Abstract
The closely related Hox transcription factors Ultrabithorax (Ubx) and Antennapedia (Antp) respectively direct first abdominal (A1) and second thoracic (T2) segment identities in Drosophila. It has been proposed that their functional differences derive from their differential occupancy of DNA target sites. Here we show that a hybrid version of Ubx (Ubx-VP16), which possesses an enhanced transcriptional activation function, no longer directs A1 denticle pattern in embryonic epidermal cells. Instead, it mimics Antp in directing T2 denticle pattern, and it can rescue the cuticular loss-of-function phenotype of Antp mutants. In cells that do not produce denticles, Ubx-VP16 appears to have largely retained its normal repressive regulatory functions. These results suggest that the modulation of Hox activation and repression functions can account for segment-specific morphological differences that are controlled by different members of the Hox family. Our results also are consistent with the idea that activity regulation underlies the phenotypic suppression phenomenon in which a more posterior Hox protein suppresses the function of a more anterior member of the Hox cluster. The acquisition of novel activation and repression potentials in Hox proteins may be an important mechanism underlying the generation of subtle morphological differences during evolution.
Keywords: Hox, Ultrabithorax, homeotic proteins, Drosophila
The Hox family of transcription factors assigns different identities to cells on the anterior–posterior body axis during animal development (1–3). Hox proteins each contain a highly homologous DNA-binding homeodomain and recognize in vitro similar DNA sequences as monomers (4). Binding specificity of Hox proteins can be enhanced in vitro by their interaction with the Exd/Pbx homeodomain proteins on specific composite DNA sites (5); this enhanced binding has been proposed to account for their functional differences in vivo. The hypothesis that selective occupancy of Hox binding sites controls the distinction of one segment morphology versus another can be termed the selective binding model for Hox functional specificity.
Alternatively, Hox proteins may bind to a highly overlapping spectrum of monomer binding sites in vivo as they do in vitro. Hox proteins may then have their activity regulated after DNA binding. Depending on the sequence context in which these common binding sites reside, each Hox protein might assume different activity states ranging from strong transcriptional activation to strong transcriptional repression. This variation in activity could be mechanistically accomplished by stable association with other proteins that act by biasing a Hox protein to be a repressor on a certain target elements, or perhaps by transient association with other proteins that covalently modify a Hox protein on certain target elements. The hypothesis that different activation/repression functions, elicited from different Hox proteins on the same binding sites, control the distinction of one segment morphology versus another can be termed the activity regulation model for Hox functional specificity (6–8). The selective binding and activity regulation mechanisms are not mutually exclusive. However, most published studies have emphasized the role of selective binding of Hox proteins in combination with Exd as the basis for functional specificity.
That activity regulation of Hox proteins exists is supported by recent experiments on Antp (9) and Deformed (Dfd) (8). For example, Antp is phosphorylated by casein kinase II (CKII) in vitro, and mutations in the CKII phosphorylation sites of Antp can modulate its biological activities in embryos (9). In the case of Dfd, its binding to monomer DNA sites in embryos may occur independently of cofactors such as Exd. However, binding site occupancy is not sufficient for gene activation in embryos (8). In addition, transfection assays show that the N-terminal region of Dfd protein contains a strong activation function (8, 10), but this function is suppressed by the homeodomain in the full-length Dfd protein (8).
Consistent with the activity regulation model, many Drosophila Hox proteins are capable of both activating and repressing gene expression. This property is well documented for Ubx, which can directly activate or repress transcription from different promoters in genetic and transfection assays (11–14). Whether Ubx mediates activation or repression appears to depend on the sequence context of Ubx binding sites. To examine how activation strengths of Hox proteins might influence their functional specificity, we altered the strength of the Ubx activation function by attachment of a potent activation domain from the VP16 viral protein, and we assayed the embryonic function of the hybrid protein.
MATERIALS AND METHODS
Generation of Transgenic UAS-Ubx-VP16 Flies.
Expression construct UAS-Ubx-VP16 was made as follows: The ORF of Ubx isoform Ia was first cloned as an EcoRI–NotI fragment into pUAST (15) to generate UAS-Ubx. A DNA fragment encoding the VP16 activation domain (codons 413–490) (16), with the addition of an optimal translation start site and ATG codon, was then amplified by PCR and cloned into the EcoRI site of the UAS-Ubx plasmid. The UAS-Ubx-VP16 construct was injected into Drosophila w1118 embryos to establish transgenic lines.
Expression of Transgenes in Drosophila Embryos.
Ubx-VP16, Ubx, or Antp proteins were expressed in Drosophila embryos by using the GAL4/UAS system (15) by crossing flies carrying UAS-Ubx-VP16, UAS-Ubx (17), or UAS-Antp with flies carrying GAL4 drivers. Cleared cuticles were subsequently prepared from these embryos (18). Two GAL4 drivers, the strong arm-GAL44 driver (19) and the weak arm-GAL44r driver (see below), were used. Both drivers gave stronger phenotypes when provided maternally. Unless otherwise noted, in the experiments described here, arm-GAL44 is provided paternally and arm-GAL44r maternally.
