Abstract
Previous studies have implicated histone deacetylation and chromatin condensation as critical mechanisms of transcription repression in yeast and mammals. A specific histone deacetylase, Rpd3, interacts with a variety of sequence-specific transcriptional repressors, including Mad–Max heterodimers and members of the nuclear receptor superfamily. Here, we present evidence that a strong hypomorphic mutation in the Drosophila Rpd3 gene causes embryonic lethality and a specific pair-rule segmentation phenotype. The analysis of a number of segmentation genes suggests that the repressor function of Even-skipped (Eve) may be diminished, causing an indirect loss of Ftz-mediated activation of engrailed. The relatively mild defects observed in Rpd3 mutants suggest that the recently identified Groucho and dCtBP corepressor proteins do not function solely through the recruitment of histone deacetylases. We discuss the possibility that Eve mediates multiple mechanisms of repression, so that Rpd3 mutants disrupt the regulation of just a subset of Eve target genes.
Transcriptional repression is essential for the segmentation of the Drosophila embryo. Anteroposterior patterning depends on a broadly distributed Hunchback repressor gradient that establishes localized patterns of other gap repressors, including Giant, Krüppel, and Knirps (1, 2). The gap repressors subsequently define the borders of segmentation stripes of pair-rule gene expression (3–5).
There are eight zygotic pair-rule genes; each appears to encode a sequence-specific transcription factor. Cross-regulatory interactions help refine the patterns of pair-rule gene expression, and the encoded products are essential for the regulation of segment polarity genes (6–8). Two homeobox-containing pair-rule genes, even-skipped (eve) and fushi-tarazu (ftz), have been shown to play a particularly important role in subdividing each segment primordium into anterior and posterior compartments through the regulation of the segment polarity genes engrailed (en) and wingless (wg) (9, 10). Eve activates en but represses wg expression in odd parasegments, whereas Ftz activates en but represses wg in even parasegments.
There is a loss of all 14 middle-body en stripes in mutant embryos that completely lack Eve function (6, 11, 12). Eve is thought to function as a dedicated transcriptional repressor that regulates en expression indirectly (7). The odd-numbered en stripes are activated by Paired and repressed by Sloppy-paired and Runt, whereas the even-numbered en stripes are activated by Ftz and repressed by Odd-skipped (Odd) (7, 8, 13). The differential repression of these target genes is thought to create small domains where the Paired and Ftz activators can initiate en expression. For example, higher concentrations of Eve are needed to repress ftz than odd (7). Consequently, a single cell within the eve stripe contains a relatively high concentration of the Ftz activator and low levels of Odd, thereby permitting the activation of the even-numbered en stripes.
Given the importance of Eve in segmentation, we have become interested in determining how it functions as a transcriptional repressor. Previous studies suggest that Eve interferes with the binding or function of TATA box-binding protein (TBP) at the core promoter (14–16). Recent studies have shown that a number of repressors in the early embryo interact with corepressor proteins, such as Groucho and dCtBP (17). It is conceivable that Eve also interacts with corepressor proteins. However, Eve activity does not appear to be impaired in mutant embryos lacking Groucho or dCtBP, although such mutants exhibit severe patterning defects because of the inactivation of other repressors such as Krüppel, Snail, and Hairy. Studies in both yeast and mammals have identified histone deacetylases as corepressors of a number of sequence-specific transcriptional repressors (18–20). The Rpd3 deacetylase is a major histone deacetylase in yeast (21), and the mammalian homolog of this protein appears to mediate repression by the thyroid hormone and retinoic acid receptors and the Mad–Max complex (20).
A Drosophila homolog of Rpd3 was identified in a genetic screen for mutations affecting position-effect variegation of white gene expression in the eye (22). However, this mutation affects Rpd3 expression only in eye imaginal discs. Searches of Drosophila genomic databases identified a P-element-induced mutation in the 5′ untranslated region of the Rpd3 deacetylase gene. This mutation causes a severe reduction in the expression of Rpd3. The analysis of embryos derived from Rpd3 germline clones revealed a pair-rule segmentation phenotype that can be attributed to a partial loss of Eve repressor function.
MATERIALS AND METHODS
Fly Stocks and Generation of Germline Clones.
