Abstract
Lon (or La) is a soluble, homooligomeric ATP-dependent protease. Mass determination and cryoelectron microscopy of pure mitochondrial Lon from Saccharomyces cerevisiae identify Lon as a flexible ring-shaped heptamer. In the presence of ATP or 5′-adenylylimidodiphosphate, most of the rings are symmetric and resemble other ATP-driven machines that mediate folding and degradation of proteins. In the absence of nucleotides, most of the rings are distorted, with two adjacent subunits forming leg-like protrusions. These results suggest that asymmetric conformational changes serve to power processive unfolding and translocation of substrates to the active site of the Lon protease.
Lon (or La) is a highly conserved ATP-dependent protease found in archaea, eubacteria, and mitochondria (1–5). In mitochondria of the yeast Saccharomyces cerevisiae, ATP-dependent proteases not only degrade abnormal proteins (6–8) but also stabilize the mitochondrial genome, regulate mitochondrial gene expression, and facilitate the assembly of oligomeric protein complexes (6, 7, 9–11). The proposed chaperone-like activity of Lon and other ATP-dependent proteases may explain the energy requirement of these enzymes. Their ATPase activity is not required for peptide bond hydrolysis per se but, rather, for unfolding or remodeling target substrates (4, 5, 12–16).
The ATP-dependent proteases ClpAP, HslUV, and the proteasome are all ring-shaped structures with 6- or 7-fold symmetry that resemble the chaperonin GroEL-GroES (17–25). This structural similarity suggests that ATP-dependent protein folding and ATP-dependent protein degradation are mechanistically related. ClpAP, HslUV, and the proteasome are two-component ATPase–protease complexes; homooligomeric Lon is unique in that its ATP-dependent chaperone-like function and its proteolytic function are combined within a single subunit. Lon is thus an attractive model to understand structure–function relationships in the more complex ATP-dependent proteases. Here, we describe the structure of the Lon (Pim1p) protease from S. cerevisiae mitochondria.
MATERIALS AND METHODS
Purification.
Active Lon carrying six C-terminal histidine residues was overexpressed in yeast, was rapidly purified to homogeneity from a isolated mitochondrial extract on a Ni2+-NTA column, and purified to homogeneity by gel filtration on a Superose 6 column as described (11); however, where indicated, 1 mM ATP or 1 mM 5′-adenylylimidodiphosphate (AMP-PNP) were present during solubilization and purification.
Analytical Ultracentrifugation.
Purified Lon (1 mg/ml) was analyzed by analytical ultracentrifugation in 20 mM Hepes (pH 8.0), 150 mM NaCl, and 10% (wt/vol) glycerol either in the absence or presence of 1 mM ATP. Sedimentation velocity and sedimentation equilibrium determinations were carried out in a Beckman Coulter Model E analytical ultracentrifuge equipped with interference and schlieren optics and in a Beckman Model XLA ultracentrifuge equipped with absorption optics. Sedimentation velocity was performed at 40,000 rpm at 20°C in a single sector 12-mm cell with schlieren optics. Sedimentation equilibrium was performed by using interference optics (5,200 rpm at 20°C in a 12-mm double sector cell) as well as by using absorption optics (scanned at 4,500 rpm at 20°C at 283 nm). Results of sedimentation equilibrium were analyzed by using a floating baseline computer program with adjusted baseline absorbance (or zero fringe) to obtain lnA versus r2 (A = the absorbance or the fringe shift in sedimentation equilibrium performed with absorption optics or interference optics, respectively; r = the radial distance). A partial specific volume of 0.73 cm3/g and a solution density of 1.03 g/cm3 was used; for the conversion of S′ to S20w, the solution viscosity was taken as 1.31 centipoise.
Transmission Electron Microscopy (TEM).
The grids for cryonegative stain TEM were prepared as described (26–28). In brief, the sample was exposed to staining solution (16% ammonium molybdate, pH 7.0) in the holes of a perforated carbon film for 30 s and was quick-frozen. These cryonegative stain grids were mounted into a Gatan (Pleasanton, CA) 626-DH Cryo-Holder operated at −180°C. Images were recorded with a Hitachi (Tokyo) H8000 transmission electron microscope operated at 100 kV at 80,000× nominal magnification under low-dose conditions. The electron dose for recording one image was ≈20 e−/Å2. Recording media were Kodak SO-163 negatives that were developed for 10 min in full-strength developer. Images were digitized with a Leafscan-45 (Scitex, Herzlia, Israel) at 1.25 Å/pixel on the specimen plane.
