Abstract
In eukaryotic cells, polypeptides are N glycosylated after passing through the membrane of the ER into the ER lumen. This modification is effected cotranslationally by the multimeric oligosaccharyltransferase (OST) enzyme. Here, we report the first cross-linking of an OST subunit to a nascent chain that is undergoing translocation through, or integration into, the ER membrane. A photoreactive probe was incorporated into a nascent chain using a modified Lys-tRNA and was positioned in a cryptic glycosylation site (-Q-K-T- instead of -N-K-T-) in the nascent chain. When translocation intermediates with nascent chains of increasing length were irradiated, nascent chain photocross-linking to translocon components, Sec61α and TRAM, was replaced by efficient photocross-linking solely to a protein identified by immunoprecipitation as the STT3 subunit of the OST. No cross-linking was observed in the absence of a cryptic sequence or in the presence of a competitive peptide substrate of the OST. As no significant nascent chain photocross-linking to other OST subunits was detected in these fully assembled translocation and integration intermediates, our results strongly indicate that the nascent chain portion of the OST active site is located in STT3.
Keywords: N glycosylation; oligosaccharyltransferase; STT3; photocross-linking; nascent protein chain
Introduction
N-linked glycosylation is one of the most common types of eukaryotic protein modification. The attachment of carbohydrates to asparagine in nascent secretory and membrane proteins occurs cotranslationally as the polypeptides are being translocated across or integrated into the membrane of the ER. The transfer of high mannose oligosaccharides from a dolichol carrier to -Asn-X-Thr/Ser- acceptor sites in the nascent chain, with X being any amino acid except proline, is catalyzed by the oligosaccharyltransferase (OST)* enzyme (Silberstein and Gilmore, 1996; Knauer and Lehle, 1999; Yan and Lennarz, 1999). N glycosylation occurs in the ER lumen, so the OST active site must reside on the lumenal side of the ER membrane. Also, because the OST enzyme functions cotranslationally, it is located near the translocon, the site of protein translocation across and integration into the ER membrane (Johnson and van Waes, 1999).
The yeast OST is composed of eight subunits (Ost1p, Ost2p, Ost3p/Ost6p, Ost4p, Ost5p, Swp1p, Wbp1p, and Stt3p), five of which (Ost1p, Ost2p, Wbp1p, Swp1p, and Stt3p) are encoded by essential genes (Silberstein and Gilmore, 1996, Karaoglu et al., 1997; Knauer and Lehle, 1999). Ost6p is a homologue of Ost3p that is incorporated in place of Ost3p into a subset of yeast OST complexes. When the OST from vertebrate organisms is resolved by PAGE in SDS and stained with Coomassie blue, only four subunits are readily detected (Kelleher et al., 1992; Kelleher and Gilmore, 1997). The mammalian OST subunits ribophorin I (RI) (66 kD), ribophorin II (RII) (63/64 kD), OST48 (48 kD), and DAD1 (10 kD) are respectively homologous to Ost1p, Swp1p, Wbp1p, and Ost2p. However, homologues of Stt3p (STT3-A and STT3-B), Ost3p/Ost6p (N33 and IAP), and Ost4p are expressed in mammalian organisms and are assembled together with RI, RII, OST48, and DAD1 into multimeric complexes that are similar to the yeast OST (unpublished data).
Why does the OST enzyme require so many different subunits? Cotranslational glycosylation of polypeptides is a very complex process. Among the mechanistic complexities are the needs to position the OST active site near the translocon, to scan the nascent polypeptide for -N-X-T/S- sites, and to direct each appropriate nascent chain sequence to the OST active site in the proper conformation. In addition, the enzyme must recognize and move or channel its other substrate, a dolichol-linked oligosaccharide, into the OST active site. Kinetic analysis indicates that the OST contains two dolichol-linked oligosaccharide binding sites, one of which allosterically regulates donor substrate selection by the catalytic site (Karaoglu et al., 2001). Thus, in addition to activating the Asn amide nitrogen for nucleophilic attack, OST must also function topographically to ensure the proper recognition and presentation of two large and very different substrates at the active site. It is therefore the prevailing view that the multiple subunits of the OST are required to accomplish the multiple recognition, binding, transport, topographical, and, finally, chemical aspects of OST function.
However, at present, the role of each of the individual subunits has not been elucidated. In particular, the OST active site has not been conclusively linked to a specific subunit. Bause et al. (1997) showed that a chemically reactive hexapeptide reacted covalently with both a [14C]oligosaccharide and either RI or OST48, thereby suggesting that the active site of OST was located in RI and/or OST48. Chemical modification of cysteine residues has suggested that Wbp1p, the yeast homologue of OST48, is involved in the recognition of the dolichol-linked oligosaccharide (Pathak et al., 1995). Yan et al. (1999) later showed that an Asn-Bpa-Thr tripeptide, where Bpa is a photoreactive p-benzoylphenylalanine residue, served as a substrate for the yeast OST and that photoactivation of this probe in the presence of microsomes abolished OST activity. As photolysis resulted in the photolabeling of Ost1p with the peptide, the authors concluded that the -N-X-S/T- sequence is recognized by Ost1p, the yeast homologue of mammalian RI. In a more recent study, Yan and Lennarz (2002) used different photoreactive peptides and observed photolabeling of Stt3p, Ost3p, and Ost1p, ultimately concluding that Stt3p contained the peptide-binding site, based on mutagenesis arguments. Another group has proposed that the STT3 subunit contains the active site, based upon the observation that archaebacterial genomes encode an Stt3p homologue (Burda and Aebi, 1999; Wacker et al., 2002). The published data are therefore somewhat conflicting and do not generate a consensus view for the identity of the subunit that contains the OST active site.
No one has yet reported the cross-linking of an OST subunit to a nascent chain that is in the process of being translocated across or integrated into the ER membrane. This is somewhat surprising because nascent chain movement through the fully assembled translocation machinery is presumably controlled to ensure that each segment of the nascent chain is exposed to the OST active site. We therefore set out to trace the path of the nascent chain after it leaves the translocon using photocross-linking. This approach would allow us to determine which OST proteins are located adjacent to (and may interact with) the nascent chain, and hence to identify which subunits of the mammalian OST are involved in the cotranslational recognition of glycosylation sites in the growing polypeptide chain. Photoreactive probes were therefore incorporated into nascent chains using modified Lys-tRNAs and procedures that have long been used by us and others to examine translocon structure and function (Johnson and van Waes, 1999). We now show that photocross-linking between nascent chains and OST occurs on the lumenal side of the ER membrane after the probe emerges from the translocon, and this cross-linking is dependent on the presence of a cryptic glycosylation sequence in the nascent chain. As STT3 is the only OST subunit that photocross-links to the nascent chain, and as this photocross-linking is blocked by a competitive peptide substrate, STT3 appears to be responsible for the recognition of the glycosylation consensus sequence in the nascent chain and hence for forming part or all of the OST active site.
Results
Experimental strategy
We, and others, have shown previously using photocross-linking that nascent secretory proteins are adjacent to translocon components during their translocation across the ER membrane (Johnson and van Waes, 1999). As nascent chains are glycosylated cotranslationally by the OST, one would predict that a nascent polypeptide could be photocross-linked to one or more of the OST subunits after the photoreactive amino acid emerges from the lumenal side of the translocon. Yet despite much effort by several groups, no one has reported any covalent reaction between an OST polypeptide and a nascent chain undergoing translocation or integration.
A likely explanation for these negative results is that once a nascent chain has been glycosylated, it no longer associates with the OST because the glycosylated product of the enzymatic reaction would have much less affinity for the OST active site. Thus, to maximize the residency time of the nascent chain in the OST active site, and hence the probability of a covalent reaction between the OST and a nascent chain, it would be desirable to identify a cryptic OST recognition sequence. Such a sequence in the nascent chain would retain sufficient elements of the legitimate recognition sequence to bind transiently to the OST active site, but would not be glycosylated. If such a cryptic sequence were to pass by the OST active site, then one might expect it to bind, dissociate, and rebind multiple times, thereby increasing the probability of a nascent chain being located in or near the active site at the time the probe is photoactivated. This would be true even if the affinity of the OST active site were very much lower for the cryptic glycosylation site than for the authentic glycosylation site.
One possible class of cryptic glycosylation sequence is -Q-X-T-, in which a glutamine replaces the asparagine in the -N-X-T- sequence. The Q and N side chains each contain an amide bond and, hence, would be expected to have similar recognition and conformational properties. For example, an enzyme active site that recognizes and binds to the N amide bond might have some affinity for the Q amide bond, even though the OST discriminates very effectively between -N-X-T/S- and -Q-X-T/S- sites in the nascent polypeptide. However, substrate binding to the OST active site is not strictly dependent upon asparagine in the -N-X-T- sequence, as peptides containing Lα,γ diaminobutyric acid in place of asparagine are competitive inhibitors of the OST (Imperiali et al., 1992; Bause et al., 1995). Hence, it is conceivable that a -Q-X-T- sequence would have some affinity for the OST active site but would not provide the stability or steric arrangement required for catalysis. In short, whatever the mechanism of OST recognition of -N-X-T- in the nascent chain, -Q-X-T- would appear to be a good candidate for a cryptic recognition sequence.