Germline clones were produced by using the FLP-dominant female sterile (DFS) technique as described (23, 24). Males heterozygous for the DFS mutation ovoD1 and containing a heat-inducible hsp70-FLP transgene were obtained by mating yw hsp70-FLP; CxD/TM3, Sb females with w; FRT3L ovoD1/TM3, Sb males. The resulting males, yw hsp70-FLP; FRT3L ovoD1/TM3, Sb, were mated with w; FRT3L l(3)04556/TM3, Sb females (kindly provided by N. Perrimon). Larvae were heatshocked for 3 hr at 37° for 3 consecutive days on days 3, 4, and 5 after egglaying to induce the expression of the FLP recombinase. Sb+ females were mated with w; FRT3L l(3)04556/TM3, Sb males, and embryos were collected and fixed for in situ hybridization. Embryos were also collected from an eveR13 stock (Bloomington Stock Center no. BL-299).
Cuticle Preparation.
Advanced-stage embryos were dechorionated in bleach, transferred to a microscopic slide, and cleared with lactic acid at 65° for 1.5 hr. They were subsequently photographed using darkfield microscopy.
In Situ Hybridization and Immunohistochemistry.
In situ hybridization assays were performed with digoxigenin-labeled eve, ftz, en, odd, and Rpd3 antisense RNA probes as described (25, 26). Embryos were double stained for Ftz protein and odd RNA as described (7). Fixed embryos were first incubated with a 1:500 dilution of a Ftz polyclonal antibody (kindly provided by H. Krause) and subsequently stained with horseradish peroxidase by using the Vectastain Elite kit (Vector Laboratories). These embryos were then hybridized with an odd antisense RNA probe and histochemically stained after incubation with an alkaline-phosphatase conjugated anti-digoxigenin antibody (Boehringer Mannheim).
RESULTS
Identification of a P-Element-Induced Mutation in Rpd3.
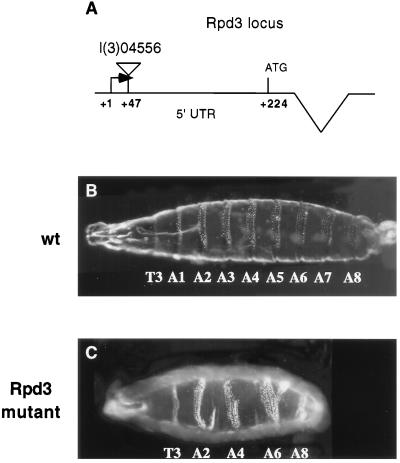
Rpd3 maps within the 64C region of chromosome 3 (22). The Berkeley Drosophila Genome Project (27) identified a P-induced lethal mutation (l(3)04556) that maps 47 bp downstream of the Rpd3 putative transcription start site (Fig. 1 and http://www.fruitfly.org/p_disrupt/). As shown previously (24), embryos derived from l(3)04556 homozygous germline clones exhibit pair-rule patterning defects that are similar to those observed in ftz− embryos (Fig. 1). In the present study, we have attempted to determine the basis for this disruption in segmentation.
Figure 1.
Summary of the Rpd3 mutation. The Berkeley Genome Project identified a P-element lethal mutation that maps within the Rpd3 gene. The location of the P-element within the 5′ UTR is summarized in A. This P-element mutation, l(3)04556, was previously examined in a large-scale screen of maternally expressed, zygotic lethals (24). Embryos derived from l(3)04556 germline clones exhibit a variable pair-rule phenotype (Rpd3 mutant, C). A wild-type embryo is shown for comparison (wt, B).
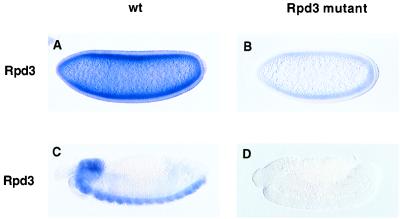
Because Rpd3 is expressed maternally, it was necessary to produce female flies homozygous for the l(3)04556 mutation. This was accomplished by making mosaic flies containing clones of homozygous germ cells by using the FLP-dominant female sterile technique (23). We first examined Rpd3 expression in mutant embryos derived from l(3)04556 germline clones (Fig. 2). Embryos were hybridized with a digoxigenin-labeled Rpd3 antisense RNA probe. There is at least a 5-fold reduction in the levels of Rpd3 transcripts in mutant vs. wild-type embryos (Fig. 2 A and B). There is no detectable expression of Rpd3 in advanced-stage mutant embryos (Fig. 2 C and D).