Function of Ubx-VP16 in Antp− Embryos.
UAS-Ubx-VP16 and arm-GAL44 on the third chromosome were separately recombined onto the Antp25 chromosome by using standard genetic procedures. The genetic compositions of the recombined chromosomes were confirmed by complementation with other Antp mutant chromosomes and by cuticle phenotypes after GAL4 induction to assay for the presence of UAS-Ubx-VP16 or the GAL4 driver on the recombinant chromosomes. All recombinant chromosomes that possessed arm-GAL44 (called arm-GAL44r) provided weaker phenotypes in combination with UAS-Ubx-VP16 than the original arm-GAL44. Immunostaining with a mouse monoclonal anti-Ubx antibody indicated that much lower protein levels of Ubx-VP16 were produced after induction by the arm-GAL44r recombinant chromosomes than by arm-GAL44 (data not shown). This lower level may be because the arm-GAL44r chromosome retains only one of the two copies of the arm-GAL4 insertions that are reported to exist on the parental chromosome (http://flybase.bio.indiana.edu). Expression of Ubx-VP16 in the Antp25 embryos was achieved by crossing of UAS-Ubx-VP16 Antp25 with arm-GAL44r Antp25 flies.
Regulation of Target Genes by Ubx-VP16.
Expression of Antp, Distalless (Dll), and decapentaplegic (dpp) were visualized by in situ hybridization with digoxigenin-UTP-labeled antisense RNA. To test the regulatory effect of Ubx-VP16 on expression of the Dll304-lacZ reporter, flies homozygous for arm-GAL44 and the Dll304 reporter were crossed with flies homozygous for UAS-Ubx-VP16, and the resulting embryos were immunostained with a mouse monoclonal anti-β-galactosidase antibody (Promega).
RESULTS AND DISCUSSION
Novel Denticle-Patterning Function of a Hyperactive Ubx Protein, Ubx-VP16.
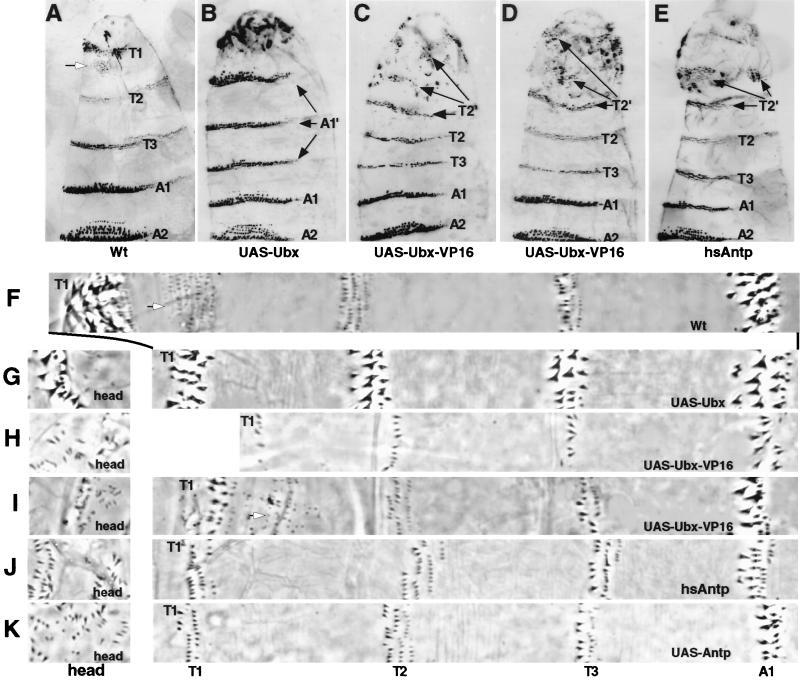
In the Drosophila embryonic epidermis, Ubx is required to specify the identity of parasegment (PS) 6. This parasegment includes the anterior compartment of the first abdominal segment (A1), which develops a unique pattern and type of denticles which differ from those of the thoracic and other abdominal segments (Fig. 1 A and F). When ectopically expressed, wild-type UbxIa protein (hereafter referred to as ectopic Ubx) is sufficient to transform head and thoracic segments into morphological replicas of A1 (17, 20, 21). The principal evidence for this transformation is the generation of A1-like denticle belts in more anterior segments (Fig. 1 B and G).
Figure 1.