Image Treatment.
Images were digitally processed by using the spider software (29). From 58 digitized negatives of crosslinked and cryonegatively stained samples, 9,500 particles were automatically and reference-free selected and boxed into 128 × 128 pixel images. From these, 4,765 particle images were selected by manual inspection, were reference-free aligned (30), and were subjected to a multivariate statistical analysis with classification (31, 32). Eighty-two percent of the particle images were classified into two classes, and particle images of each class were separately reference-free aligned and averaged. No symmetry was imposed at any step during the whole image treatment.
RESULTS
Stability.
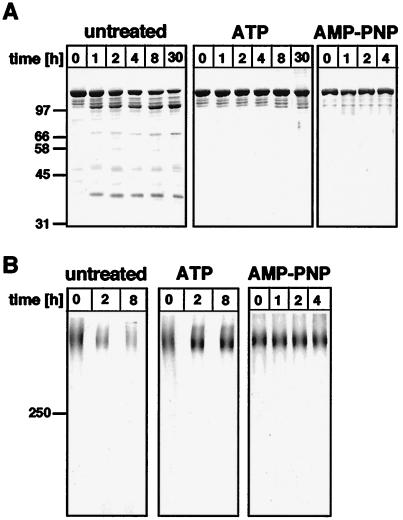
The pure active Lon was unstable at 30°C, presumably because it cleaved itself, but degradation was prevented by 1 mM ATP or AMP-PNP (Fig. 1). These nucleotides stabilized both the individual subunits (Fig. 1A) as well as the oligomeric enzyme (Fig. 1B), suggesting that stabilization required binding but not hydrolysis of ATP.
Figure 1.
Lon is stabilized by ATP or AMP-PNP. (A) Stability of the Lon subunit. Lon (2–5 μg) purified and kept in the absence (untreated) or in the presence of 1 mM ATP or 1 mM AMP-PNP was incubated at 30°C for the indicated times and then was analyzed by 10% SDS/PAGE and staining with Coomassie blue. Shown at left are molecular weight standards in kDa. (B) Stability of the Lon oligomer. Lon (5–8 μg) purified and kept in the absence (untreated) or the presence of 1 mM ATP or 1 mM AMP-PNP was incubated at 30°C for the indicated times, was crosslinked with 0.1% glutaraldehyde for 30 min on ice, and was analyzed by 3.3% SDS/PAGE and Coomassie blue staining (34). At left is a recombinant 250-kDa molecular weight marker provided by Amersham Pharmacia.
Scanning Transmission Electron Microscopy (STEM) Mass Analysis.
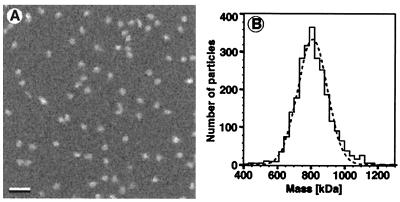
To determine the number of subunits in the holoenzyme, the mass of pure, active Lon was measured by STEM (17). When Lon purified in the presence of ATP was crosslinked with 0.1% glutaraldehyde, adsorbed onto thin carbon film, freeze-dried, and imaged in the STEM, circular particles of uniform brightness were seen (Fig. 2A). The measured mass values had a Gaussian distribution with a single peak at 804 kDa (Fig. 2B). As each Lon subunit has a mass of 117 kDa, these data identify Lon as a heptamer. STEM analysis required crosslinking of the Lon complex to prevent dissociation of the oligomer during the grid preparation. Lon purified in the presence of ADP or without nucleotides had essentially the same mass as Lon prepared in the presence of ATP, provided that crosslinking was carried out immediately after purification.
Figure 2.
Mass of Lon determined by STEM. (A) Crosslinked, freeze-dried, and unstained Lon, purified in the presence of ATP, was recorded in the dark-field mode at a dose of 200–400 electrons/nm2 with a Vacuum Generator STEM HB5 operated at 80 kV. (Bar = 50 nm.) (B) Mass evaluation of 2,641 particles yielded a histogram with a Gaussian envelope. After correction for the electron dose-associated mass loss (17), the Gauss peak was centered at 804 kDa and had a standard deviation of 76 kDa. The standard mean error was 3 kDa, which, together with the absolute calibration error of 5%, yielded a total error of 43 kDa.