A photoreactive probe in the nascent chain maximizes the chances of detecting a close juxtaposition of a nascent chain and an OST subunit because, upon photolysis, such a probe will react covalently with any adjacent protein. Thus, our strategy was to incorporate a photoreactive N ɛ-(5-azido-2-nitrobenzoyl)-lysine (ɛANB-Lys) probe into the X position of the cryptic glycosylation sequence because such a probe would be close to the catalytic site of the OST during nascent chain recognition and selection. Previous work has shown that a lysine in the X position or on either side of the NXT sequence does not influence the extent of glycosylation (Shakin-Eshleman et al., 1996; Mellquist et al., 1998). As for the position of the cryptic glycosylation sequence within the nascent chain, we have previously shown that the number of residues required to span the distance between the ribosomal P site and the active site of the OST complex can be measured by a glycosylation mapping assay (Whitley et al., 1996). This assay showed that glycosylation of the ribosome-bound nascent chain is observed when the glycosylation site is 65–75 residues away from the ribosomal P site.
A homogeneous population of ribosome–nascent chain complexes (RNCs) can be prepared by the translation of truncated mRNAs that lack a stop codon and encode a specific length of a secretory protein. Ribosomes initiate normally on such mRNAs and translation continues until the ribosome reaches the end of the truncated mRNA. As the mRNA lacks a stop codon, the ribosome does not dissociate from the mRNA but instead remains bound to the truncated mRNA and peptidyl-tRNA to yield RNCs in which the length of the nascent chain is dictated by the length of the truncated mRNA. When such nascent chains with signal sequences are translated in the presence of signal recognition particle (SRP) and ER microsomes, a homogeneous sample of membrane-bound RNCs is created (Fig. 1 A). All of the nascent chains examined here were long enough to be targeted efficiently to the translocon in the microsomal membrane and to be processed by signal peptidase.
Figure 1.
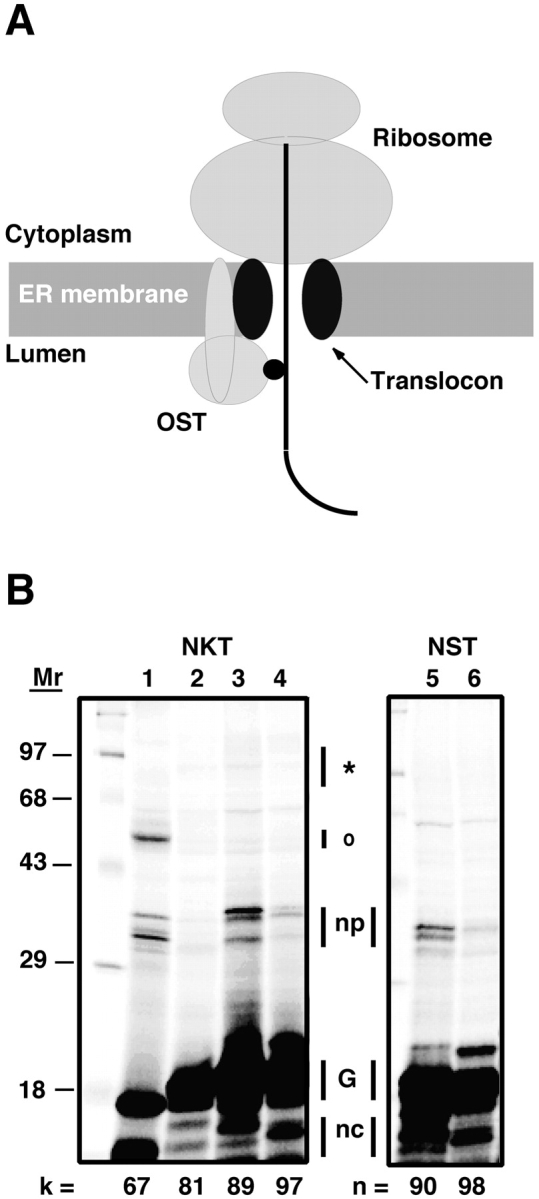
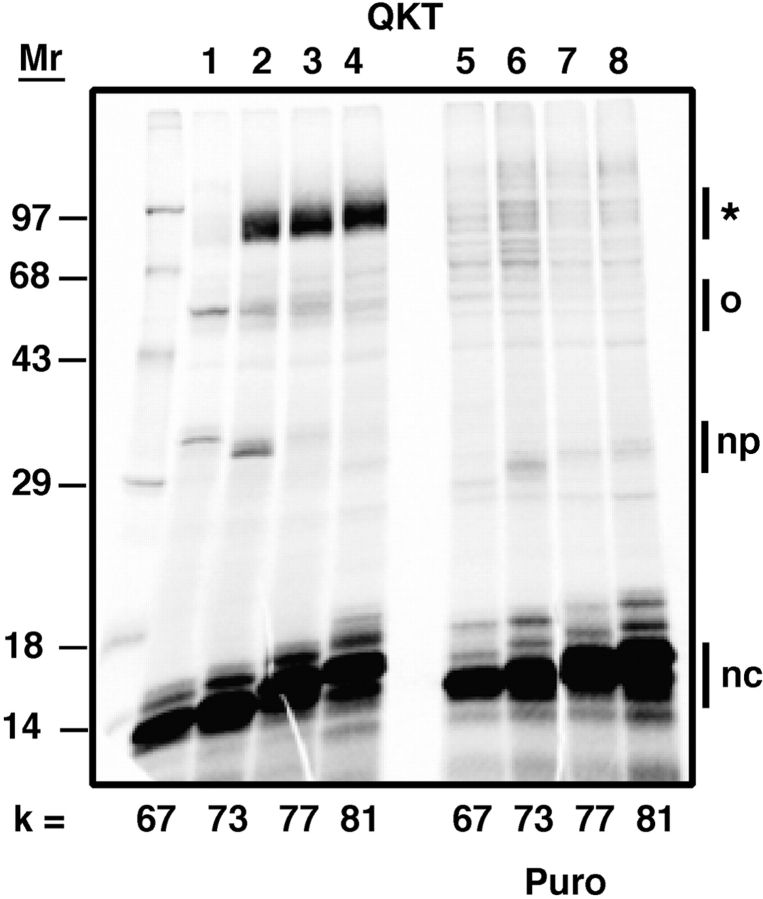
Glycosylation and photocross-linking of nascent chains containing an authentic glycosylation sequence. Translocation intermediates containing pPL-sK(NKT) or pPL-sK(NST) nascent chains of various lengths were prepared, photolyzed, and analyzed as detailed in the Materials and methods. (A) Approximate location of photoreactive probe (small black circle) in a translocation intermediate in which the probe is located 70–90 residues from the COOH-terminal end of the nascent chain (the ribosomal P site). (B) SDS-PAGE analysis of translocation intermediates after photolysis. The number of nascent chain residues between the probe and the tRNA are given by k, and the number of nascent chain amino acids between the asparagine residue and the tRNA are given by n. The asterisk indicates the size range expected for photoadducts between these nascent chains and STT3, RI, or RII, while o indicates the size range expected for photocross-links of these nascent chains to TRAM and Sec61α. The bands indicated by np are present in samples lacking the ɛANB probe (lanes 5 and 6) and in samples that have not been photolyzed (not depicted), so these radioactive species are not nascent chain photoadducts. The glycosylated nascent chains are indicated by G, while the nonglycosylated nascent chains are indicated by nc.
Cross-linking of nascent polypeptides to a 60–70-kD protein
An authentic NKT glycosylation sequence or the cryptic glycosylation sequence QKT was engineered into the mature portion of the model secretory protein preprolactin (pPL) in place of the YAQ sequence at position 66–68 in pPL-sK, a previously characterized derivative of pPL (Crowley et al., 1994). The resulting constructs were designated pPL-sK(NKT), etc. Translocation intermediates were prepared in vitro in the presence of SRP, ER microsomes, and the photoreactive ɛANB-Lys-tRNA.
Efficient N glycosylation of an authentic glycosylation sequence was observed whenever the nascent chain was long enough for the NXT sequence to reach the OST active site (n ≥ 67) (Fig. 1 B; unpublished data). The presence of ɛANB-Lys in place of lysine in the NKT nascent chains did not inhibit glycosylation. As expected, photocross-linking of NKT-containing nascent chains to two translocon components, Sec61α and TRAM, was observed when the probe was inside the translocon (k up to 67–69) (e.g., Fig. 1 B, lane 1). However, no photocross-linking of these NKT-containing nascent chains to a polypeptide with the molecular mass of an OST subunit was observed for k values between 53 and 97 residues (Fig. 1 B; unpublished data).
When the authentic NKT glycosylation sequence was replaced by a cryptic QKT glycosylation sequence, the nascent chains were no longer glycosylated (Fig. 2). Thus, as expected, the QKT sequence cannot substitute for NKT in the glycosylation reaction. Photocross-linking to translocon proteins was again observed when the probe was inside or close to the translocon (k ≤ 67) (Fig. 2 A; Fig. 2 B, lane 1). A major new photoadduct appeared when the nascent chain became long enough for the probe to emerge from the translocon (k > 67) (Fig. 2 B, lanes 2–4). This broad photoadduct band contained, in addition to the nascent chain, a protein that migrated with an apparent molecular mass of 60–70 kD. Interestingly, the appearance of this photoadduct coincided with the disappearance of the photocross-links to Sec61α and TRAM (Fig. 2 B, compare lanes 1 and 2). The presence of the cryptic QKT glycosylation sequence in the nascent chain therefore appears to have positioned the nascent chain adjacent to a protein not previously identified in cross-linking studies.