Cuticular transformations generated by ubiquitous expression of Ubx-VP16 are similar to that generated by Antp and unlike those induced by Ubx. (A and F) Wild-type embryos. Notice the differences in denticle morphology in the three thoracic (T1, T2, and T3), and first abdominal (A1) segments. White arrows point to the T1 denticle beard. (B and G) UAS-Ubx/arm-GAL44 embryos. Head and all three thoracic segments are transformed to an A1-like identity (marked A1′ in B). (C and H) UAS-Ubx-VP16/arm-GAL44 embryos. (D and I) UAS-Ubx-VP16/arm-GAL44r embryos. In these embryos (C, D, H, I), head and T1 segments are transformed to a T2-like identity (marked by T2′ in C and D). Ubx-VP16 expression is driven by a weaker GAL4 driver in D and I than in C and H. (E and J) hsAntp embryos. (K) UAS-Antp/arm-GAL44 embryos. Head and T1 segments are transformed to a T2-like identity (marked as T2′ in E). Cuticles in A to E were shown from the anterior (top) to A2 in ventral view. Stripes of cuticles in F to K are close-ups of the embryos, all at the same magnification (×166.)
This patterning function of Ubx is not retained when the VP16 activation domain (16) is attached to the N terminus of the UbxIa protein. When the Ubx-VP16 protein is uniformly expressed in embryos by using the arm-GAL44 driver (hereafter referred to as ectopic Ubx-VP16), head and thoracic segments develop epidermal features (Fig. 1 C and H) that are strikingly different from the A1-like structures induced by ectopic Ubx (Fig. 1 B and G). First, individual denticle morphologies in the second and third thoracic (T2 and T3) segments of ectopic Ubx-VP16 embryos appear normal although the numbers of denticles are reduced. Second, the first thoracic (T1) segment in ectopic Ubx-VP16 embryos develops without the characteristic patch of beard denticles. This is also observed in ectopic Ubx embryos. However, in contrast to ectopic Ubx, the remaining T1 denticles in ectopic Ubx-VP16 embryos have a size and shape that are indistinguishable from those of T2 type denticles (compare Fig. 1 H with G). This observation is consistent with the idea that the T1 epidermis has been transformed to a T2-like identity. Third, the head segments fail to involute and remain external in ectopic Ubx-VP16 embryos, but in contrast to ectopic Ubx embryos, the type of denticles generated ectopically in these head segments is T2-like (compare Fig. 1 H with G, head). In these ectopic Ubx-VP16 and Ubx embryos, both proteins are localized in nuclei and produced at comparable levels as revealed by immunostaining with antibodies against UbxIa (data not shown), so the patterning differences between the two cannot be attributed to differing expression levels or subcellular localizations.
The ability of Ubx-VP16 to specify T2-like denticles remains even at low concentrations. When UAS-Ubx-VP16 is activated by the weaker arm-GAL44r driver, the level of ectopic Ubx-VP16 is barely detectable by immunostaining (data not shown). In such embryos (Fig. 1 D and I), T2 and T3 denticle belts remain wild type, T1 beard denticles are reduced, and the remaining T1 denticles at the anterior border of the segment are fewer and smaller. Some of these denticles in T1 now resemble T2 denticles. Very importantly, the partially involuted head segments develop ectopic patches of denticles with a size and shape indistinguishable from those of T2 denticles (Fig. 1 D and I, head). In contrast, ectopic Ubx induced by the same driver does not result in any visible developmental abnormality of thoracic and head segments (data not shown). Thus, Ubx requires higher threshold level than Ubx-VP16 to generate transformation. As reported by Mann and Hogness (21), at the threshold level for transformation, the denticles produced by ectopic Ubx in thoracic and head segments are A1 type. Taken together, these results demonstrate that the function of Ubx-VP16 and Ubx with respect to specifying denticle development is qualitatively different. While Ubx confers A1 denticle patterning function, the addition of the strong VP16 activation domain provides the hybrid protein with a T2 denticle patterning function.
Functional Similarity of Ubx-VP16 and Antp.
The T2 denticle patterning function of Ubx-VP16 is markedly similar to that of Antp (22) in that ectopic expression of Antp results in a denticle transformation of the head and T1 segments to T2-like morphologies (Fig. 1 E, J, and K). This is most obvious in comparing the size and shape of denticles produced ectopically in the head segments of these embryos (compare Fig. 1 H and I with J and K, head). The only detectable cuticular difference between Ubx-VP16 and Antp is that ectopic expression of Antp has no influence on Keilin’s organ development in thoracic segments (22), whereas Ubx-VP16, like Ubx, can suppress the formation of this sensory structure in high dosages (data not shown).