Figure 2.
The l(3)04556 mutation causes a substantial loss in Rpd3 expression. Embryos were hybridized with an Rpd3 digoxigenin-labeled antisense RNA probe and are oriented with dorsal up and anterior to the left. (A and C) Cellularizing (A) and retracted (C) wild-type embryos. Maternal Rpd3 products are ubiquitously distributed throughout early embryos (A). In contrast, zygotic products appear to exhibit a more localized distribution, with peak expression in the central nervous system, including both the brain and the ventral nerve cord (C). (B and D) Cellularizing (B) and retracted (D) l(3)04556 mutants. Embryos were collected from germline clones after mating with l(3)04556/+ males. Half the embryos lack both maternal and zygotic Rpd3 activity, and presumably, the embryo shown in D corresponds to such an example. There is at least a 5-fold reduction in the levels of maternal Rpd3 products (B) as compared with wild-type (A). There is a virtual loss in detectable products in advanced-stage l(3)04456/l(3)04556 homozygotes.
Most Repressors Are Active in Rpd3 Mutant Embryos.
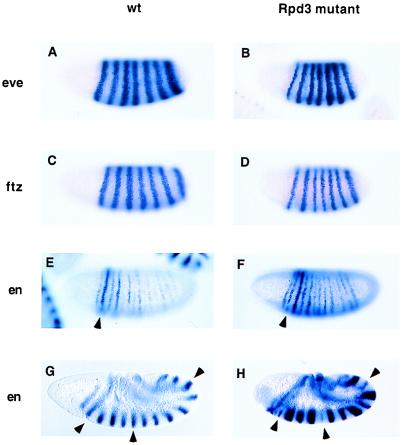
The primary pair-rule genes eve (Fig. 3 A and B), hairy, and runt (not shown) exhibit essentially normal patterns of expression in mutant embryos, although sharpening of the eve stripes might be delayed in gastrulating embryos (not shown). This result indicates that all of the gap gene repressors are active in the Rpd3 mutant. The secondary pair-rule gene ftz also exhibits a normal pattern of expression in mutant embryos (Fig. 3 C and D), although there may be a delay in the refinement of the stripes as seen for eve (not shown). These results suggest that the Hairy and Runt repressors function normally in Rpd3 mutants.
Figure 3.
eve, ftz, and en expression in Rpd3 mutants. Embryos were hybridized with the indicated digoxigenin-labeled antisense RNA probes and are oriented with anterior to the left and dorsal up. Rpd3 mutant embryos were derived from l(3)04556 germline clones. (A and B) Wild-type (wt) and mutant (Rpd3 mutant) embryos undergoing cellularization after hybridization with an eve probe. The mutant embryo is reduced in size, but the seven-stripe eve pattern appears essentially normal. (C and D) Wild-type and mutant embryos undergoing cellularization after hybridization with a ftz probe. The mutant embryo exhibits an essentially normal seven-stripe pattern. (E and F) Wild-type and mutant embryos at the onset of gastrulation after hybridization with an en probe. In wild-type embryos, the even-numbered en stripes are stronger than the odd-numbered stripes. The arrowhead indicates en stripe 2. Note that it is stained more intensely than the adjacent stripe 3. In mutant embryos, there is a relative loss in the even-numbered en stripes so that en stripe 2 (arrowhead) is significantly weaker than stripe 3. (G and H) Wild-type and mutant embryos undergoing the rapid phase of germband elongation. The normal en pattern includes 14 stripes that extend along the length of the germband (G). The mutant embryos show a variable loss or reduction in the even-numbered en stripes. For example, the arrowheads in H indicate en stripes 2, 6, and 12. These stripes are reduced in the mutant, but are expressed at the same levels as the other stripes in wild-type embryos (see arrowheads in G).