Figure 2.
Photocross-linking of nascent chains containing a cryptic glycosylation sequence to the translocon and a new protein. Translation intermediates containing pPL-sK(QKT) nascent chains with an ɛANB-Lys probe positioned k residues from the tRNA were prepared as in Fig. 1. (A) Photocross-linking of the nascent chains to translocon components when k is 67 or less. Radioactive species in the total sample are shown in lanes 1–6, whereas lanes 7–9 and 10–12 show the material immunoprecipitated by antibodies specific for Sec61α and TRAM, respectively. (B) Photocross-linking to a new protein with an apparent molecular mass of 60–70 kD as the nascent chain lengthens and the cryptic glycosylation sequence emerges from the translocon. (C) Cross-linking to both the translocon proteins and the new 60–70-kD protein target is light dependent.
Nascent chain photocross-linking to the 60–70-kD protein was observed whenever the nascent chain primary sequence contained a QKT sequence (as long as k > 67). However, the efficiency of photoadduct formation varied when the QKT sequence was positioned at seven different sites between residues 65 and 95 in the nascent chain (unpublished data). Thus, photocross-linking required the presence of a tripeptide sequence that functioned as a cryptic glycosylation sequence, but its location in the nascent chain did affect, to some extent, the efficiency of cross-linking. This variation presumably results from the slight differences in nascent chain conformation elicited by different flanking sequences. In all of our experiments, nascent chain photocross-linking to translocon components or to the 60–70-kD protein required both light (Fig. 2 C) and the ANB probe (unpublished data).
It is important to note that the nascent chain length dependence of photocross-linking to the 60–70-kD protein is nearly equivalent to the length dependence of glycosylation. We have shown previously (Whitley et al., 1996) that the NXT site in a nascent chain must be located at least 65 residues from the tRNA in the ribosomal P site to be glycosylated, and that glycosylation is maximal when the NXT is located 70–75 residues from the P site. In this study, the QKT was positioned relative to the OST complex at every second k value between 53 and 81 (unpublished data), and no photocross-linking to the 60–70-kD protein was observed until the K was 69 residues from the P site for this secretory protein. The efficiency of photocross-linking to the 60–70-kD protein reached a maximum when QKT was positioned 70–100 amino acids from the P site.
Nascent chain sequence dependence of photocross-linking to new target protein
To ascertain whether cross-linking to the 60–70-kD protein was dependent on the putative cryptic glycosylation sequence, we replaced the QKT sequence with a selection of related sequences (AKT, GKT, RKT, WKT, IKT, LKT, DKT, QKS, AKS, AKA, QKQ, and NKQ) in the pPL-sK(QKT) protein. When RNCs with these nascent chain sequences were prepared, targeted to microsomes, and photolyzed with UV light, no photocross-linking to a 60–70-kD protein was observed for 8 of the 12 sequences (Table I). This lack of photocross-linking was not due to inactive photoprobes because positive controls done in parallel showed that these eight nascent chains each photocross-linked to translocon components when k < 69 (unpublished data). It is also worth noting the numerous negative controls done previously by us and others who have used photocross-linking to examine the translocon and nascent chain processing without ever identifying a nascent chain photoadduct to an OST subunit (Johnson and van Waes, 1999).
Table I. Sequence dependence of nascent chain glycosylation and photocross-linking to STT3.
| Construct | Photocross-linking | Glycosylation |
|---|---|---|
| NKT | − | + |
| NST | − | + |
| QKT | +++ | − |
| AKT | ++ | − |
| GKT | +++ | − |
| RKT | + | − |
| WKT | − | − |
| IKT | − | − |
| LKT | − | − |
| DKT | − | − |
| QKS | ++ | − |
| AKS | − | − |
| AKA | − | − |
| QKQ | − | − |
| NKQ | − | − |
| PQKT | + | − |
| QKTP | ++ | − |
| QKQSTKQ | − | − |
| KRQSTGK | − | − |
RNCs with various sequences in place of the NKT in the pPL-sK(NKT) nascent chain were prepared, targeted to microsomes, and photolyzed. In each case, translocation intermediates with different lengths of nascent chain were examined so that the probe was incorporated 67, 73, 81, 89, or 97 residues from the tRNA in parallel samples, except that NST nascent chains had an Asn (not a probe) located 68, 74, 82, 90, or 98 residues from the tRNA. KRQSTGK nascent chains had probes at 64 + 70, 70 + 76, 78 + 84, 86 + 92, or 94 + 100 residues from the P site, whereas KQSTK nascent chains had probes at 65 + 69, 71 + 75, 79 + 83, 87 + 91, or 95 + 99.
Interestingly, when QKT was replaced by either AKT or GKT, the nascent chain still photocross-linked efficiently to the 60–70-kD protein (Table I). Because an amino acid with a small side chain can replace Q without markedly reducing the extent of nascent chain photocross-linking to the 60–70-kD protein, the interaction of the nascent chain threonine with the 60–70-kD protein appears to be most important in aligning the two polypeptides adjacent to each other. This result further suggests that steric constraints may prevent most nascent chains with an -X-X-T- sequence from binding (incorrectly) to the putative OST active site. Similarly, the low, but detectable, yield of photocross-linked product observed with a nascent chain containing an RKT sequence suggests that the flexible arginine side chain can bend out of the way sufficiently to be transiently accommodated in the putative nascent chain binding site of the 60–70-kD protein. The importance of threonine in nascent chain recognition and binding by the 60–70-kD protein is further shown by the fact that replacing T in an AKT cryptic sequence with S eliminated the photocross-linking of the nascent chain to the 60–70-kD protein, and, hence, must have reduced its affinity for the nascent chain (compare AKT with AKS in Table I, as well as QKT with QKS). This observation is consistent with previous in vitro work showing that an NXT sequence is glycosylated 40-fold more efficiently than an NXS sequence (Bause, 1983), and that -N-L-T-X sites are more efficiently glycosylated than N-L-S-X sites in the context of a nascent polypeptide (Mellquist et al., 1998).
It is important to note that the placement of a proline residue either before or after the cryptic glycosylation sequence significantly reduces the extent of nascent chain photocross-linking to the 60–70-kD protein (Table I). These results are consistent with earlier studies that showed that a Pro at the glycosylation site interferes with OST function (Bause, 1983; Gavel and von Heijne, 1990; Shakin-Eshleman et al., 1996; Mellquist et al., 1998). Thus, the conformation of the nascent chain also appears to affect its recognition by, and binding to, the 60–70-kD protein and, hence, both its glycosylation and photocross-linking efficiency.
The most important conclusion from the data of Table I is that nascent chain photocross-linking to the 60–70-kD protein is dependent upon the sequence of the nascent chain. This observation rules out the possibility that the observed photocross-linking results simply from a topographical proximity of the 60–70-kD protein to the nascent chain as it passes through the translocation machinery (if this were true, each nascent chain would cross-link to the 60–70-kD protein with the same efficiency). Instead, the sequence dependence of the photocross-linking demonstrates that the proximity of the nascent chain to the 60–70-kD protein is dictated by the sequence of the nascent chain. This in turn can only be explained if the 60–70-kD protein binds to the nascent chain in a sequence-dependent manner. By positioning a photoreactive probe in the nascent chain adjacent to each of the two residues that are recognized (or not) by the 60–70-kD protein, we are able to detect whether or not the nascent chain is associating with, and hence in proximity to, the 60–70-kD protein. The photocross-linking results therefore reveal that nascent chain proximity to the 60–70-kD protein requires the presence of a cryptic glycosylation sequence.
Only ribosome-bound nascent chains react covalently with the 60–70-kD protein
To further characterize the photocross-linking between nascent chains containing a cryptic glycosylation sequence and the 60–70-kD protein, we treated the membrane-bound translocation intermediates with puromycin to release the nascent chains from the ribosomes before photolysis. No photocross-linking was observed after the nascent chain was released from the ribosome into the ER lumen by puromycin (Fig. 3). As the nascent chain is no longer adjacent to either the translocon proteins or to the 60–70-kD protein after being released from the ribosome and the membrane, it appears that the nascent chain interacts with the 60–70-kD protein cotranslationally, in the context of the translocon and its associated proteins.
Figure 3.
Photoadduct formation requires ribosome-bound nascent chains. Translocation intermediates containing a cryptic glycosylation sequence in their nascent chains were incubated without (lanes 1–4) or with (lanes 5–8) puromycin before photolysis. The asterisk identifies photoadducts containing the 60–70-kD protein. Radioactive species are identified as in Fig. 1.