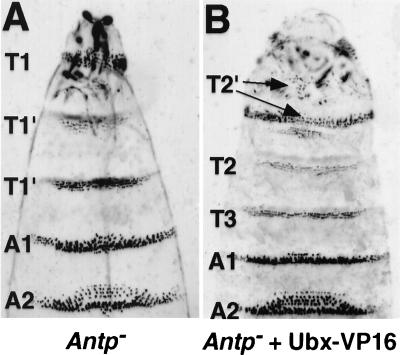
To corroborate that Ubx-VP16 has gained an Antp-like function in T2 denticle patterning, we tested whether expression of Ubx-VP16 can rescue the cuticular phenotype caused by the loss of Antp function. The UAS-Ubx-VP16 gene was recombined with an Antp null mutation, Antp25, and then tested for its function in Antp mutant embryos. Controls without arm-GAL44r driver in the genetic background (homozygous Antp25, UAS-Ubx-VP16) displayed the expected loss-of-function Antp segmental transformation of T2 and T3 toward T1 (Fig. 2A). However, when Ubx-VP16 expression was induced in the Antp25 embryos, denticle belts of the T2 and T3 segments were rescued to their normal appearances, and some head segments developed small patches of T2-like denticles (Fig. 2B). This Ubx-VP16 phenotype is nearly identical to that provided in a wild-type genetic background (Fig. 1D). We conclude that Ubx-VP16 can functionally substitute for Antp in specifying T2 denticle patterns. To do so, Ubx-VP16 presumably activates the transcription of target genes (whose identities are not currently known) that would normally be activated by Antp to generate the T2 type and pattern of denticles.
Figure 2.
Rescue of the Antp cuticular function by Ubx-VP16. (A) Antp25 mutant embryos. Notice that the T2 and T3 segments possess multiple rows of irregular T1-like denticle belts (marked as T1′). (B) UAS-Ubx-VP16/arm-GAL44r embryos in the Antp25 mutant background. The T2 and T3 segments are rescued to near normality with respect to their denticle belt morphology. In addition, T2-like denticles develop in the head region.
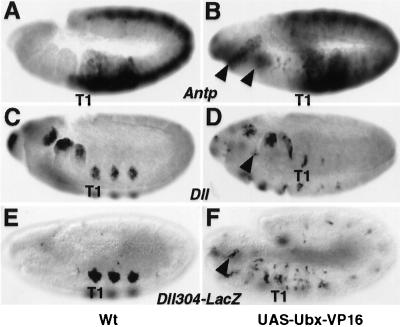
As an Antp-like T2 denticle patterning function of Ubx-VP16 is gained concomitant with the addition of the VP16 activation domain to Ubx, we wanted to verify that Ubx-VP16 indeed has increased activation function while retaining the ability to regulate the same target sites. To this end, we examined the regulation of normal Ubx target genes by ectopic Ubx-VP16. One such target gene is Antp, whose transcription normally is repressed by Ubx. In contrast to ectopic Ubx (23), ectopic Ubx-VP16 embryos exhibit activation of Antp transcription in patches of cells in the head segments (Fig. 3 A and B). The ectopic Antp protein might account for the ectopic Ubx-VP16 function in these segments. However, as shown in Fig. 2, Ubx-VP16 can induce T2 denticle development, even in the head, independently of Antp. Transcription from some other Ubx target genes and response elements is also activated in ectopic locations (see below). These results suggest that Ubx-VP16 regulates the same targets as Ubx in embryos and the VP16 activation domain enhances the activation function of Ubx on those targets.
Figure 3.
Distinct regulatory roles of Ubx-VP16 on different target genes. (A and B) Expression of Antp in wild-type (A) and UAS-Ubx-VP16/arm-GAL44 (B) embryos. Arrowheads indicate the ectopic expression of Antp in a few head segments after ubiquitous Ubx-VP16 expression (B). (C–F) Expression of Dll (C and D) and the Dll304-lacZ reporter (E and F) in wild-type (C and E) and UAS-Ubx-VP16/arm-GAL44 (D and F) embryos. Expression of both Dll (D) and Dll304-lacZ (F) is partially repressed in thoracic segments, and weakly activated in a few ectopic epidermal positions (arrowheads), by Ubx-VP16. Antp and Dll transcripts were visualized by in situ hybridization with an Antp (A and B) or a Dll (C and D) antisense RNA probe; β-galactosidase was detected by immunostaining with a mouse monoclonal anti-β-galactosidase antibody (E and F). The position of the first thoracic segment primordia (T1) is indicated. Embryos are shown at stage 11 in lateral view. In embryos where UAS-Ubx-VP16 was expressed (B, D, and F) arm-GAL44 was provided maternally.
Interestingly, a deletion of the Ubx N terminus that removes an activation domain that functions in tissue culture cells (13) does not impair its ability to specify A1-type denticles (21). Thus, the known activation domains of Ubx apparently are not important for it to perform its normal A1 denticle patterning function. This function is likely caused by Ubx repression activity.
Involvement of Activity Regulation in Hox Functional Specificity.