We next examined the expression of the segment polarity gene en (Fig. 3 E–H). In normal embryos, the even-numbered en stripes are initially stronger than the odd-numbered stripes. However, these stripes are weaker than the odd-numbered stripes in Rpd3 mutants (Fig. 3F). This effect becomes more dramatic in older embryos (compare Fig. 3 G with H) where some of the even-numbered en stripes are very weak or completely missing. There is a variable loss of the even-numbered en stripes in early embryos and a corresponding variation in the cuticular defects observed in older embryos (data not shown).
Rpd3 Mutants Alter Eve Repressor Function.
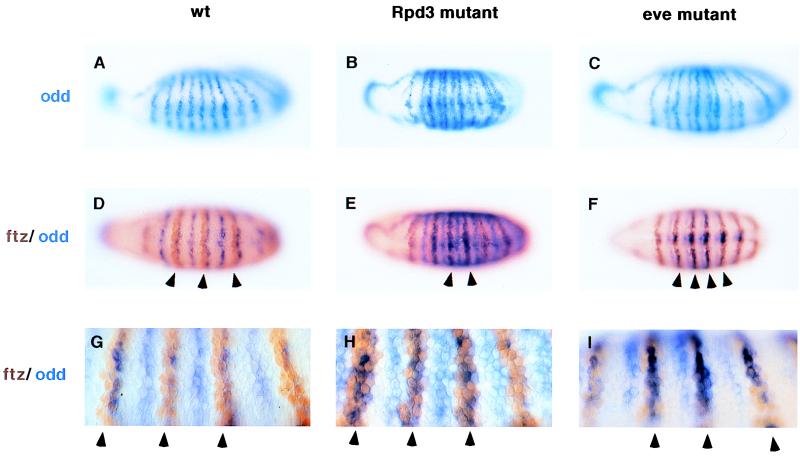
The preceding results suggest that the Rpd3 mutation might impair the Ftz or Ftz-F1 activators because these are required for the expression of the even-numbered en stripes (28–30). Alternatively, the loss of Rpd3 might lead to a change in the expression pattern of a repressor, which in turn inhibits Ftz activity. To distinguish between these possibilities, we examined the expression of odd, a known repressor of en (8, 13, 31, 32) (Fig. 4 A and B). odd is initially expressed in a pair-rule pattern of seven stripes, but during gastrulation seven additional secondary stripes are formed to generate a 14-stripe expression pattern (33). In normal embryos, these stripes are evenly spaced (Fig. 4A), whereas in Rpd3 mutants they are not (Fig. 4B). In the mutant embryos there is a partial pair-wise alignment of adjacent odd stripes. A similar change is observed in eve− embryos (Fig. 4C).
Figure 4.
Altered patterns of ftz and odd expression in Rpd3 mutants. Gastrulating embryos were stained with an odd RNA probe (A–C) or with a mixture of an odd RNA probe and anti-Ftz antibodies (D–I). (A) Wild-type embryo (wt) that was hybridized with the odd RNA probe. odd is initially expressed in a seven-stripe pattern during cellularization (not shown), but is expressed in 14 stripes during gastrulation. Note that the stripes are evenly spaced along the germ band. (B) Same as A except that the mutant embryo was derived from l(3)04556 germline clones (Rpd3 mutant). There is a pairwise alignment of adjacent odd stripes, including stripes 2 + 3, 4 + 5, 6 + 7, etc. (C) Same as A and B except that the embryo is homozygous for a null mutation in eve (eveR13). As seen in the Rpd3 mutant, there is a pairwise alignment of adjacent stripes, including 2 + 3, 4 + 5, 6 + 7, etc. (D) Wild-type embryo stained to show both odd RNAs (blue) and Ftz protein (brown). Note that each Ftz stripe is shifted just anterior of each odd-numbered odd stripe. Consequently, odd and Ftz are coexpressed in posterior regions of each Ftz stripe, whereas Ftz alone is expressed in anterior regions. The latter sites are thought to correspond to the locations of the even-numbered en stripes (arrowheads). This overlap in the two patterns is more easily seen at high magnification (G, arrowheads). (E) Same as D, except that the embryo was obtained from an l(3)04556 germline clone. The Ftz and odd patterns largely coincide, so that the anterior portion of each Ftz stripe also expresses odd (arrowheads, see high-magnification view in H). (F) Same as D and E, except that the embryo is homozygous for the eve null mutation. There is a similar failure of the Ftz and odd patterns to resolve (arrowheads, see high-magnification view in I).
