Abstract
We have purified a complex from Saccharomyces cerevisiae containing the spindle components Ndc80p, Nuf2p, Spc25p, and Spc24p. Temperature-sensitive mutants in NDC80, SPC25, and SPC24 show defects in chromosome segregation. In spc24-1 cells, green fluorescence protein (GFP)-labeled centromeres fail to split during spindle elongation, and in addition some centromeres may detach from the spindle. Chromatin immunoprecipitation assays show an association of all four components of the complex with the yeast centromere. Homologues of Ndc80p, Nuf2p, and Spc24p were found in Schizosaccharomyces pombe and GFP tagging showed they were located at the centromere. A human homologue of Nuf2p was identified in the expressed sequence tag database. Immunofluorescent staining with anti–human Nuf2p and with anti-HEC, the human homologue of Ndc80p, showed that both proteins are at the centromeres of mitotic HeLa cells. Thus the Ndc80p complex contains centromere-associated components conserved between yeasts and vertebrates.
Keywords: Ndc80p, Nuf2p, Spc25p, Spc24p, centromere
Introduction
A large number of components of the mitotic spindle of budding yeast Saccharomyces cerevisiae have been identified by a combination of genetic and biochemical approaches (Skibbens and Hieter 1998; Saunders 1999). An increasing proportion of these components have vertebrate homologues which appear to have a similar function such as kinesins, or in the case of less well-characterized proteins, locate to the equivalent position in the vertebrate spindle, suggesting that they might share some functions. Thus, components of the yeast Tub4p complex (Schiebel 2000) are homologous to all the components of the Drosophila small γTuRC complex (Oegema et al. 1999), and to a subset of the components of the large γTuRC complex in vertebrates (Wiese and Zheng 1999). All of these complexes are located at the spindle pole. Similarly Cdc31p, homologous to centrins (Schiebel and Bornens 1995), and calmodulin (Geiser et al. 1993; Li et al. 1999), are located at the spindle poles of both yeast and vertebrate cells. The yeast kinetochore components Cse4p (Stoler et al. 1995; Meluh et al. 1998) and Mif2p (Brown 1995; Meluh and Koshland 1995, Meluh and Koshland 1997) are homologous to the vertebrate kinetochore components CENP-A (Sullivan et al. 1994) and CENP-C (Saitoh et al. 1992). Also vertebrate homologues of the yeast spindle checkpoint genes Mad1p (Chen et al. 1998), Mad2p (Chen et al. 1996; Li and Benezra 1996), and Bub1p (Taylor and McKeon 1997; Taylor et al. 1998) localize to unattached vertebrate kinetochores. Very recently the Xenopus inner centromere protein INCENP which binds the Aurora-related kinase AIRK2 (Adams et al. 2000) was shown to be homologous to yeast Sli15p which binds the yeast Aurora-like kinase Ilp1p (Kim et al. 1999). These results suggest that there might be a basic conservation of mechanism in eukaryote mitosis, with budding yeast cells undertaking a simplified version of mitosis compared with vertebrates.
We have taken a biochemical approach towards identification of components of the budding yeast spindle. We prepared highly enriched spindle poles and used matrix-assisted laser desorption/ionization (MALDI) mass spectrometry to identify components (Wigge et al. 1998). This approach identified a substantial number of novel spindle pole components, which were located by immunoEM to different parts of the spindle pole body (SPB) and the spindle (Wigge et al. 1998). In this paper we further define the relationship between some of these components and show that three of them, Ndc80p, Spc25p, and Spc24p (Rout and Kilmartin 1990; Wigge et al. 1998), together with the previously described protein Nuf2p (Osborne et al. 1994) are present in a complex associated with the yeast centromere, and that they have a function in chromosome segregation. Three of the components, Ndc80p, Nuf2p, and Spc24p, have Schizosaccharomyces pombe homologues, and two, Ndc80p and Nuf2p, have human homologues. All of these homologues localize to the centromere.