In our experiments the strength of activation function in Ubx is artificially varied. However, the partial change in segmental identity conferred by the Ubx-VP16 protein suggests that regulating the activity state of Ubx may modulate its functional specificity in denticle patterning. The fact that the Ubx-VP16 denticle patterning function is Antp-like suggests that the functional difference between the Ubx and Antp proteins in diversifying denticle patterns may reside in differences in activation and repression strengths on similar target genes rather than in differences in target occupancy. This suggestion is consistent with previous results indicating that Ubx and Antp recognize identical DNA sequences in vitro (4) and regulate several common target genes in embryos (24, 25). Note that our evidence indicates that the segment identity functions of Ubx and Ubx-VP16 are distinct, but it does not eliminate the possibility that the VP16 domain increases activation function by altering the binding selectivity of the hybrid protein in developing embryos. We believe this is unlikely because the specific Ubx targets such as dpp, Antp, and Dll that we tested are all regulated, and thus presumably occupied at similar Hox sites, by both Ubx and Ubx-VP16.
How does regulation of activation and repression functions contribute to Hox specificity? A simple explanation is that a Hox response element is regulated in a manner dependent on the strength of these functions that are produced by Hox proteins and other transcriptional regulators that bind nearby. Depending on what type of function is supplied, the Hox target element can be activated, repressed, or unaffected. In this scenario, both Ubx and Antp can bind to Hox binding sites near genes that promote T2 denticle patterning, but only Antp elicits an activation function sufficient to activate these genes, whereas Ubx exerts either no effect or repressive effects on them. However, when the VP16 activation domain is added to Ubx, the hybrid protein has the ability to activate these genes and thus mimics Antp to promote T2 denticle development. Activation of Antp expression by Ubx-VP16 (Fig. 3 A and B) but not by Ubx is consistent with such an explanation.
A switch of Ubx denticle patterning function from A1 to T2 was previously achieved by swapping the homeodomain of Ubx with that of Antp (21, 26). Similar homeodomain swap experiments involving other Hox proteins have indicated that residues in and around the homeodomain are responsible for the functional differences between them (27–30). None of the discriminatory homeodomain region residues directly interact with DNA in cocrystal structures (30). In light of the results reported here, we suggest that these residues are involved in regulating the activation and repression functions of Hox proteins. How this occurs is not yet clear. However, in the Hox protein Dfd, the homeodomain is required both to repress the Dfd activation function in tissue culture cells and to mediate interaction with the Exd cofactor in vitro (8). Exd has been proposed to convert Hox proteins into transcriptionally activated states (7, 8). Recently, a balance of activation and repression functions has been found to be crucial for the activity of the POU-type homeodomain protein Pit-1, and in cultured pituitary cells this balance appears to be mediated by protein–protein binding competition between coactivator and corepressor complexes for the homeodomain region of the protein (31).
Overlapping Hox Functions and Regulated Activation/Repression Activities.
Interestingly, although Ubx-VP16 acquires an Antp-like ability in denticle patterning, it preserves the Ubx ability to repress Keilin’s organ development in thoracic segments (data not shown). Therefore, Ubx-VP16 displays a mix of Antp-like and Ubx-like functions, dependent on tissue types and cell positions.
As development of Keilin’s organs requires the appendage-promoting gene Distalless (Dll), we examined the regulation of Dll by Ubx-VP16. The expression of Dll in thoracic appendage primordia cells is repressed by Ubx by means of the Dll304 element (11), presumably by eliciting the Ubx repression function on the element. In ectopic Ubx-VP16 embryos, both Dll expression (Fig. 3 C and D) and the activity of the Dll304 element (Fig. 3 E and F) are partially repressed. However, unlike ectopic Ubx, Ubx-VP16 is capable of activating Dll304 in other cells outside the appendage primordia (Fig. 3 E and F). Thus, the Ubx-like function of Ubx-VP16 in repressing Keilin’s organ development stems from retaining the Ubx repressive function on Dll transcription. As this repression appears specific for appendage primordia cells, the repression function of Ubx-VP16 is not constitutive but rather generated in a regulated manner. Taken together, the above results suggest that Ubx-VP16 functions are due to normal Ubx repressive effects on some targets (e.g., Dll), despite the attached VP16 activation domain, as well as a novel activation function on other targets (e.g., Antp) caused by the VP16 domain.
The mix of functions that Ubx-VP16 exhibits is also often observed for natural Hox proteins (6). For example, Ubx is like Antp in activating expression of the teashirt (tsh) gene and repressing expression of the Sex comb reduced (Scr) gene (24, 25). On such targets Ubx and Antp apparently provide similar states of activation function. We suggest that the differences in their states of activity on other target genes explain their distinct morphological functions. For instance, although Ubx but not Antp represses Dll expression, Antp is apparently able to occupy the Dll promoter, since a hybrid Ubx protein containing the Antp homeodomain can repress Dll expression (26). Presumably, Ubx protein sequences outside the homeodomain provide a repressive function on the Dll appendage enhancer. Consistent with this idea, when the activity of Antp is altered by mutations in its casein kinase II phosphorylation sites, it acquires the ability to repress Dll expression and thus Keilin’s organ development (9).