Analytical Ultracentrifugation.
We also measured the mass of the pure Lon complex by analytical ultracentrifugation. When purified in the presence of ATP, Lon had a mass of 818 ± 40 kDa as determined by sedimentation velocity and a mass of 838 ± 40 kDa as determined by sedimentation to equilibrium. Adding ATP was essential for preserving the Lon oligomer during ultracentrifugation. If no ATP was added, TEM of negatively stained samples showed that Lon dissociated during the extended time required for centrifugation to equilibrium. Ultracentrifugation thus showed Lon to be a heptamer, in excellent agreement with the STEM analysis.
TEM.
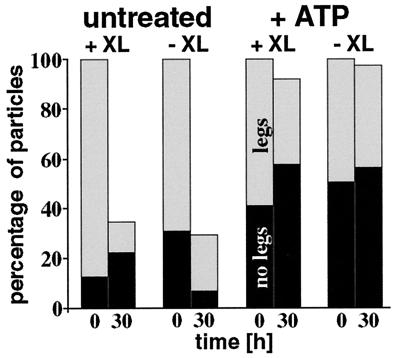
The projected three-dimensional structure of purified Lon was analyzed by TEM. Lon that had been purified and stored on ice in the absence of added nucleotides and without crosslinking appeared as a heterogeneous particle population with many small fragments. This result was obtained both with negatively stained samples and with unstained samples that had been vitrified in the holes of a perforated carbon-film grid (26, 27). In contrast, Lon samples examined immediately after purification appeared as uniform circular particles that frequently had leg-like protrusions. Systematic analysis of negatively stained Lon samples that had been purified and stored for different times in the absence or presence of ATP revealed that ATP increased the fraction of legless particles 2- to 7-fold over controls without added ATP (Fig. 3). ATP-treated Lon retained its cylindrical ring-like appearance for as long as 30 hr at 30°C even without being cross-linked before examination.
Figure 3.
Effect of ATP and crosslinking on the frequency of leg-less Lon particles. Lon purified and maintained either in the absence (untreated) or presence of 1 mM ATP was negatively stained with uranyl formate and was air dried and viewed in the transmission electron microscope either immediately after purification or after incubation at 30°C for 30 h. Samples were placed onto the grid without further treatment (−XL) or after having been crosslinked with 0.1% glutaraldehyde for 30 min on ice (+XL). From 37 negatives with images of the different samples, 22,249 particles were manually scored as particles without legs (black bars), particles with legs (gray bars), or unassigned particles (no bars). The sum of Lon particles with and without legs at the 0 h time point was taken as 100%.
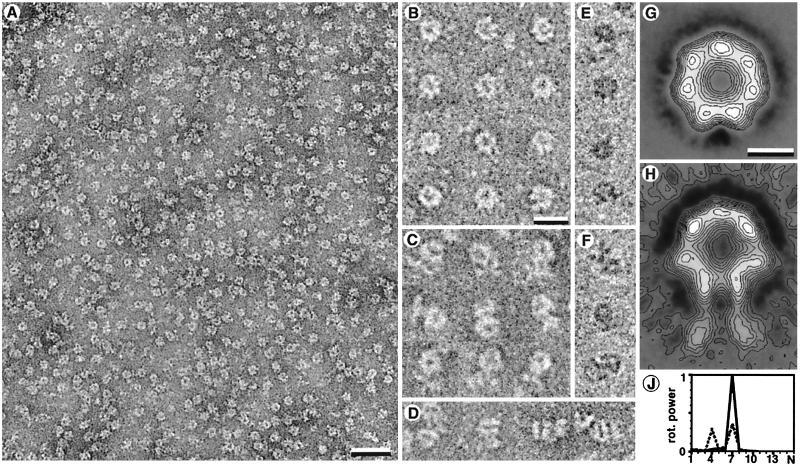
A better resolution was achieved by cryonegative staining TEM (28). When applied to crosslinked Lon, this method revealed the ring-shaped particles even more clearly (Fig. 4 A–D) than examination of vitrified unstained preparations (Fig. 4 E and F). Also, these images revealed a mixture of legless and leg-containing particles. Noncrosslinked Lon preparations disassembled during the 30-s staining with 16% ammonium molybdate (pH 7.0).
Figure 4.