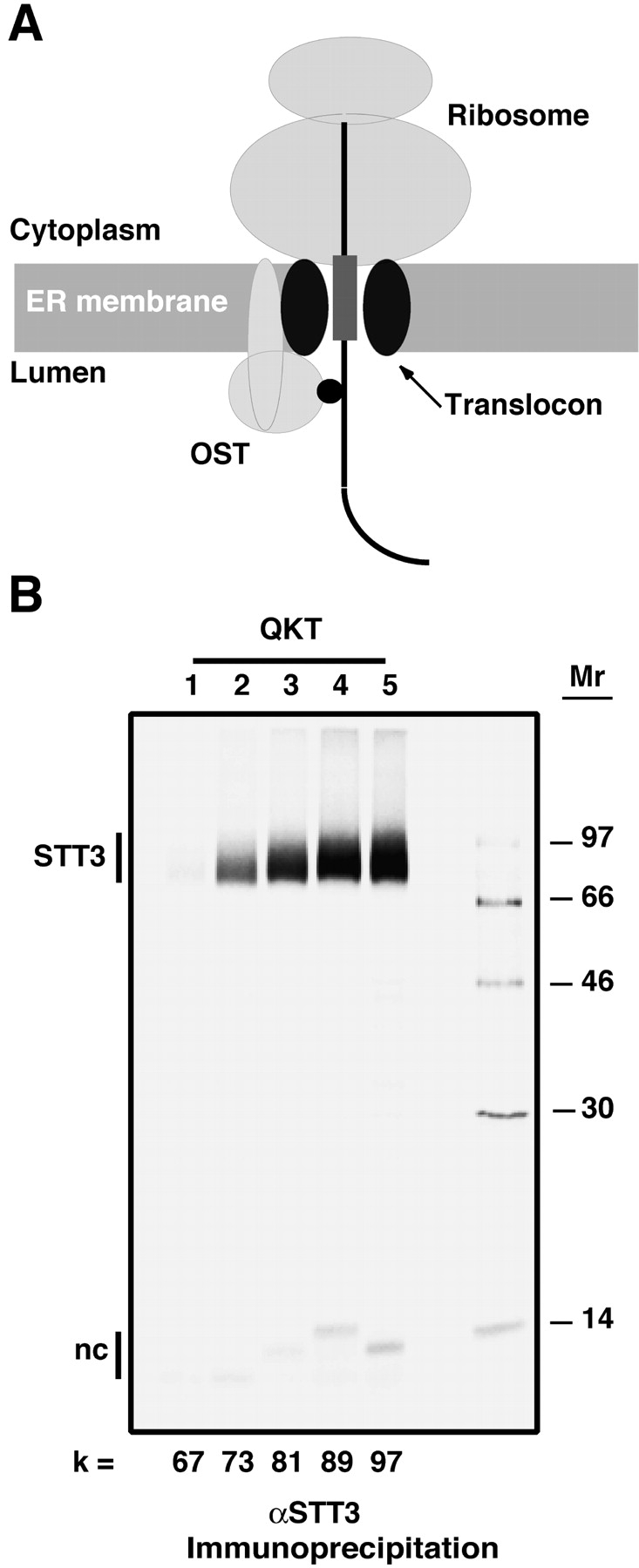
STT3 is photocross-linked to the nascent chain in the fully assembled translocation complex
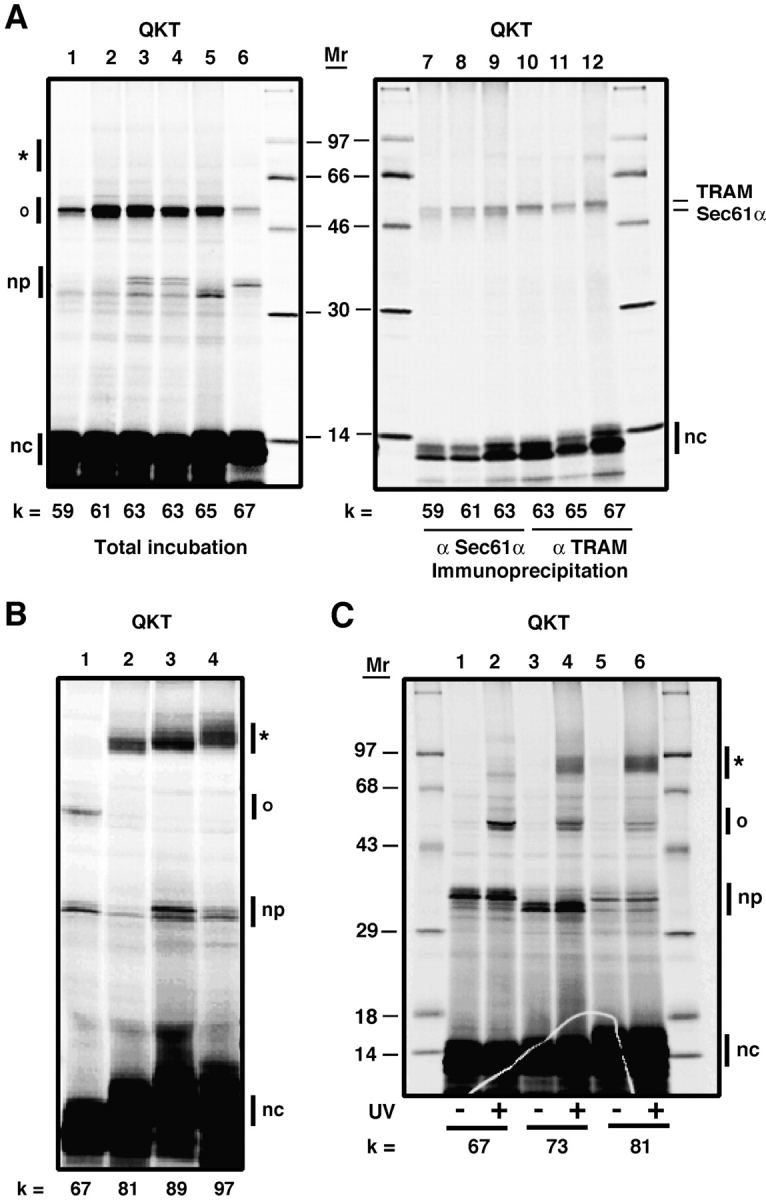
The identity of the 60–70-kD protein that photocross-links to ribosome-bound nascent chains with appropriately positioned cryptic glycosylation sequences was determined by immunoprecipitation with specific antibodies. A large sample containing pPL-sK(QKT) nascent chains with the probe 81 residues from the ribosomal P site was photolyzed and split into five equal aliquots that were then immunoprecipitated with antibodies specific for STT3-A, RI, RII, OST48, or Dad1 (Fig. 4 A). It is clear from lanes 2–6 of Fig. 4 A that the prominent photocross-linked product is almost exclusively derived from STT3-A. There is also a very small amount of photocross-linking to RI in lane 3 that becomes evident only upon prolonged exposure of the gel to the phosphorimager plate. When translocation intermediates containing a range of nascent chain lengths were photolyzed and analyzed, photoadducts containing STT3-A were found as soon as the cryptic glycosylation sequence emerged from the translocon (Fig. 4 B). Thus, the cryptic glycosylation sequence in the nascent chain appears to be recognized by, and interact solely with, STT3-A. Antibodies specific for the less-abundant STT3-B were not tested in this experiment because the 94-kD STT3-B protein would yield less rapidly migrating cross-linked products in a molecular mass range where no photoadducts were seen in our experiments.
Figure 4.

Immunoprecipitation of the photoadduct containing the 60–70-kD protein. (A) A large sample of photoreactive pPL-sK(QKT) translocation intermediates was prepared as in Fig. 1 (lane 1). Five equal aliquots of this sample were then immunoprecipitated with antisera specific for STT3-A (lane 2), RI (lane 3), RII (lane 4), OST48 (lane 5), or Dad1 (lane 6) before SDS-PAGE. (B) Translation interme- diates with nascent chains of different lengths were photolyzed and immunoprecipitated with STT3-A antiserum. Bands are identified as in the legend to Fig. 1.
An acceptor peptide substrate blocks nascent chain photocross-linking to STT3
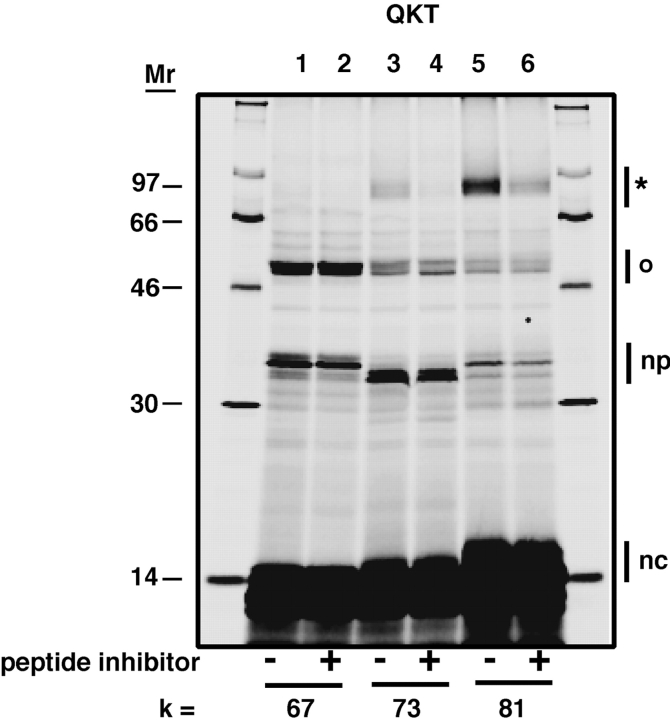
To assess whether nascent chain photocross-linking to STT3 was occurring at the active site of OST, we prepared translocation intermediates in the presence of an acceptor peptide, N-benzoyl-Asn-Leu-Thr-methylamide, that functions as a competitive inhibitor of glycosylation. When these samples were photolyzed, photocross-linking to STT3 was greatly reduced (Fig. 5). As the acceptor peptide is a competitive inhibitor of both glycosylation and photocross-linking, the observed photocross-linking of a nascent chain to STT3 must require nascent chain binding to the OST active site or its immediate vicinity.