Activity Regulation and the Phenotypic Suppression Phenomenon.
Regulation of activation and repression functions may also be the mechanism that underlies the phenomenon of phenotypic suppression, in which one Hox protein can dominantly suppress the function of other coexpressed Hox proteins (20, 32). It has been proposed that competition of Hox proteins for DNA binding sites is responsible for this phenomenon (20, 32).
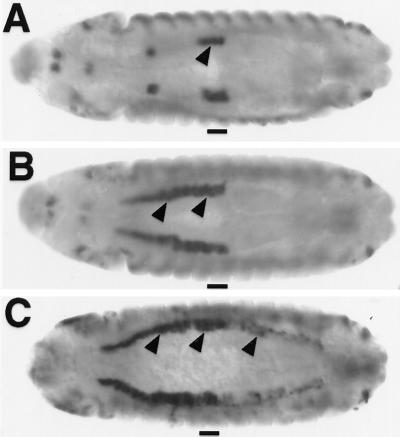
A well studied example of phenotypic suppression is the parasegment-specific transcription of the decapentaplegic (dpp) gene in the visceral mesoderm (VM). dpp is directly activated by Ubx protein in PS7 but is repressed by Abdominal-A (Abd-A) protein in PS8–12 of the VM, even when Ubx protein is ectopically expressed in PS8–12 (12, 33) (Fig. 4 A and B). Recently, it was found that the repression conferred by Abd-A and the activation conferred by Ubx involves separate clusters of Hox binding sites within the dpp674 element (34). This suggests that Abd-A does not compete with Ubx for binding to the same DNA sites to antagonize Ubx activation on dpp. Instead, Abd-A and Ubx proteins can occupy many sites on the dpp674 element in PS8–12 (34), but only Abd-A is capable of conferring repression from one of the clusters of Hox sites. The Abd-A repression function can then override the Ubx activation function that is produced from another cluster of Hox binding sites on dpp674.
Figure 4.
Ubx-VP16 overrides the repression effect of abd-A on dpp. (A–C) dpp expression in the midgut mesoderm (arrowheads) of wild-type (A), UAS-Ubx/arm-GAL44 (B), and UAS-Ubx-VP16/arm-GAL44 (C) embryos. PS7 is marked as a bar. Notice that Ubx does not activate dpp posterior to PS7, because of repression of dpp by Abd-A (33). Ubx-VP16 activates dpp posterior to PS7, even in abd-A+ embryos. dpp transcripts were visualized by in situ hybridization with a dpp antisense RNA probe. Embryos are shown at stage 13 in dorsal view.
Assuming that Abd-A repression function suppresses the Ubx activation function on dpp674, it might be possible to reverse the dominant suppression by enhancing Ubx activation function. Indeed, when Ubx-VP16 is ubiquitously expressed, transcription of dpp is activated in PS8–12 (Fig. 4C), whereas ectopic Ubx has no detectable influence on dpp in these cells (Fig. 4B). The dpp674 element is also activated in PS8–12 in a manner similar to the dpp transcription unit (data not shown). This result suggests the regulatory specificity of Hox response elements such as dpp674 is ultimately determined on the cumulative balance of Hox-dependent activation and repression functions.
Activity Regulation and Evolution of Body Plans.
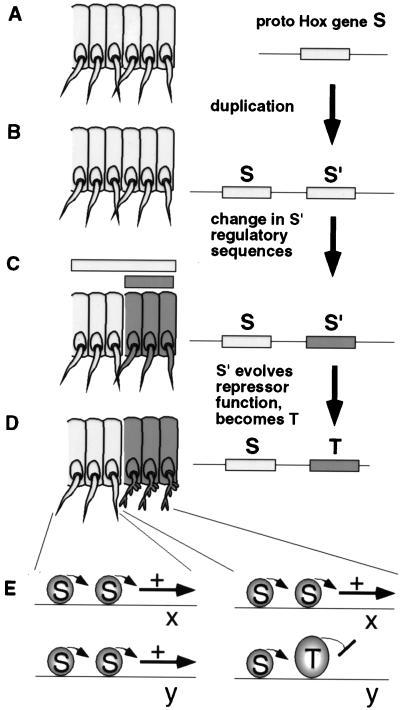
The role of activity regulation in Hox segmental specificity may provide new insight into understanding how the Hox patterning system evolved (Fig. 5). At an early point in metazoan evolution, prototypes of Hox genes such as Ubx and Antp were generated by the duplication of a common ancestral gene (Fig. 5 A and B). After the duplication event, one or both of the two copies accumulated mutations and evolved distinct functions. One evolutionary event that altered function was changes in regulatory sequence that altered expression patterns compared with the ancestral copy (35–37) (Fig. 5 B and C). From the study of extant Hox genes, we also know that changes in coding sequence during evolution have generated functional distinctions between adjacent Hox genes (labeled S and T in Fig. 5). We propose that coding region changes that resulted in different functional specificities did so by altering activation/repression strengths on a largely common set of downstream genes. One reason for this proposal is that mutations in coding sequences that altered binding specificity would presumably influence target occupancy on all or most downstream genes. Therefore, the newly evolved Hox protein T would no longer regulate many of the genes under the control of Hox protein S, and protein T would also immediately acquire a novel battery of downstream genes. We imagine that these events would result in striking morphological changes in the body plan that would have a low chance of surviving and being selected (“hopeless monsters”; cf. ref. 38).