Materials and Methods
Saccharomyces cerevisiae Strains
All S. cerevisiae strains were prepared in Nasmyth's (IMP) K699 background or the isogenic diploid K842; ndc10-1 (Goh and Kilmartin 1993) was backcrossed to K699 six times and nuf2-61 (Osborne et al. 1994) backcrossed three times. All vectors used were the pRS series (Sikorski and Hieter 1989). Yeast strains containing COOH-terminal protein A (prA) fusions (Aitchison et al. 1995) with Ndc80p, Spc25p, Spc24p, Spc34p (Wigge et al. 1998), Spc110p (Kilmartin et al. 1993), and Nuf2p (Osborne et al. 1994) were prepared by recombinant PCR (Wach et al. 1997; Wigge et al. 1998).
SPC24 and SPC25 were cloned by PCR and temperature-sensitive (ts) mutants were prepared by error-prone PCR between −150 and 852 for SPC24 and −156 and 841 for SPC25 (all bases are numbered from the A of the presumed initiator ATG). The PCR fragments were transformed into the covered knockout strains (Wigge et al. 1998) together with the gapped plasmids cut between SnaBI (−75) and EspI (690) for SPC24 and MscI (2) and a BamHI (841) site in the polylinker for SPC25. Plasmids were recovered from ts strains and sequenced to confirm the presence of amino acid changes. The plasmids were then amplified from −300 to 1270 for spc24-1 and −230 to 890 for spc25-1 using 80-mer oligos to give the maximum overlap and the PCR fragments were transformed into the covered knockout strains to give the ts mutants spc24-1 and spc25-1 integrated at the wild-type (wt) locus (Tyers et al. 1993). The region between −306 and 1349 in spc24-1 and −472 and 894 in spc25-1 was amplified and sequenced, and the following changes were found for spc24-1: R19G, I24S, I68M, K72R, L77S, I127T, N196Y, and Y206C; and for spc25-1: L25P. In addition, spc24-1 had the change T651C in the 3′ noncoding sequence 10 bases from the stop codon. Removal of this change had no effect on the ts phenotype. All of the changes found were present in the original plasmids recovered from the ts screen.
Purification of the Ndc80p Complex
One liter of cells harvested at 4 × 107 cells/ml was spheroplasted (Grandi et al. 1993) and lysed with a Polytron in 30 ml of 7.5% glycerol, 50 mM Tris-Cl, pH 7.8, 0.1 M NaCl, 5 mM EGTA, 1 mM EDTA, 1% Triton, and 1:150 solution Q. Solution Q contains 174 mg PMSF, 157 mg benzamidine, 10 mg pepstatin, 10 mg leupeptin, 10 mg aprotinin, 10 mg antipain, 10 mg chymostatin (resuspended) in 10 ml of absolute ethanol, and was subsequently added to all buffers at 1:1,000. After pelleting (Grandi et al. 1993), the lysate was supplemented with NaCl (5.6 ml 1 M NaCl/32 ml lysate), applied to a 0.2-ml column of IgG-Sepharose (Amersham Pharmacia Biotech) and washed. Before elution in fractions with acid (Grandi et al. 1993), the top half of the column was removed and eluted separately; fractions were concentrated by freeze drying.