Figure 5.
A competitive peptide substrate of OST greatly reduces nascent chain photocross-linking to STT3. Translocation intermediates containing a cryptic QKT sequence were photocross-linked in the presence (+) or absence (−) of a peptide substrate, N-benzoyl-Asn-Leu-Thr-methylamide. This peptide functions as a competitive inhibitor of N glycosylation. Bands are identified as in Fig. 1.
We also observed that the tripeptide inhibitor greatly reduced the cross-linking of STT3 to nascent chains containing either GKT or AKT (unpublished data). This result indicates that the GKT and AKT probes are also cross-linking to STT3 from the nascent chain portion of the OST active site.
How large is the OST nascent chain recognition site?
In the above experiments, each nascent chain contained a single ɛANB-Lys probe located in the middle of the QKT sequence. This probe location would be expected to yield maximum photocross-linking to any protein that bound the cryptic sequence. We therefore wondered whether the OST active site was only large enough to recognize and bind to three consecutive amino acids in the nascent chain, or whether the nascent chain was instead threaded through an extended groove that channeled the nascent chain into the OST recognition site.
To address this issue, we examined nascent chains with a photoreactive probe on one side or the other of a nonlysine-containing cryptic sequence. Thus, pPL-sK constructs coding for our standard cryptic glycosylation sequence -Q-R-Q-K-T-G-Q- (the positive control) and two additional derivatives, -Q-K-Q-S-T-K-Q- and -K-R-Q-S-T-G-K-, were transcribed and then translated in the presence of ɛANB-Lys-tRNA to yield, in the last case, -(ɛANB-Lys)-R-Q-S-T-G-(ɛANB-Lys)- (actually, due to competition from endogenous Lys-tRNAs in the translation, most nascent chains contain either one or no ɛANB-Lys residues; Krieg et al., 1989). When translocation intermediates containing these nascent chains were photolyzed, no photocross-linking to STT3-A was detected for nascent chains containing a probe on either side of the QST sequence (Table I). It therefore appears that the nascent chain does not move through the active site of OST via an extended channel that guides the nascent chain. Instead, the site in STT3 that recognizes and binds to the cryptic glycosylation sequence, and presumably the authentic glycosylation sequence, appears to contact only three nascent chain residues. The interaction between STT3 and the nascent chain therefore appears to be very limited in scope.
STT3 also interacts with membrane proteins
As OST glycosylates both secretory and membrane proteins in vivo, we wished to determine whether a photoreactive nascent membrane protein containing a QKT sequence would react covalently with STT3. We therefore introduced leucines into the nascent chain sequence to convert the secretory pPL-sK protein into a membrane protein. This approach has been used previously to show that a stretch of as few as eight leucines can function as a stop-transfer sequence and anchor a polypeptide in the ER membrane (Kuroiwa et al., 1991; Nilsson et al., 1994). Here, we substituted two, four, or six leucines into the mature pPL sequence adjacent to a naturally occurring stretch of 10 nonpolar residues that were located COOH terminal of the NKT or QKT sequences in the nascent chain. To assess the stop-transfer efficiency of these constructs, we positioned a second glycosylation site COOH terminal of the potential transmembrane (TM) segment. The ratio of polypeptides translocated across the ER membrane to polypeptides integrated into the membrane is then given by the ratio of diglycosylated to monoglycosylated proteins (Fig. 6 A), an approach that we have used successfully before to evaluate TM insertion efficiency (Sääf et al., 1998). As is evident from the data of Fig. 6 B, the lengthening of the nonpolar stretch in pPL-sK by only four leucines converts the polypeptide into a membrane protein by creating a TM sequence with sufficient hydrophobicity and length to insert into the ER membrane.
Figure 6.
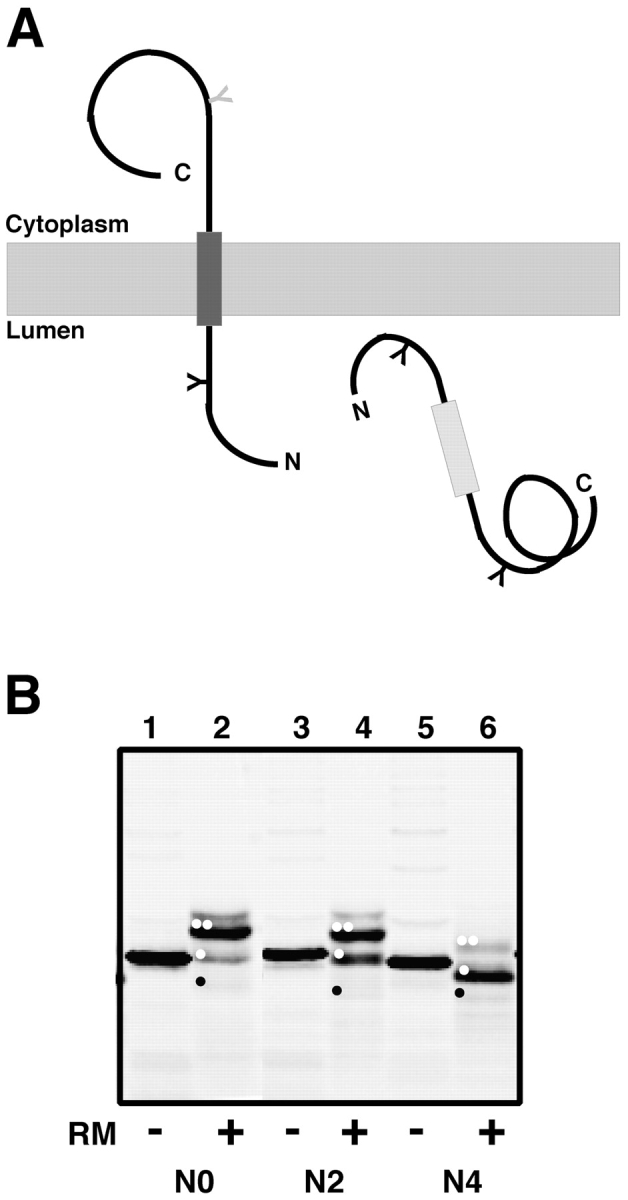
Conversion of the pPL-sK secretory protein into a membrane protein. (A) By introducing an NST sequence into the nascent chain behind the putative TM sequence, the extent of nascent chain integration into the ER membrane can be assessed (Sääf et al., 1998). If the potential TM segment does not function as a stop-transfer sequence (light gray rectangle), the entire protein will be translocated into the ER lumen and will be glycosylated at two sites. But if the TM segment is inserted into the bilayer (dark gray rectangle), only one of its glycosylation sites will be modified (black Y). (B) Full-length polypeptides containing zero (pPL-sK[NKT], which is the same as pPL-sK[N0]), two (pPL-sK[N2]), or four (pPL-sK[N4]) leucine substitutions in addition to an extra glycosylation site in the COOH-terminal portion of the protein were translated in reticulocyte lysate in the absence (−) or presence (+) of rough microsomes (RM). Signal sequence–cleaved diglycosylated, monoglycosylated, and nonglycosylated molecules are indicated by two white dots, one white dot, and one black dot, respectively. Very little cleaved, nonglycosylated product is seen in this experiment (black dot), but translation of the same construct lacking both glycosylation sites confirms the identity of this band (not depicted).
Integration intermediates were prepared with photoreactive nascent chains of different lengths containing six leucine substitutions and a cryptic QKT sequence (Fig. 7 A). After photolysis and analysis by SDS-PAGE, the nascent chains were found to photocross-link to STT3 after the QKT sequence emerged from the translocon (Fig. 7 B). No photocross-linking to STT3 was observed when the nascent chains contained the authentic NKT glycosylation sequence instead of the cryptic QKT sequence (unpublished data). As expected, the NKT-containing nascent chains were glycosylated, whereas the QKT-containing nascent chains were not. Because these nascent chains were inserted into the bilayer (Fig. 6 B), the photocross-linking of a nascent chain to STT3 is dependent upon the presence of a cryptic glycosylation site, not upon the type of nascent chain (secretory or membrane protein). Thus, the OST interacts with nascent secretory and membrane proteins in a similar fashion.
Figure 7.
The extent of photocross-linking to STT3 is unaffected by nascent chain conversion from a secretory protein to a membrane protein. (A) An integration intermediate with a TM segment (dark gray rectangle) in the nascent chain and a photoreactive probe (small black circle) positioned ∼80 residues from the ribosomal P site. The first residue of the artificial TM span is located 28 residues on the COOH-terminal side of the lysine residue in the -Q-K-T sequence. (B) Integration intermediates containing pPL-sK(Q6) nascent chains of different lengths, each with six leucine substitutions and a cryptic QKT sequence, were photolyzed and analyzed by SDS-PAGE after immunoprecipitation with STT3-A–specific antiserum.
