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr. Biol. 11:537–549. [DOI] [PubMed] [Google Scholar]
- Niedenthal, R.K., M. Sen-Gupta, A. Wilmen, and J.H. Hegemann. 1993. Cpf1 protein induced bending of yeast centromere DNA element I. Nucleic Acids Res. 21:4726–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, J., O. Stemmann, S. Rank, and J. Lechner. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13:1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, N., A. Poddar, A. Lohia, and P. Sinha. 1997. The mcm17 mutation of yeast shows a size-dependent segregational defect of a mini-chromosome. Curr. Genet. 32:182–189. [DOI] [PubMed] [Google Scholar]
- Severin, F., B. Habermann, T. Huffaker, and T. Hyman. 2001. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 153:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger, P.K., F.F. Severin, and A.A. Hyman. 1994. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J. Cell Biol. 127:995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, B.M., and A.W. Murray. 2001. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 11:1462–1467. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U., N. Rachidi, C. Janke, G. Pereira, M. Galova, E. Schiebel, M.J.R. Stark, and K. Nasmyth. 2002. Evidence that the Ipl1-Sli15 (aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., O. Nureki, H. Kurumizaka, S. Fukai, S. Kawaguchi, M. Ikuta, J. Iwahara, T. Okazaki, and S. Yokoyama. 2001. Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 20:6612–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A.H.Y., M. Evangelista, A.B. Parsons, H. Xu, G.D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C.W.V. Hogue, H. Bussey, et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294:2364–2368. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., and J.V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, H.J., and J. Carbon. 1999. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. USA. 96:13208–13213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., J.A. Kahana, P.A. Silver, M.K. Morphew, J.R. McIntosh, I.T. Fitch, J. Carbon, and W.S. Saunders. 1999. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146:415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]