Mass Spectrometry
Proteins in SDS gel bands were identified by mass spectrometry after increasing the loading 10-fold so that the silver-stained bands in Fig. 1 were just visible by Coomassie staining. Gel bands were digested with trypsin (Wigge et al. 1998) and the tryptic peptide masses were determined by MALDI mass spectrometry in a PerSeptive Biosystems Voyager-DE STR mass spectrometer using external standards or matrix peaks and trypsin peptides as internal standards. The National Center for Biotechnology Information nonredundant database of about half a million proteins was searched using MS-fit (available at http://prospector.ucsf.edu) set at 50 parts per million, 0–300 kD. All of the proteins identified in Fig. 1 were the top match with MOWSE (Pappin et al. 1993) scores (P factor 0.4) of between 3 × 105 and 4 × 1017 apart from the Spc25p–prA track where Spc24p and prA had scores of 9 × 104 and 2 × 103. A second search allowing methionine oxidation, protein NH2-terminal acetylation, and two missed tryptic cleavages was then carried out to match further peptides. For the identifications in this paper the first number in brackets is the number of tryptic peptides identified followed by the percentage of sequence covered; these were (going from left to right across the gel in Fig. 1 for each individual protein) Ndc80p: 19, 38% (vector: 9, 43%); 23, 39% (second track); 30, 46% (third track); 28, 47% (fourth track); Nuf2p: 20, 45%; 13, 34% (vector: 6, 36%); 14, 37%; 16, 42%; Spc24p: 13, 58%; 9, 55%; 13, 66% (vector: 10, 46%); 13, 69%; Spc25p: 7, 33%; 11, 59%; 12, 60%; 10, 49% (vector: 8, 37%); and prA: 8, 35%.
Figure 1.
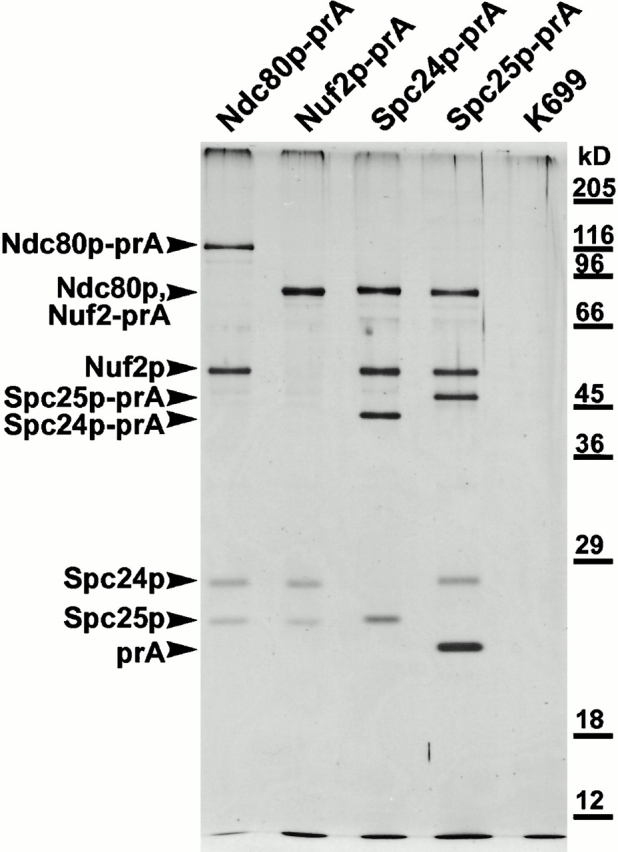
Silver-stained gel of the purified Ndc80p complex, prepared by tagging each of the individual components with prA and affinity purification on an IgG-Sepharose column. Components were identified by MALDI mass spectrometry (see Materials and Methods). Note that in the second lane, Ndc80p and Nuf2p-prA comigrate. The lane marked K699 shows an untagged strain.
Schizosaccharomyces pombe Strains and Cytology
S. pombe strains were constructed by recombinant PCR: transformation with a PCR fragment followed by homologous recombination (Bahler et al. 1998), in either a wt strain IH365 (Bridge et al. 1998) or JPJ415 (Bernard et al. 1998) which has HA-tagged Bub1. Strains were checked by Southern blotting and backcrossing to show 2:2 segregation of the kanamycin and other markers. In addition, because the 3′ divergently transcribed open reading frames adjacent to both SpNuf2 and SpSpc24 are separated by <30 bp of noncoding sequence, the genomic regions of the tagged strains corresponding to the 3′ oligo were amplified and sequenced to select transformants, ∼80% of those examined, which did not contain deletions introduced by the oligos.