Discussion
This study reports the first cross-linking of an OST subunit to a nascent protein chain that is undergoing translocation or integration at the ER membrane. Although we, and many others, have tried for a number of years to cross-link the nascent chain to an OST subunit in a fully assembled nascent chain–ribosome–translocon–OST complex, it is now clear that those efforts were unsuccessful largely because of the stringent requirements for OST interaction with the nascent chain. In the end, only a few nascent chain sequences are recognized as cryptic glycosylation sequences by the OST (Table I), and only a few residues of the nascent chain are adjacent to STT3 at any one time. The discovery of the experimental requirements necessary to detect nascent chain photocross-linking to STT3 has provided a novel perspective on the N glycosylation of nascent chains by OST.
Both nascent secretory and membrane proteins photocross-link to only one OST component, STT3 (Figs. 4 and 7). Because this photocross-linking was completely dependent upon the primary sequence of the nascent chain, the observed photocross-linking did not result from random encounters between STT3 and a photoreactive nascent chain (Table I). Instead, a recognition event and/or selection process must have positioned nascent chains with cryptic glycosylation sequences in close proximity to STT3. Moreover, as the nascent chain sequences that generated cross-links to STT3 were related to the authentic -N-X-T/S- glycosylation sequences (Table I), it appears that the close approach of the nascent chain to STT3 was dictated by its affinity for potential glycosylation sites in the nascent chain. Although the cryptic sequences were not glycosylated and, hence, were not recognized as substrates by the OST, the cryptic sequences did interact sufficiently well to position the nascent chain at the OST active site.
This conclusion was confirmed by the discovery that a competitive inhibitor of glycosylation greatly reduced nascent chain photocross-linking to STT3 (Fig. 5). Because the competitive peptide substrate is small (only a tripeptide), can be glycosylated, and, hence, is directed to the OST active site, one explanation for the inhibition of nascent chain photocross-linking to STT3 is that the peptide competes with the nascent chain for interaction at the OST nascent chain binding site. Another possible explanation for tripeptide-dependent inhibition of STT3 photolabeling is suggested by a recent kinetic analysis of the OST (Karaoglu et al., 2001). The addition of a membrane-permeable tripeptide substrate to canine microsomes leads to a rapid depletion of the dolichol-linked oligosaccharide donor. Kinetic experiments indicate that the OST has two nonequivalent binding sites for dolichol-linked oligosaccharides. Binding of a dolichol-linked oligosaccharide to an activator site is required for subsequent binding of both the donor and acceptor substrates to the catalytic site (Karaoglu et al., 2001). Consequently, depletion of the donor oligosaccharide pool is predicted to reduce the interaction between the OST and the cryptic glycosylation sites. This may explain why a tripeptide substrate with a low binding affinity for the OST (K d ≈ 20 μM) can inhibit photocross-linking of the OST to a nascent chain that is positioned near the active site. In either case, the tripeptide interacts directly with the OST active site, and this in turn blocks the photocross-linking between STT3 and a cryptic sequence in a nascent chain. We therefore conclude that STT3 is responsible for cotranslational nascent chain recognition and binding, and that part or all of the OST active site is located in STT3.
The data reported here therefore provide direct experimental support for the proposal by Burda and Aebi (1999), based on evolutionary grounds, that STT3 contains the active site of OST because STT3 is the only OST subunit found in archaebacteria. More recently, Aebi and colleagues have demonstrated that point mutations in the Camphylobacter jejuni homologue of Stt3p (PglB) blocks asparagine-linked glycosylation in that organism (Wacker et al., 2002). Here we have reached the same conclusion using an approach that allows nascent chain recognition site–directed photocross-linking of the mammalian OST.
Previous studies by others have provided evidence that the OST active site may be composed of multiple subunits. For example, hexapeptides can be glycosylated and chemically cross-linked to RI and to OST48 (Bause et al., 1997). OST48 was suggested to be responsible for shepherding the dolichol-bound oligosaccharide substrate to the active site (Pathak et al., 1995). Although the initial studies by Yan et al. (1999) identified Ost1p as the active site of the OST using photoligands that have a benzophenone residue at the X position of an NXT acceptor substrate, a more recent report indicates that three OST subunits (Stt3p, Ost3p, and Ost1p) can all be photolabeled when the photoreactive amino acid is positioned at different locations relative to the NXT consensus sequence (Yan and Lennarz, 2002). Based upon mutagenesis arguments, the Lennarz lab has concluded that Stt3p contains the tripeptide binding site or catalytic site of the OST. Although it is certainly conceivable that the catalytic site of the OST is formed by the close juxtaposition of two or more OST subunits, the absence of any nascent chain photocross-links to other OST subunits (Fig. 4 A) strongly suggests that the acceptor substrate binding site is located on STT3. Although we do not yet have an explanation for the discrepancy between our data and the previous studies, we suspect that the different results may originate from the use of peptide substrates instead of nascent chain substrates. Whereas nascent chains in intact translocation and integration intermediates are presumably presented to the OST as they are in vivo, the peptide analogues rely only on their affinity for the active site to direct them to the proper OST site, and this affinity is relatively low (10–25 μM; Welply et al., 1983; Karaoglu et al., 2001; unpublished data). Hence, even minor changes in the small peptides, such as the presence of a probe moiety, may alter its binding affinity and location.
In contrast, the nascent chain is maintained adjacent to the OST active site via its attachment to the tRNA and the constraints placed on its movement by the ribosome and translocon. In fact, the maintenance of the location of the cryptic glycosylation sequence near the OST active site by the nascent chain dramatically increases the local concentration of the QKT sequence and hence promotes its association with the OST active site. As a rough estimate, if a single QXT acceptor site in a translocating nascent chain is assumed to be confined to a cube with sides of 20 Å near the OST active site, the local concentration of the QXT sequence would be ∼0.2 M. This nascent chain–dictated high local concentration may explain why the QXT sequence in nascent chains leads to photocross-linking but QLT tripeptides are not effective as competitive inhibitors of N-linked glycosylation (Welply et al., 1983).
One major advantage of using photoreactive nascent chains rather than peptides to examine substrate interactions with the OST is the opportunity to track the movement of the nascent chain as it passes through the translocon and into the OST active site. As OST is a multisubunit enzyme and the functional roles of its components have yet to be established, it is conceivable that one or more of the OST subunits may be required to direct or channel the nascent chain to the OST active site to facilitate its scanning of the nascent chain for glycosylation sites. If this were the case, then one would predict that the photoreactive nascent chain should cross-link to such a channeling OST subunit after the probe leaves the translocon and before it reaches STT3. However, when we examined multiple intermediates with nascent chains of different lengths (Figs. 2 and 4), we did not detect additional novel photoadducts with any of our nascent chains. Instead, the disappearance of nascent chain photocross-linking to translocon proteins coincided with the appearance of nascent chain photocross-linking to STT3 (Figs. 2 and 4). Thus, our results do not support the hypothesis that one of the other OST subunits directs the nascent polypeptide toward the OST active site.
STT3 interacts with the nascent chain cotranslationally, as shown by the absence of STT3–nascent chain photocross-linking if the nascent chain is released from the ribosome by puromycin before the sample is irradiated (Fig. 3). The rate of translocation, therefore, must be sufficiently slow and/or the space in which the nascent chain can diffuse must be sufficiently restricted to ensure that each portion of the nascent chain encounters the OST active site and provides an opportunity for STT3 recognition of a glycosylation sequence as the nascent chain passes by the OST active site. N-linked oligosaccharides are added in a highly synchronized manner to elongating nascent chains (Chen et al., 1995; Daniels et al., 2003). Perhaps equally important for ensuring complete glycosylation of a nascent chain, our data demonstrate that a (cryptic) nascent chain glycosylation sequence can access (photocross-link) the STT3 binding site for a prolonged period of time; the extent of nascent chain photocross-linking to STT3 does not decrease while the nascent chain is elongated by >30 amino acids (Fig. 2; unpublished data).
Our data indicate that STT3 itself interacts with only a limited number of nascent chain residues at any given time. Even when photoreactive probes were positioned on either side of the QST sequence in the QKQSTKQ construct, no photocross-linking to STT3 was observed (Table I). This result contrasts dramatically with the efficient photocross-linking of STT3 that occurs when the photoprobe is located in the middle of the cryptic QKT glycosylation sequence. It therefore appears that the nascent chain binding site on STT3 extends over only approximately three residues of the nascent chain.
The discovery of the experimental conditions necessary to obtain photocross-linking of STT3 now allows us to examine OST structure and function using a new approach. Specifically, by replacing the photoreactive probe in the above translocation and integration intermediates with a fluorescent probe, one can characterize nascent chain interactions with STT3 spectroscopically. Among other things, one could determine the environment and accessibility of the STT3 active site using a fluorescent probe in a cryptic nascent chain sequence (e.g., Crowley et al., 1994) and directly measure its height above the membrane surface using fluorescence resonance energy transfer (e.g., Yegneswaran et al., 1999).