Figure 5.
How acquisition of novel activation and repression functions in Hox proteins might lead to incremental morphological changes in evolution. A shows a region of arthropod trunk with six segments. Proto-Hox gene S is expressed in all. In B, a tandem genomic duplication has occurred, to generate proto-Hox genes S and S′. In C, S′ regulatory sequences have undergone mutations so that S′ transcripts are restricted to the three posterior segments. S transcripts still accumulate in all six segments. In D and E, gene S′ is transformed to gene T by accumulating amino acid substitutions (or other deletions or insertions) that alter its balance of transcriptional activation/repression functions. When accompanied by changes in gene y regulatory sequences that permit the binding of T-specific corepressors, proto-Hox T protein is capable of repressing gene y in the three posterior segments. Other downstream genes (x) could still be activated by proto-Hox S (or proto-Hox T) in all six segments. Such subtle changes in downstream gene regulation would sometimes result in subtle tissue-specific morphological differences between the anterior and posterior groups of segments (see the appendages in D).
However, differences in coding sequences controlling activation/repression strengths could subtly or drastically vary the amount of gene expression from one or a few members of a common set of downstream genes (represented by genes x and y in Fig. 5E). In Fig. 5, the difference that evolves in Hox T is depicted as an adoption of a novel repression ability on gene y. This mode of Hox protein evolution would be more likely to result in subtle changes in metameric morphology compatible with survival. Occasionally some of these subtle morphological changes would result in slight advantages in natural selection for certain niches. This model also requires changes (either preexisting or acquired) in downstream gene regulatory sequences that are near Hox binding sites, so that factors that regulate the repression strength of Hox T could switch it into a repressive mode. This model of evolving diverse Hox functions by subtle changes in activation/repression strengths is not meant to discount the importance of evolutionary variation in downstream genes in morphological variation.
How can we examine whether this process has occurred in evolution? In the embryo of the crustacean Artemia, the Antp, Ubx, and abd-A homologs are coexpressed in a trunk region that is composed wholly of appendage-bearing segments (39). In contrast to Drosophila, the Artemia Ubx and Abd-A homologs do not repress Dll transcription and do not repress appendage development. There are a variety of reasons why the Artemia Ubx and Abd-A proteins might be incapable of repressing appendages, but one possibility is that sequence motifs within the proteins that would allow them to repress the appendage enhancer of Dll are missing. This possibility may be testable by placing the Artemia versions of Ubx and Abd-A proteins in the context of Drosophila early embryonic cells and assaying their effects on appendage development.
Acknowledgments
We thank K. Harding for providing the hsAntp cuticle preparations; R. White for a monoclonal anti-Ubx antibody; S. Cohen for the Dll304-lacZ flies; T. Kaufman for the UAS-Antp flies; the Bloomington stock center for various fly stocks; and Ethan Bier and Brian Florence for comments on the manuscript. This work was supported by National Institute of Child Health and Human Development Grant 28315 (W.M).