For observations of green fluorescence protein (GFP) in live S. pombe, cells were grown in EMM2 (Moreno et al. 1991) and observed on 2% agarose/EMM2 pads with a video camera (RTEA/CCD-1800-Y; Princeton Instruments). Exposures were initially 0.2 s and then 1 s as cells passed through anaphase A. For immunofluorescence (Hagan and Hyams 1988), we initially did a time course of fixation (3, 9, and 30 min) using the sorbitol–paraformaldehyde procedure (Ekwall et al. 1996). This was because both we (Rout and Kilmartin 1990) and others (Osborne et al. 1994) had experienced accessibility problems in staining Ndc80p and Nuf2p in S. cerevisiae. At 3 min of fixation all the antigens gave bright staining, but cytoplasmic microtubules were not well preserved. At 30 min both GFP fluorescence and anti-GFP immunofluorescence were invisible by eye and too faint to record with the video camera. We used 9 min fixation because, although antigen staining was less bright than at 3 min, a reasonable intensity of the high resolution direct GFP image remained and cytoplasmic microtubules were well preserved.
Cloning and Antibodies to Mouse HEC and Human Nuf2R
Mouse HEC had been identified by us from the EST database as a potential homologue of Ndc80p (Wigge et al. 1998) before the appearance of the human HEC paper (Chen et al. 1997). It is 84% identical to human HEC and both have 643 amino acids. The mouse HEC sequence was derived from mouse ESTs. Image 1295296 (Genbank/EMBC/DDBJ accession no. AA896240; GenBank identifiers are in brackets) has amino acids 26–643 and Image 605203 (AA152570) has amino acids 41–643. Amino acids 1–25 were deduced from Image 1194045 (AA726711) which could not be recovered. A full length clone of human HEC was obtained from Image 196139 (R92253). Human Nuf2R (465 amino acids) was assembled from Image 1626830 (AI018298) which had amino acids 1–257 together with 131 bases of 5′ noncoding sequence, Image 1476616 which had amino acids 29-255, and EST 33951 (AA330249) which had amino acids 203–465 and 289 bases of 3′ noncoding sequence ending in a poly A tail. A polymorphism (L229S) was found among the different ESTs. A full length clone was assembled from EST 33951 and Image 1626830 whose sequence was extended with oligos to include the unique Hind III site at 788. Recently an identical protein, apart from K113S or E (this difference is either at the end or the beginning of an intron) and D170E, was found in 13 exons within ∼30 kb of DNA in three contigs of a 175-kb fragment of human genomic DNA from chromosome 1 (AC034278). Mouse Nuf2R (464 amino acids) was sequenced from Image 2352862 (AW3188543) which contains a full length insert. These sequence data are available from GenBank/EMBL/DDBJ under accession nos. AF326730 (mouse HEC), AF326731 (human Nuf2R), and AF326732 (mouse Nuf2R).
Antibodies were prepared in rabbits against glutathione S-transferase (GST) fusions of mouse HEC (residues 235–409) and hNuf2R (residues 22–215). Sera were absorbed first with GST-Sepharose then affinity purified with the GST fusion coupled to Sepharose. Affinity-purified anti–mouse HEC reacted with a band of 77 kD in HeLa cell extracts similar to the results reported earlier (Chen et al. 1997).
Other Methods
ImmunoEM, immunofluorescence, EM, and flow cytometry were done as described previously (Wigge et al. 1998; Adams and Kilmartin 1999). Chromatin immunoprecipitation (ChIP) assays were carried out by PCR as described in Meluh and Koshland 1997 with the same relative template dilutions, and the same regions were amplified for CEN3 and an AT-rich region near PGK1. The regions amplified for 1 kb on either side of CEN3 were 112956–113196 and 115112–115349.