Materials and methods
Materials
Unless otherwise stated, all enzymes, plasmid pGEM1, and rabbit reticulocyte lysate were from Promega or New England Biolabs, Inc. T7 DNA polymerase, [35S]Met, 14C-methylated marker proteins, ribonucleotides, deoxyribonucleotides, dideoxyribonucleotides, and the cap analogues m7G(5′)ppp(5′)G and G(5′)ppp(5′)G were from Amersham Biosciences. Ex Taq polymerase was from TaKaRa Biomedicals. Protein A–Sepharose and puromycin were from Sigma-Aldrich. Rabbit antiserum to the COOH-terminal 14 amino acids of Sec61α and affinity-purified rabbit antiserum to the COOH-terminal 15 amino acids of TRAM were obtained from Research Genetics. The rabbit anti-RI and -RII antibodies were raised against polypeptides corresponding to a COOH-terminal domain of rat RI (residues 564–583) and a lumenal domain of rat RII (residues 1–22), respectively (Yu et al., 1990). The anti-OST48 antibody was obtained by immunizing rabbits with a polypeptide corresponding to the entire lumenal domain of canine OST48 (Fu and Kreibich, 2000). The rabbit anti-Dad1 antibody was raised against a polypeptide (residues 76–91) of the hamster protein (Nikonov et al., 2002). The antibody to STT3-A was raised against the COOH terminus (residues 693–705) of human STT3 (Kelleher et al., 2003). The competitive glycosylation inhibitor, benzoyl-Asn-Leu-Thr-methylamide, was purchased from Quality Controlled Biochemicals. Dog pancreas column-washed rough microsomes (CRMs), SRP, and wheat germ extract were prepared as before (Walter and Blobel, 1983; Liao et al., 1997). ɛANB-Lys-tRNA was prepared as detailed previously (Krieg et al., 1986, 1989).
DNA plasmids
Bovine pPL glycosylation constructs were made from the previously described pPL construct (pPL-sK) and were cloned into the pVW1 plasmid behind the SP6 promoter (Crowley et al., 1994). The pPL-sK polypeptide lacks residues 2–9 of natural pPL and therefore does not have any lysines in its signal sequence. An authentic NST glycosylation sequence was introduced by site-specific mutagenesis in place of the YAQ sequence at position 66–68 in the mature part of pPL-sK. The amino acid sequence ..K64RYAQGK70.. at position 64–70 was replaced by the amino acid sequence ..QRNSTGQ.. to create the pPL-sK(NST) construct. The authentic NST glycosylation sequence in pPL-sK(NST) was then changed by PCR mutagenesis to make a series of pPL-sK(NST) derivatives in which the NST was replaced by NKT, QKT, AKT, GKT, RKT, WKT, IKT, LKT, DKT, QKS, AKS, AKA, QKQ, or NKQ. The two glutamine residues surrounding the authentic glycosylation sequence ..QRNSTGQ.. were changed to lysine residues in the pPL-sK(KRNSTGK) construct, and the asparagine residue in this construct was changed to a glutamine in the pPL-sK(KRQSTGK) construct. In another construct, the lysines were positioned adjacent to the QST sequence to yield pPL-sK(QKQSTKQ). A proline residue was introduced instead of R or G in the cryptic glycosylation sequence ..QRQKTGQ.. in the pPL-sK(QKT) construct to yield pPL-sK(PQKT) and pPL-sK(QKTP), respectively. When nascent chains containing only a single photoreactive probe were required, the lysine codons at position 91, 128, 146, 164, and 181 in pPL-sK were replaced with a nonlysine codon.
Membrane proteins were created from the above pPL derivatives by converting two, four, or six amino acids into leucines at positions 99–100, 97–100, or 95–100, respectively, adjacent to a natural hydrophobic sequence in mature pPL to yield a potential TM sequence: ..QRQKTGQGFITMALNSCHTSSLPTPED90QEQAQQTHHE VLMSLILGLLRSWND.. (cryptic glycosylation sequence is underlined, the residues replaced by leucines are shown in bold, and the natural hydrophobic sequence is both bold and underlined). The construct formed by substituting six leucines into pPL-sK(QKT) was designated pPL-sK(Q6), and the other two constructs were termed Q2 and Q4. Constructs formed by Leu substitution into pPL-sK(NKT) were designated N2, N4, and N6. To assess the extent of TM insertion into the ER membrane, an NST glycosylation sequence was introduced COOH terminal of the putative TM sequence at position 164–166 in the NKT, N2, and N4 derivatives by PCR mutagenesis.
Site-specific mutagenesis was performed using the QuikChangeTM Site-Directed Mutagenesis kit from Stratagene. All mutants were confirmed by the sequencing of plasmid DNA. All cloning steps were done according to standard procedures.
Transcription in vitro
Transcription of full-length pPL-sK mRNAs from the pPL plasmid using SP6 RNA polymerase was performed as described previously (Nilsson et al., 2001). The DNA template for in vitro transcription of truncated pPL-sK mRNAs was prepared using PCR to amplify a fragment from the plasmid. The 5′ primer was situated 100 bases upstream of the translation start, and the amplified fragment thus contained the SP6 transcriptional promoter. The 3′ primer was chosen to produce a truncated fragment ending at the desired codon, and no stop codon was included. Truncated mRNAs were transcribed from the amplified DNA fragments as previously described (Liao et al., 1997), except that the GTP concentration was raised to 0.5 mM after 90 min to ensure completion of all transcripts.
Translation in vitro
In vitro translations (usually 25 μl total volume, but 50 μl for immunoprecipitations) of mRNA in wheat germ cell-free extract were incubated at 22°C for 30 min in the presence of 40 nM canine SRP, [35S]Met (usually 0.2 μCi/μl, but 2 μCi/μl for immunoprecipitations), and eight equivalents of CRMs and 15 pmol of ɛANB-Lys-tRNA per 25 μl of incubation (Do et al., 1996; Liao et al., 1997). Samples were photolyzed on ice for 15 min using a 500-W mercury arc lamp and filters that only transmit light with a wavelength >300 nm. After photolysis, membranes were sedimented through a 0.5 M sucrose cushion in a Beckman Coulter airfuge at 4°C for 5 min at 20 psi or a Beckman Coulter TLA ultracentrifuge at 4°C for 4 min at 100,000 rpm in a TLA100 rotor.
For immunoprecipitation, microsome pellets were resuspended in buffer (100 mM Tris-HCl [pH 7.6] for Sec61α- and TRAM-specific antibodies, and 50 mM Tris-HCl [pH 7.6], 95 mM NaCl, 3 mM EDTA for STT3-A-, RI-, RII-, OST48-, and Dad1-specific antibodies) containing detergent (0.25% [wt/vol] SDS for Sec61α-specific antibodies, 1% [wt/vol] SDS for TRAM-specific antibodies, and 2% [wt/vol] SDS for STT3-A-, RI-, RII-, OST48-, and Dad1-specific antibodies) and placed at 55°C for a minimum of 30 min. The volume was increased with buffer A (140 mM NaCl, 10 mM Tris-HCl [pH 7.6], 2% [vol/vol] Triton X-100, 0.2% [wt/vol] SDS) for Sec61α antibodies, buffer B (150 mM NaCl, 50 mM Tris-HCl [pH 7.6], 2% [vol/vol] Triton X-100, 0.2% [wt/vol] SDS) for TRAM antibodies, and buffer C (50 mM Tris-HCl [pH 7.6], 95 mM NaCl, 3 mM EDTA, 1.25% [vol/vol] Triton X-100, 0.2% [wt/vol] SDS) for STT3-A, RI, RII, OST48, and Dad1 antibodies. Samples were then precleared by rocking with protein A–Sepharose at room temperature for 60 min before the Sepharose beads were removed by sedimentation.
Sec61α-, TRAM-, STT3-A-, RI-, RII-, OST48-, or Dad1-specific antiserum was added to a precleared sample, and the samples were rocked overnight at 4°C. Protein A–Sepharose was then added to each sample and incubated for a minimum of 2 h at 4°C. The immunoprecipitates were recovered by sedimentation, washed twice with buffer A, B, or C, and then washed a final time with the same buffer containing no detergent. The immunoprecipitated proteins were analyzed by SDS-PAGE and visualized using a Bio-Rad Laboratories FX Molecular Imager using the Quantity One Quantification software.
To determine the puromycin dependence of the cross-linking, truncated pPL-sK(QKT) mRNA was translated at 22°C in a total volume of 25 μl. After a 30-min incubation, 1.5 μl of 30 mM puromycin was added, and the incubation was continued at 22°C for another 10 min before photolysis and analysis as above.
To demonstrate inhibition of cross-linking in the presence of a competitive peptide substrate of OST, translation/translocation reactions of truncated pPL-sK(QKT) mRNA were performed at 22°C in a total volume of 25 μl. After a 25-min incubation, 0.6 μl of 30 mM glycosylation inhibitor peptide (N-benzoyl-Asn-Leu-Thr-methylamide) was added, and the incubation was continued at 22°C for another 5 min before photolysis and analysis as above.
Translations of full-length pPL-sK mRNAs were performed at 30°C for 60 min as previously described (Liljeström and Garoff, 1991) in a total volume of 14 μl containing nuclease-treated reticulocyte lysate, 40 U of RNase inhibitor, [35S]Met (0.7 μCi/μl), 70 μM of each amino acid except Met, and four equivalents of CRMs. Samples were then analyzed by SDS-PAGE, and proteins were visualized as above.
Acknowledgments
We gratefully thank Brian Mosteller and Kristen L. Pfeiffer for their excellent technical assistance, and Stephanie Etchells, Andrey Karamyshev, and Rajesh Ramachandran for their advice and for critical comments on the manuscript.