Previous studies suggest that both ftz and odd stripes are under the control of the Eve repressor (7). Differential repression of ftz and odd resolves the two patterns, so that each ftz stripe is normally shifted anterior of each odd-numbered odd stripe (Fig. 4D). In Rpd3 mutants, the ftz and odd patterns fail to resolve (Fig. 4 E and H), so that odd-numbered odd stripes mostly coincide with the ftz stripes. We suggest that this failure in ftz–odd resolution is responsible for the pair-rule phenotype observed in Rpd3 mutant embryos (see Discussion). A prediction of this proposal is that eve mutants should exhibit similar alterations in the ftz and odd expression patterns. Double staining assays reveal that eve−/eve− embryos exhibit a similar failure to resolve the ftz and odd expression patterns (Fig. 4 F and I).
DISCUSSION
We have analyzed mutant embryos derived from germline clones of a P-element mutation in the Rpd3 gene. Although the mutation does not eliminate Rpd3+ gene activity, there is at least a 5-fold reduction in maternal Rpd3 products. This reduction causes a pair-rule phenotype that is similar to the one observed in ftz− mutants (34), whereby the odd-numbered abdominal segments are lost. This phenotype demonstrates a surprisingly specific role of the Rpd3 histone deacetylase, even though maternal Rpd3 products are ubiquitously distributed throughout the early embryo. It would appear that the majority of the embryonic repressors are active in mutant embryos, including the gap repressors Hunchback, Giant, Krüppel, and Knirps. This situation contrasts with the patterning defects caused by mutations in two other corepressors present in the early embryo, Groucho and dCtBP (35–38). Mutations in either gene cause compound patterning defects because of the loss of multiple repressors. For example, removal of maternal dCtBP products causes a reduction or loss in the repression activities of Krüppel and Knirps (38) and a corresponding disruption in both thoracic and abdominal segments. An implication of these observations is that neither Groucho nor dCtBP functions solely by the recruitment of Rpd3.
Given the importance of Rpd3 as a corepressor of both yeast and mammalian transcriptional repressors, we anticipated that the Rpd3 mutants would exhibit more severe patterning defects. Instead, it would appear that this histone deacetylase does not represent a major pathway of repression in the early embryo. Of course, it is conceivable that the complete loss of Rpd3 products would cause more severe patterning defects. Unfortunately, it might not be possible to produce germline clones for a null mutation in the Rpd3 gene because the present hypomorphic allele produces very few eggs and mutations in genes that encode associated proteins such as Sin3 (not shown) and Mi-2 (39) fail to produce viable germline clones. An alternative explanation for the relatively mild Rpd3 patterning defects is that there is redundancy among different deacetylases. Indeed, two additional histone deacetylases are maternally expressed and ubiquitously distributed throughout the early embryo (M.M., unpublished results).
There are several possible explanations for impaired Ftz function in Rpd3 mutants. It is conceivable that the Rpd3 mutation disrupts Ftz-mediated activation. However, we favor the idea that Rpd3 functions as a corepressor of Eve. The similarities in the Rpd3 and ftz mutant phenotypes may be caused by the coincident odd and ftz expression patterns observed in embryos derived from l(3)04556 germline clones (summarized in Fig. 5). The Odd repressor is thought to block Ftz-mediated activation of en (8, 13, 40). We have presented evidence that this expansion in Odd might result from an inability of Eve to repress odd expression in Rpd3 mutant embryos (Fig. 5). Consistent with this proposal, we find that in vitro translated Eve interacts with a glutathione S-transferase-Rpd3 fusion protein (data not shown). Because the Eve repressor is required for both the odd- and even-numbered en stripes, it would appear that the Rpd3 mutation does not cause a general loss of Eve function. For example, eve hypomorphs cause the loss of odd-numbered en stripes, whereas null mutations cause a loss of all en stripes (6, 12). It would therefore appear that Eve fails to repress certain promoters (e.g., odd and possibly ftz) in Rpd3 mutant embryos, but retains repressor function on other promoters (e.g., paired and sloppy-paired). This selectivity in the regulation of different target promoters is consistent with the notion that Eve mediates repression through multiple mechanisms, including the recruitment of corepressors and direct interactions with TBP (14–16). Multiple modes of repression may be mediated by other transcriptional repressors, such as Hairy, which appears to interact with different classes of corepressors (37, 41, 42).