(A–D) Images of vitrified negatively stained crosslinked Lon particles (26–28). (A) Bar = 30 nm. (B) Legless Lon particles as predominating in the presence of ATP or AMP-PNP. (C) Leg-containing Lon particles. (D) Rare side views of leg-containing particles. (E and F) Leg-less (E) and leg-containing (F) crosslinked particles visualized by cryo-TEM without staining. Although the signal to noise ratio is lower, the images resemble those of negatively stained samples. [Bars = 15 nm (B–F).] (G) The nonsymmetrized average image of the first class containing 2,571 images. (H) The nonsymmetrized average of the images of the second class containing 1,377 images. The leg-like structures are less contrasted in the average image because of their high flexibility. [Bars = 5 nm (G and H).] (J) Angular power analysis of the gray-value distribution within a circular radial interval on the average images of the two classes. The abscissa is given in arbitrary units. The legless class of images clearly reveals a 7-fold symmetry (continuous line). The circular core of the average image of the second class indicates a predominant 7-fold symmetry and a weaker contribution of a 4-fold symmetry (dotted line). The latter is attributable to the deformation of the circular core of the particles.
Image Treatment.
The high signal-to-noise ratio of Lon preparations that had been negatively stained and quick-frozen allowed us to select particles automatically, align and classify them by reference-free algorithms, and calculate an average projection map at a resolution of 1.9 nm (Fig. 4 G and H). We identified two classes of particles. The average image of the most prominent particle class was a 7-fold symmetric ring with an outer diameter of 11.5 nm and an inner hole of diameter 2.5 nm. As seen in the selected particles in Fig. 4B, some of the holes had an inclusion in the center. The second class of particles yielded an average image resembling that of the first class except for two leg-like protrusions extending from one side. The overall length of this extended average particle was 17 nm. This average image had a higher variance in the image parts occupied by the leg-like protrusions, suggesting that the legs are flexible. The angular power spectra of the nonsymmetrized average images of the two classes, calculated from the ring-shaped structures, are reproduced in Fig. 4J. The leg-less average image had a 7-fold symmetry. In contrast, the leg-containing average image had a 7-fold symmetry together with a 4-fold symmetric contribution, probably because protrusion of the adjacent legs leads to ring deformation. As the preparation of samples for TEM yielded highly oriented Lon molecules, different orientations such as side-views were too rare for image averaging. The few particles seen from the side had an overall length of 17 nm, and their core was 12 nm long and 11 nm wide (Fig. 4D).
Manual inspection of the particles with leg-like protrusions (Fig. 4C) excluded the possibility that a leg pair belongs to a second adjacent Lon complex. To exclude that the legs represent contaminants or degraded Lon subunits, we examined purified Lon by SDS/PAGE followed by either silver staining or immunoblotting with Lon antiserum. None of these methods revealed contaminating proteins or degraded Lon subunits that would have been sufficient to account for the leg-like extensions (data not shown).
DISCUSSION
The flexible leg-like protrusions of the Lon complex may be the structural correlate of a common mechanistic feature of ATP-dependent proteases. Flexible linkages have been observed between the 19S ATPase cap complexes and the 20S core of the eukaryotic proteasome, and within the cap complexes themselves (33). A flexibility requirement also may underlie the “symmetry mismatch” between the heptameric rings of the Clp protease and the hexameric ring of the ClpA ATPase (18, 24), and between the hexameric rings of the HslV protease and the heptameric or hexameric ring of the HslU ATPase (22). The ATP-dependent, asymmetric conformational changes observed in the Lon complex may be indicative of a ratcheting or rotational mechanism by which protein substrates are unfolded and processively translocated to the active site of the protease.
Acknowledgments
We thank D. Perec̆ko for help with the statistical evaluation of the particles and S. Müller for help with the evaluation of the STEM data. H.S. is grateful to M. Adrian for helpful discussions pertaining to the cryonegative staining technique. This work was supported by the M.E. Müller Foundation of Switzerland (to A.E. and H.S.), the Swiss National Science Foundation (to G.S. and E.K.), the Human Frontier Science Program Organization (to G.S.), the Human Capital and Mobility Program of the European Economic Community (to G.S.), and the Scientific Grant Agency of the Ministry of Education of the Slovak Republic and the Slovak Academy of Sciences (to E.K.).