This work was supported by grants from the Swedish Research Council and the Swedish Foundation for International Cooperation in Research and Higher Education (STINT) (I.M. Nilsson), by grant RPG-92-CB from the American Cancer Society (G. Kreibich), by National Institutes of Health grants GM 43768 (R. Gilmore) and GM 26494 (A.E. Johnson), by grants from the Swedish Cancer Foundation and the Swedish Research Council (G. von Heijne), and by the Robert A. Welch Foundation (A.E. Johnson).
Footnotes
Abbreviations used in this paper: ɛANB-Lys, N ɛ-(5-azido-2-nitrobenzoyl)-lysine; CRM, column-washed rough microsome; OST, oligosaccharyltransferase; pPL, preprolactin; RI and -II, ribophorin I and II; RNC, ribosome–nascent chain complex; SRP, signal recognition particle; TM, transmembrane.
References
- Bause, E. 1983. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem. J. 209:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause, E., B. Breuer, and S. Peters. 1995. Investigation of the active site of the oligosaccharyltransferase using synthetic peptides as tools. Biochem. J. 312:979–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause, E., M. Wesemann, A. Bartoschek, and W. Breuer. 1997. Epoxyethylglycyl peptides as inhibitors of oligosaccharyltransferase: double-labeling of the active site. Biochem. J. 322:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda, P., and M. Aebi. 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta. 1426:239–257. [DOI] [PubMed] [Google Scholar]
- Chen, W., J. Helenius, I. Braakman, and A. Helenius. 1995. Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc. Natl. Acad. Sci. USA. 92:6229–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley, K.S., S. Liao, V.E. Worrell, G.D. Reinhart, and A.E. Johnson. 1994. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 78:461–471. [DOI] [PubMed] [Google Scholar]
- Daniels, R., B. Kurowski, A.E. Johnson, and D.N. Hebert. 2003. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol. Cell. 11:79–90. [DOI] [PubMed] [Google Scholar]
- Do, H., D. Falcone, J. Lin, D.W. Andrews, and A.E. Johnson. 1996. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 85:369–378. [DOI] [PubMed] [Google Scholar]
- Fu, J., and G. Kreibich. 2000. Retention of subunits of the oligosaccharyltransferase complex in the endoplasmic reticulum. J. Biol. Chem. 275:3984–3990. [DOI] [PubMed] [Google Scholar]
- Gavel, Y., and G. von Heijne. 1990. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites - implications for protein engineering. Protein Eng. 3:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiali, B., K.L. Shannon, M. Unno, and K.W. Rickert. 1992. A mechanistic proposal for asparagine-linked glycosylation. J. Am. Chem. Soc. 114:7944–7945. [Google Scholar]
- Johnson, A.E., and M.A. van Waes. 1999. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15:799–842. [DOI] [PubMed] [Google Scholar]
- Karaoglu, D., D.J. Kelleher, and R. Gilmore. 1997. The highly conserved Stt3 protein is a subunit of the yeast oligosaccharyltransferase and forms a subcomplex with Ost3p and Ost4p. J. Biol. Chem. 272:32513–32520. [DOI] [PubMed] [Google Scholar]
- Karaoglu, D., D.J. Kelleher, and R. Gilmore. 2001. Allosteric regulation provides a molecular mechanism for preferential utilization of the fully assembled dolichol-linked oligosaccharide by the yeast oligosaccharyltransferase. Biochemistry. 40:12193–12206. [DOI] [PubMed] [Google Scholar]
- Kelleher, D.J., and R. Gilmore. 1997. DAD1, the defender against apoptotic cell death, is a subunit of the mammalian oligosaccharyltransferase. Proc. Natl. Acad. Sci. USA. 94:4994–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher, D.J., G. Kriebich, and R. Gilmore. 1992. Oligosaccharyltransferase activity is associated with a protein complex composed of ribophorins I and II and a 48 kd protein. Cell. 69:55–65. [DOI] [PubMed] [Google Scholar]
- Kelleher, D.J., D. Karaoglu, E. Mandon, and R. Gilmore. 2003. Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol. Cell. In press. [DOI] [PubMed] [Google Scholar]
- Knauer, R., and L. Lehle. 1999. The oligosaccharyltransferase complex from yeast. Biochim. Biophys. Acta. 1426:259–273. [DOI] [PubMed] [Google Scholar]
- Krieg, U.C., A.E. Johnson, and P. Walter. 1989. Protein translocation across the endoplasmic reticulum membrane: identification by photocross-linking of a 39 kD integral membrane glycoprotein as part of a putative translocation tunnel. J. Cell Biol. 109:2033–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg, U.C., P. Walter, and A.E. Johnson. 1986. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl. Acad. Sci. USA. 83:8604–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa, T., M. Sakaguchi, K. Mihara, and T. Omura. 1991. Systematic analysis of stop-transfer sequence for microsomal membrane. J. Biol. Chem. 266:9251–9255. [PubMed] [Google Scholar]
- Liao, S., J. Lin, H. Do, and A.E. Johnson. 1997. Both lumenal and cytosolic gating of the aqueous ER translocon pore is regulated from inside the ribosome during membrane protein integration. Cell. 90:31–41. [DOI] [PubMed] [Google Scholar]
- Liljeström, P., and H. Garoff. 1991. Internally located cleavable signal sequences direct the formation of semliki forest virus membrane proteins from a polyprotein precursor. J. Virol. 65:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellquist, J.L., L. Kasturi, S.L. Spitalnik, and S.H. Shakin-Eshleman. 1998. The amino acid following an Asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry. 37:6833–6837. [DOI] [PubMed] [Google Scholar]
- Nikonov, A.V., E. Snapp, J. Lippincott-Schwartz, and G. Kreibich. 2002. Active translocon complexes labeled with GFP-Dad1 diffuse slowly as large polysome arrays in the endoplasmic reticulum. J. Cell Biol. 158:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, I., P. Whitley, and G. von Heijne. 1994. The COOH-terminal ends of internal signal and signal-anchor sequences are positioned differently in the ER translocase. J. Cell Biol. 126:1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, I., H. Ohvo-Rekila, J.P. Slotte, A.E. Johnson, and G. von Heijne. 2001. Inhibition of protein translocation across the endoplasmic reticulum membrane by sterols. J. Biol. Chem. 276:41748–41754. [DOI] [PubMed] [Google Scholar]
- Pathak, R., T.L. Hendrickson, and B. Imperiali. 1995. Sulfhydryl modification of the yeast Wbp1p inhibits oligosaccharyl transferase activity. Biochemistry. 34:4179–4185. [DOI] [PubMed] [Google Scholar]
- Sääf, A., E. Wallin, and G. von Heijne. 1998. Stop-transfer function of pseudo-random amino acid segments during translocation across prokaryotic and eukaryotic membranes. Eur. J. Biochem. 251:821–829. [DOI] [PubMed] [Google Scholar]
- Shakin-Eshleman, S.H., S.L. Spitalnik, and L. Kasturi. 1996. The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J. Biol. Chem. 271:6363–6366. [DOI] [PubMed] [Google Scholar]
- Silberstein, S., and R. Gilmore. 1996. Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB J. 10:849–858. [PubMed] [Google Scholar]
- Wacker, M., D. Linton, P.G. Hitchen, M. Nita-Lazar, S.M. Haslam, S.J. North, M. Panico, H.R. Morris, A. Dell, B.W. Wren, and M. Aebi. 2002. N-linked glycosylation in C. jejuni and its functional transfer to E. coli. Science. 298:1790–1793. [DOI] [PubMed] [Google Scholar]
- Walter, P., and G. Blobel. 1983. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 96:84–93. [DOI] [PubMed] [Google Scholar]
- Welply, J.K., P. Shenbagamurthi, W.J. Lennarz, and F. Naider. 1983. Substrate recognition by oligosaccharyltransferase. Studies on glycosylation of modified asn-x-thr/ser tripeptides. J. Biol. Chem. 258:11856–11863. [PubMed] [Google Scholar]
- Whitley, P., I. Nilsson, and G. von Heijne. 1996. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 271:6241–6244. [DOI] [PubMed] [Google Scholar]
- Yan, Q., and W.J. Lennarz. 1999. Oligosaccharyltransferase: a complex multisubunit enzyme of the endoplasmic reticulum. Biochem. Biophys. Res. Commun. 266:684–689. [DOI] [PubMed] [Google Scholar]
- Yan, Q., and W.J. Lennarz. 2002. Studies on the function of the oligosaccharyltransferase subunits. Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 277:47692–47700. [DOI] [PubMed] [Google Scholar]
- Yan, Q., G.D. Prestwich, and W.J. Lennarz. 1999. The Ost1p subunit of yeast oligosaccharyl transferase recognizes the peptide glycosylation site sequence, -Asn-X-Ser/Thr-. J. Biol. Chem. 274:5021–5025. [DOI] [PubMed] [Google Scholar]
- Yegneswaran, S., M.D. Smirnov, O. Safa, N.L. Esmon, C.T. Esmon, and A.E. Johnson. 1999. Relocating the active site of activated protein C eliminates the need for its protein S cofactor. A fluorescence resonance energy transfer study. J. Biol. Chem. 274:5462–5468. [DOI] [PubMed] [Google Scholar]
- Yu, Y.H., D.D. Sabatini, and G. Kreibich. 1990. Antiribophorin antibodies inhibit the targeting to the ER membrane of ribosomes containing nascent secretory polypeptides. J. Cell Biol. 111:1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]