Results
A Complex Containing Ndc80p, Nuf2p, Spc25p, and Spc24p
Highly enriched spindle poles from budding yeast contain several components that colocalize with the spindle microtubules close to the nuclear side of the SPB. These components were identified either by a monoclonal screen (Rout and Kilmartin 1990) or by MALDI mass spectrometric analysis of the enriched spindle poles (Wigge et al. 1998). A particularly interesting set were components such as Ndc80p, Spc25p, and Spc24p which by immunoEM appear to associate with a subset of nuclear microtubules (Rout and Kilmartin 1990; Wigge et al. 1998). This was in contrast to components such as Spc34p and Spc19p which apparently associated with all of the nuclear microtubules (Wigge et al. 1998). The similarities in the immunoEM staining pattern shown by Ndc80p, Spc25p, and Spc24p suggested they might associate. We tested for this using prA tagging, which has been successfully used to identify subcomplexes associated with the nuclear pore (Grandi et al. 1993) and the SPB (Knop and Schiebel 1997). When Ndc80-prA was purified from yeast cell lysates, three other bands were present (Fig. 1). These were identified by MALDI mass spectrometry as Nuf2p (Osborne et al. 1994), Spc25p, and Spc24p (Wigge et al. 1998). This suggests that all four proteins are associated in a complex. To confirm the existence of the complex, we tagged Nuf2p, Spc25p, and Spc24p in turn with prA and purified them. In each case, the other three components of the complex were also purified (Fig. 1). We call this complex the Ndc80p complex.
ImmunoEM of Nuf2p-GFP
Ndc80p, Spc25p, and Spc24p are all spindle-associated proteins, localizing to a subset of nuclear microtubules (Rout and Kilmartin 1990; Wigge et al. 1998), whereas Nuf2p has been described as associated with the nuclear face of the SPB (Osborne et al. 1994; Kahana et al. 1995). Some potential spindle localization of Nuf2p was observed by immunofluorescence (Osborne et al. 1994), but because of antibody accessibility problems necessitating the use of spheroplasts, the precise relationship between Nuf2p, the SPB, and the nuclear microtubules was difficult to establish by light microscopy. We reinvestigated this relationship by immunoEM (Adams and Kilmartin 1999), and avoided accessibility problems by using GFP as a tag. Two types of staining patterns were observed. In short spindles (0.5–0.8 μm, n = 8), staining was observed all along the spindle (Fig. 2, a and b), whereas in longer spindles (1–2.4 μm, n = 4), staining was observed close to the SPB (Fig. 2, c1–c3). The number of spindles examined was small because the cells tend to break up during processing (Adams and Kilmartin 1999), and in order to measure spindle length (estimated as the distance between the nuclear edges of the two central plaques), it was necessary for the spindle to be in, or very close to, the plane of the section. In addition, all single SPBs (n = 30), some of which may have been produced by breakup of longer spindles, had staining close to the nuclear face of the SPB (as in Fig. 2, c1 and c3). Very similar results were found for GFP-tagged Ndc80p (data not shown), confirming the earlier immunoEM results (Rout and Kilmartin 1990). These results show that all four components of the Ndc80p complex are localized to a subset of spindle microtubules.
Figure 2.

ImmunoEM of Nuf2p-GFP in short spindles (a and b) and a longer spindle in three consecutive serial sections (c1–c3). Bars, 0.1 μm.
Phenotype of spc24-1 and spc25-1
Ts mutants in both NUF2 and NDC80 show defects in mitosis. Nuf2-61 cells arrest in mitosis (Osborne et al. 1994) and ndc80-1 cells segregate SPBs but not chromosomes (Wigge et al. 1998), suggesting a specific defect in chromosome segregation. We examined ts mutants in both SPC25 and SPC24 and found that they had a similar phenotype to ndc80-1. We looked at both asynchronous and synchronized cells because some aspects of the phenotype are easier to interpret in synchronized cells. After an asynchronous block for 4 h at 36°C, about half the cells in both spc24-1 (45%) and spc25-1 (50%) are apparently aploid containing SPBs but little DNA (arrowheads in Fig. 3b and Fig. e). All of the postanaphase spindles examined in both mutants (19% of the cells in spc24-1, 3% in spc25-1) showed segregation of SPBs but not DNA (arrows in Fig. 3, a and d). However, we could not be sure that these were spindles because most had a break in the microtubules in the neck region. The rest of the cells in both mutants were cells containing SPBs with DNA (36% in spc24-1 and 40% in spc25-1) and cells containing short spindles with DNA (1% in spc24-1 and 7% in spc25-1).