ABBREVIATIONS
- STEM
scanning transmission electron microscopy
- TEM
transmission electron microscopy
- AMP-PNP
5′-adenylylimidodiphosphate
References
- 1.Maurizi M R. Experientia. 1992;48:178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- 2.Kutejová E, Durcova G, Surovka E, Kuzela S. FEBS Lett. 1993;329:47–50. doi: 10.1016/0014-5793(93)80190-6. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg A L, Moerschell R P, Chung C H, Maurizi M R. Methods Enzymol. 1994;244:350–375. doi: 10.1016/0076-6879(94)44027-1. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman S, Wickner S, Maurizi M R. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki C K, Rep M, Maarten van Dijl J, Suda K, Grivell L A, Schatz G. Trends Biochem Sci. 1997;22:118–123. doi: 10.1016/s0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki C K, Suda K, Wang N, Schatz G. Science. 1994;264:273–276. doi: 10.1126/science.8146662. [DOI] [PubMed] [Google Scholar]
- 7.van Dyck L, Pearce D A, Sherman F. J Biol Chem. 1994;269:238–242. [PubMed] [Google Scholar]
- 8.Wagner I, Arlt H, Dyck L V, Langer T, Neupert W. EMBO J. 1994;13:5135–5145. doi: 10.1002/j.1460-2075.1994.tb06843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rep M, van Dijl J M, Suda K, Schatz G, Grivell L A, Suzuki C K. Science. 1996;274:103–106. doi: 10.1126/science.274.5284.103. [DOI] [PubMed] [Google Scholar]
- 10.Arlt H, Steglich G, Perryman R, Guiard B, Neupert W, Langer T. EMBO J. 1998;17:4837–4847. doi: 10.1093/emboj/17.16.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Dijl J M, Kutejová E, Suda K, Perecko D, Schatz G, S. C K. Proc Natl Acad Sci USA. 1998;95:10584–10589. doi: 10.1073/pnas.95.18.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Melderen L, Thi M H D, Lecchi P, Gottesman S, Couturier M, Maurizi M R. J Biol Chem. 1996;271:27730–27738. doi: 10.1074/jbc.271.44.27730. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman S, Maurizi M R, Wickner S. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 15.Pak M, Wickner S. Proc Natl Acad Sci USA. 1997;94:4901–4906. doi: 10.1073/pnas.94.10.4901. , 10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoskins J R, Pak M, Maurizi M R, Wickner S. Proc Natl Acad Sci USA. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller S, Goldie K N, Bürki R, Häring R, Engel A. Ultramicroscopy. 1992;46:317–334. [Google Scholar]
- 18.Kessel M, Maurizi M R, Kim B, Kocsis E, Trus B L, Singh S K, Steven A C. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 19.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 20.Bochtler M, Ditzel L, Groll M, Huber R. Proc Natl Acad Sci USA. 1997;94:6070–6074. doi: 10.1073/pnas.94.12.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik H D, Huber R. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 22.Rohrwild M, Pfeifer G, Santarius U, Müller S A, Huang H C, Engel A, Baumeister W, Goldberg A L. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 24.Beuron F, Maurizi M R, Belnap D M, Kocsis E, Booy F P, Kessel M, Steven A C. J Struct Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- 25.Sigler P B, Xu Z, Rye H S, Burston S G, Fenton W A, Horwich A L. Annu Rev Biochem. 1998;67:581–608. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- 26.Adrian M, Dubochet J, Lepault J, McDowall A W. Nature (London) 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 27.Dubochet J, Adrian M, Chang J-J, Homo J-C, Lepault J, McDowall A W, Schultz P. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 28.Adrian M, Dubochet J, Fuller S D, Harris J R. Micron. 1998;29:145–160. doi: 10.1016/s0968-4328(97)00068-1. [DOI] [PubMed] [Google Scholar]
- 29.Walz J, Erdmann A, Kania M, Typke D, Koster A J, Baumeister W. J Struct Biol. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- 30.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 31.Penczek P, Radermacher M, Frank J. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- 32.Van Heel M. Ultramicroscopy. 1984;13:165–184. doi: 10.1016/0304-3991(84)90066-4. [DOI] [PubMed] [Google Scholar]
- 33.Frank J, Bretaudiere J P, Carazo J M, Verschoor A, Wagenknecht T. J Microsc (Oxford) 1988;150:99–115. doi: 10.1111/j.1365-2818.1988.tb04602.x. [DOI] [PubMed] [Google Scholar]
- 34.Azem A, Weiss C, Goloubinoff P. Methods Enzymol. 1998;290:253–268. doi: 10.1016/s0076-6879(98)90024-6. [DOI] [PubMed] [Google Scholar]