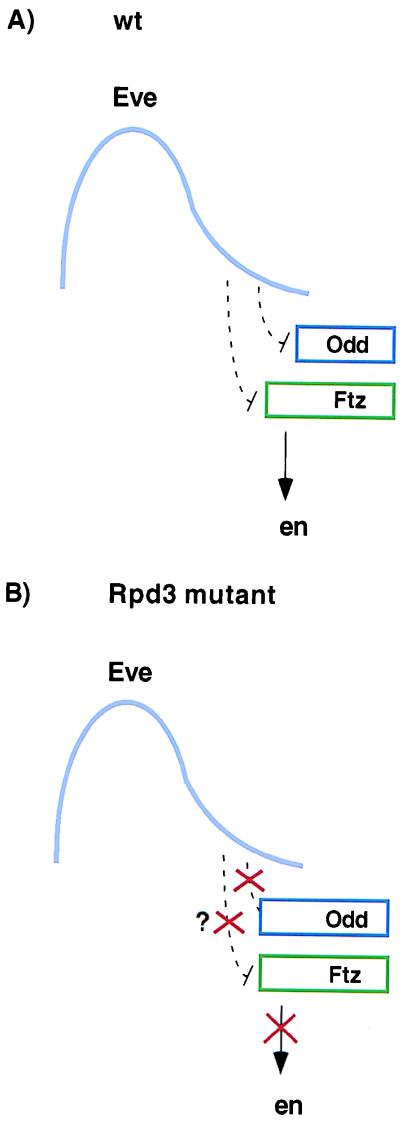
Figure 5.
Summary of segmentation defects in Rpd3 mutants. (A) The diagram represents a single Eve stripe in a wild-type gastrulating embryo. The stripe is asymmetric, with peak levels of Eve protein in anterior regions and progressively lower levels in posterior regions (6). Genetic studies suggest that this Eve gradient differentially represses the expression of ftz and odd, thereby generating overlapping but noncoincident ftz and odd expression patterns (7, 13). Ftz activates en in anterior regions lacking the Odd repressor (arrow). Odd restricts the expression of the even-numbered en stripes to a single cell. (B) In Rpd3 mutants, the ftz and odd patterns fail to resolve. Consequently, anterior cells in each Ftz stripe also express the Odd repressor, thereby attenuating the even-numbered en stripes. It is possible that Eve fails to repress odd, whereas ftz expression is unaffected. Alternatively, both the odd and ftz patterns might shift into more anterior regions. In either case, the two patterns coincide so that the Odd repressor blocks Ftz activity.
Acknowledgments
We thank Norbert Perrimon for providing FRT l(3)04556 flies, Henry Krause for Ftz antibody, Hilary Ashe for comments on the manuscript, and Armen Manoukian for sharing unpublished results. M.M. was supported by a fellowship from the Swedish Foundation for International Cooperation in Research and Higher Education (STINT). This work was funded by a grant from the National Institutes of Health (GM 34431).