References
- 1.Lawrence P A, Morata G. Cell. 1994;78:181–189. doi: 10.1016/0092-8674(94)90289-5. [DOI] [PubMed] [Google Scholar]
- 2.Manak J R, Scott M P. Development (Cambridge, U.K.) 1994. , Suppl., 61–71. [Google Scholar]
- 3.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 4.Ekker S, Jackson D, Kessler D, Sun B, Young K, Beachy P. EMBO J. 1994;13:3551–3560. doi: 10.1002/j.1460-2075.1994.tb06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mann R S, Chan S K. Trends Genet. 1996;12:259–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 6.Biggin M D, McGinnis W. Development (Cambridge, UK) 1997;124:4425–4433. doi: 10.1242/dev.124.22.4425. [DOI] [PubMed] [Google Scholar]
- 7.Pinsonneault J, Florence B, Vaessin H, McGinnis W. EMBO J. 1997;16:2032–2042. doi: 10.1093/emboj/16.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Murre C, McGinnis W. EMBO J. 1999;18:198–211. doi: 10.1093/emboj/18.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaffe L, Ryoo H, Mann R S. Genes Dev. 1997;11:1327–1340. doi: 10.1101/gad.11.10.1327. [DOI] [PubMed] [Google Scholar]
- 10.Zhu A, Kuziora M A. Development (Cambridge, UK) 1996;122:1577–1587. doi: 10.1242/dev.122.5.1577. [DOI] [PubMed] [Google Scholar]
- 11.Vachon G, Cohen B, Pfeifle C, McGuffin M E, Botas J, Cohen S M. Cell. 1992;71:437–450. doi: 10.1016/0092-8674(92)90513-c. [DOI] [PubMed] [Google Scholar]
- 12.Capovilla M, Brandt M, Botas J. Cell. 1994;76:461–475. doi: 10.1016/0092-8674(94)90111-2. [DOI] [PubMed] [Google Scholar]
- 13.Krasnow M A, Saffman E E, Kornfeld K, Hogness D S. Cell. 1989;57:1031–1043. doi: 10.1016/0092-8674(89)90341-3. [DOI] [PubMed] [Google Scholar]
- 14.Saffman E E, Krasnow M A. Proc Natl Acad Sci USA. 1994;91:7420–7424. doi: 10.1073/pnas.91.16.7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 16.Triezenberg S J, LaMarco K L, McKnight S L. Genes Dev. 1988;2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- 17.Castelli-Gair J, Greig S, Micklem G, Akam M. Development (Cambridge, UK) 1994;120:1983–1995. doi: 10.1242/dev.120.7.1983. [DOI] [PubMed] [Google Scholar]
- 18.Wieschaus E, Nüsslein-Volhard C. In: Drosophila: A Practical Approach. Roberts D B, editor. Oxford: IRL; 1986. pp. 199–227. [Google Scholar]
- 19.Sanson B, White P, Vincent J-P. Nature (London) 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Reyes A, Urquia N, Gehring W J, Struhl G, Morata G. Nature (London) 1990;344:78–80. doi: 10.1038/344078a0. [DOI] [PubMed] [Google Scholar]
- 21.Mann R S, Hogness D S. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- 22.Gibson G, Gehring W J. Development (Cambridge, UK) 1988;102:657–675. [Google Scholar]
- 23.Gonzalez-Reyes A, Morata G. Cell. 1990;61:515–522. doi: 10.1016/0092-8674(90)90533-k. [DOI] [PubMed] [Google Scholar]
- 24.Andrew D J, Horner M A, Petitt M G, Smolik S M, Scott M P. EMBO J. 1994;13:1132–1144. doi: 10.1002/j.1460-2075.1994.tb06362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick A, Core N, Kerridge S, Scott M. Development (Cambridge, UK) 1995;121:2799–2812. doi: 10.1242/dev.121.9.2799. [DOI] [PubMed] [Google Scholar]
- 26.Chan S, Mann R S. Genes Dev. 1993;7:796–811. doi: 10.1101/gad.7.5.796. [DOI] [PubMed] [Google Scholar]
- 27.Lin L, McGinnis W. Genes Dev. 1992;6:1071–1081. doi: 10.1101/gad.6.6.1071. [DOI] [PubMed] [Google Scholar]
- 28.Zeng W, Andrew D J, Mathies L D, Horner M A, Scott M P. Development (Cambridge, UK) 1993;118:339–352. doi: 10.1242/dev.118.2.339. [DOI] [PubMed] [Google Scholar]
- 29.Furokubo-Tokunaga K, Flister S, Gehring W J. Proc Natl Acad Sci USA. 1993;90:6360–6364. doi: 10.1073/pnas.90.13.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kornberg T B. J Biol Chem. 1993;268:26813–26816. [PubMed] [Google Scholar]
- 31.Xu L, Lavinsky R M, Dasen J S, Flynn S E, McInerney E M, Mullen T M, Heinzel T, Szeto D, Korzus E, Kurokawa R, et al. Nature (London) 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- 32.Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. Cell. 1991;66:1105–1119. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- 33.Sun B, Hursh D A, Jackson D, Beachy P A. EMBO J. 1995;14:520–535. doi: 10.1002/j.1460-2075.1995.tb07028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capovilla M, Botas J. Development (Cambridge, UK) 1998;125:4949–4957. doi: 10.1242/dev.125.24.4949. [DOI] [PubMed] [Google Scholar]
- 35.Carroll S B. Nature (London) 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 36.Akam M. Phil Trans R Soc London. 1995;349:313–319. doi: 10.1098/rstb.1995.0119. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Noll M. Nature (London) 1994;367:83–87. doi: 10.1038/367083a0. [DOI] [PubMed] [Google Scholar]
- 38.Goldschmidt R. The Material Basis of Evolution. New Haven, CT: Yale Univ. Press; 1940. [Google Scholar]
- 39.Averof M, Akam M. Nature (London) 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]