Figure 3.
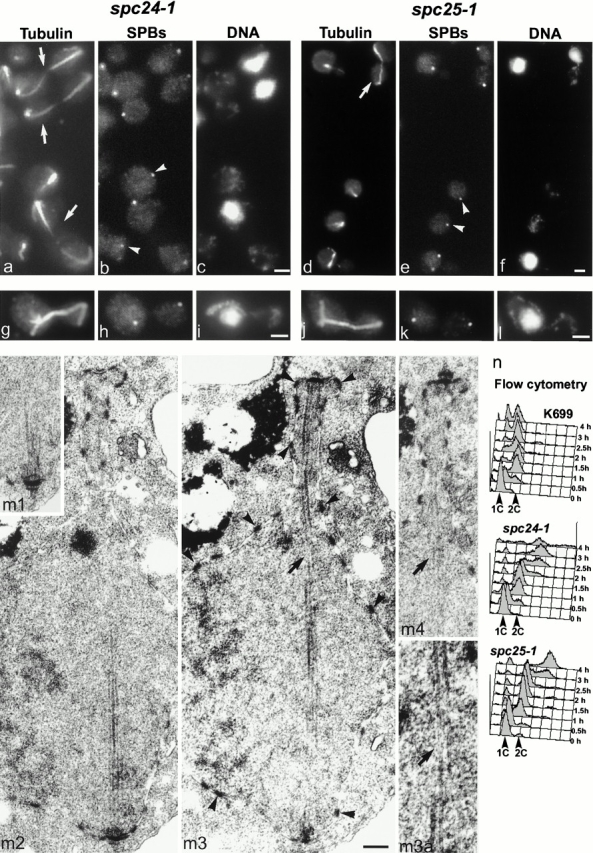
Phenotype of spc24-1 and spc25-1. Immunofluorescent staining of unsynchronized spc24-1 (a–c) and spc25-1 cells (d–f) after 4 h at 36°C with antitubulin (a and d), anti-Tub4p (b and e), and DAPI (c and f). Arrowheads show aploid cells with SPBs, arrows postanaphase cells. spc24-1 (g–i) and spc25-1 (j–l) cells synchronized in G1 with α-factor and released at 36°C for 1.5 h were stained similarly. (m) EM of the same synchronized spc24-1 cells at 1.5 h. Four consecutive serial sections are shown (m1–m4), and overlapping microtubules are present between the two SPBs. The same SPB is shown in m1 and m2. Arrowheads show some of the nuclear pore complexes in m3, and the arrow shows an apparent discontinuity in the spindle in m3 caused by the microtubules being slightly out of the plane of the section. The microtubules can be seen in the equivalent position in the next section (arrow in m4) and even followed if viewed end on at twice the magnification of m3 (m3a). (n) Flow cytometry of K699 (wt), spc24-1, and spc25-1 cells synchronized in G1 and released at 36°C. Bars: (a–l) 2 μm; (m 1–4) 0.2 μm.
We confirmed these phenotypes by EM of complete serial sections of nuclei. Eight large-budded spc24-1 cells were sectioned and seven had an identical phenotype to ndc80-1 (Wigge et al. 1998), that is one SPB associated with the bulk of the nucleus and one in the opposite cell at the end of a narrow isthmus of nuclear envelope. 11 spc25-1 cells were sectioned and 6 were like ndc80-1, while the remaining cells in both mutants had one or two unconnected SPBs associated with the bulk of the nucleus (data not shown). However, it was still very difficult to follow the microtubules between the SPBs in the ndc80-1–like cells and know that these cells had indeed passed through anaphase B, possibly because the central region of the spindle had begun to break down. Therefore, we decided to examine the anaphase spindles at an earlier stage using cells synchronized in G1 with α-factor and released at 36°C. At 1 h after release, immunofluorescence showed cells had formed short spindles; at 1.5 h after release about a third of the cells in each mutant had anaphase spindles, all of which had segregated SPBs but not DNA (Fig. 3, g–l). The immunofluorescence results indicated that the SPBs were connected by nuclear microtubules in two thirds of the spindles (Fig. 3g and Fig. j). This was confirmed by EM in spc24-1 where overlapping microtubules passed between the two SPBs (Fig. 3 m). These results show that SPB separation does indeed result from anaphase B. Again the EM shows that spc24-1 cells show a similar phenotype to ndc80-1, with one SPB in a narrow isthmus of nuclear envelope (Fig. 3, m3), suggesting it is associated with little DNA.