ABBREVIATION
- TBP
TATA box-binding protein
References
- 1.Hulskamp M, Pfeifle C, Tautz D. Nature (London) 1990;346:577–580. doi: 10.1038/346577a0. [DOI] [PubMed] [Google Scholar]
- 2.Struhl G, Johnston P, Lawrence P A. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- 3.Carroll S B. Cell. 1990;60:9–16. doi: 10.1016/0092-8674(90)90711-m. [DOI] [PubMed] [Google Scholar]
- 4.Gray S, Cai H, Barolo S, Levine M. Philos Trans R Soc Lond B. 1995;349:257–262. doi: 10.1098/rstb.1995.0111. [DOI] [PubMed] [Google Scholar]
- 5.Rivera-Pomar R, Jäckle H. Trends Genet. 1996;12:478–483. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- 6.Frasch M, Warrior R, Tugwood J, Levine M. Genes Dev. 1988;2:1824–1838. doi: 10.1101/gad.2.12b.1824. [DOI] [PubMed] [Google Scholar]
- 7.Manoukian A S, Krause H M. Genes Dev. 1992;6:1740–1751. doi: 10.1101/gad.6.9.1740. [DOI] [PubMed] [Google Scholar]
- 8.Fujioka M, Jaynes J B, Goto T. Development (Cambridge, UK) 1995;121:4371–4382. doi: 10.1242/dev.121.12.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence P A, Johnston P, Macdonald P, Struhl G. Nature (London) 1987;328:440–442. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- 10.Ingham P W, Baker N E, Martinez-Arias A. Nature (London) 1988;331:73–75. doi: 10.1038/331073a0. [DOI] [PubMed] [Google Scholar]
- 11.Macdonald P M, Ingham P, Struhl G. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- 12.DiNardo S, O’Farrell P H. Genes Dev. 1987;1:1212–1225. doi: 10.1101/gad.1.10.1212. [DOI] [PubMed] [Google Scholar]
- 13.Manoukian A S, Krause H M. Development (Cambridge, UK) 1993;118:785–796. doi: 10.1242/dev.118.3.785. [DOI] [PubMed] [Google Scholar]
- 14.Um M, Li C, Manley J L. Mol Cell Biol. 1995;15:5007–5016. doi: 10.1128/mcb.15.9.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Austin R J, Biggin M D. Mol Cell Biol. 1995;15:4683–4693. doi: 10.1128/mcb.15.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, Manley J L. Mol Cell Biol. 1998;18:3771–3781. doi: 10.1128/mcb.18.7.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mannervik M, Nibu Y, Zhang H, Levine M. Science. 1999;284:606–609. doi: 10.1126/science.284.5414.606. [DOI] [PubMed] [Google Scholar]
- 18.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 19.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 20.Torchia J, Glass C, Rosenfeld M G. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 21.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. Nature (London) 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- 23.Chou T B, Perrimon N. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perrimon N, Lanjuin A, Arnold C, Noll E. Genetics. 1996;144:1681–1692. doi: 10.1093/genetics/144.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 26.Jiang J, Kosman D, Ip Y T, Levine M. Genes Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- 27.Spradling A C, Stern D M, Kiss I, Roote J, Laverty T, Rubin G M. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Florence B, Guichet A, Ephrussi A, Laughon A. Development (Cambridge, UK) 1997;124:839–847. doi: 10.1242/dev.124.4.839. [DOI] [PubMed] [Google Scholar]
- 29.Guichet A, Copeland J W, Erdelyi M, Hlousek D, Zavorszky P, Ho J, Brown S, Percival-Smith A, Krause H M, Ephrussi A. Nature (London) 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 30.Yu Y, Li W, Su K, Yussa M, Han W, Perrimon N, Pick L. Nature (London) 1997;385:552–555. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]
- 31.Mullen J R, DiNardo S. Dev Biol. 1995;169:295–308. doi: 10.1006/dbio.1995.1145. [DOI] [PubMed] [Google Scholar]
- 32.Saulier-Le Drean B, Nasiadka A, Dong J, Krause H M. Development (Cambridge, UK) 1998;125:4851–4861. doi: 10.1242/dev.125.23.4851. [DOI] [PubMed] [Google Scholar]
- 33.Coulter D E, Swaykus E A, Beran-Koehn M A, Goldberg D, Wieschaus E, Schedl P. EMBO J. 1990;9:3795–3804. doi: 10.1002/j.1460-2075.1990.tb07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wakimoto B T, Kaufman T C. Dev Biol. 1981;81:51–64. doi: 10.1016/0012-1606(81)90347-x. [DOI] [PubMed] [Google Scholar]
- 35.Fisher A L, Caudy M. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 36.Nibu Y, Zhang H, Levine M. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 37.Poortinga G, Watanabe M, Parkhurst S M. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kehle J, Beuchle D, Treuheit S, Christen B, Kennison J A, Bienz M, Muller J. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- 40.Benedyk M J, Mullen J R, DiNardo S. Genes Dev. 1994;8:105–117. doi: 10.1101/gad.8.1.105. [DOI] [PubMed] [Google Scholar]
- 41.Paroush Z, Finley R L, Jr, Kidd T, Wainwright S M, Ingham P W, Brent R, Ish-Horowicz D. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, Levine M. Proc Natl Acad Sci USA. 1999;96:535–540. doi: 10.1073/pnas.96.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]