Both mutants fail to arrest the cell cycle. Cells have two to three buds after 4 h at 36°C, and DNA replication as measured by flow cytometry starts at about the normal time but then continues, producing apparently tetraploid cells (Fig. 3 n). The aploid cells seen in immunofluorescence of the asynchronous cells (see above) were not detected by flow cytometry probably because the buds of the multiply budded cells are partly detached during processing for immunofluorescence. The most likely interpretation of these phenotypes is that there is a defect in chromosome attachment, either direct or indirect, to one of the SPBs and the mitotic spindle checkpoint does not fully operate in the mutants. Cells replicate DNA normally, enter anaphase, and segregate one SPB without DNA attached and the other with DNA, giving one aploid cell and one diploid cell. A further round of DNA replication and anaphase may occur giving more aploid cells and a tetraploid cell.
We also examined the behavior of GFP-labeled centromeres and SPBs (He et al. 2000; Tanaka et al. 2000) in live synchronized spc24-1 cells. At 23°C and 1.5 h after release from the α-factor block centromeres had split (Fig. 4 a). They then oscillated normally back and forth between the two Spc42p spots (He et al. 2000; Tanaka et al. 2000) before finally dissociating to the poles during anaphase. At 36°C, however, centromeres did not split during mitosis (Fig. 4 b, 1–3 and c 1–2), as was also observed in ndc10-1 (Tanaka et al. 2000). There were relative movements of the unsplit centromeres and the poles (Fig. 4b 1–2), but it was difficult to tell whether this was microtubule based or whether it reflected the constant movement of the spindle, possibly mediated in part by the cytoplasmic microtubules relative to a stationary unconnected centromere. There also appeared to be lateral displacements of the centromeres from the spindle axis, suggesting a detachment of centromeres from the spindle. This was quantified in cells with all three dots in focus, which would actually bias against detecting lateral displacement. At 1.25–1.75 h the average lateral displacement (measured as distance r in He et al. 2000) was 0.3 ± 0.2 μm (n = 67), which is within the range for wild-type cells (He et al. 2000). However, at 2–2.75 h the lateral displacement was 0.6 ± 0.35 μm (n = 63). These cells were examined by immunofluorescence with antitubulin and anti-GFP to check whether microtubules were connected to individual centromeres that showed a large lateral displacement. For antitubulin we used the mAb YOL1/34 which can detect individual microtubules by immunofluorescence in 3T3 cells (Kilmartin et al. 1982); no connecting microtubules were detected (data not shown). The nuclear microtubules remained in the tight bundles seen by immunofluorescence. These results might suggest detachment of the centromeres from the spindle in spc24-1 cells. However, the detachment is only seen later in the block and thus may be indirect, and also microtubules connecting such centromeres to the pole could be particularly unstable during processing for immunofluorescence and difficult to detect.
Figure 4.
Live cell imaging of spc24-1 cells containing GFP-Spc42p, an SPB component, and GFP-CEN5. CEN5 was labeled by integration of tet operators (which bind GFP-tet repressors) 1.4 kb from CEN5 (Tanaka et al. 2000). Cells were synchronized in G1 with α-factor and released at 23°C for 1.5 h (a) or at 36°C (b1–b3, c1–c2) for the times indicated; arrowheads show the SPBs which are brighter than the centromeres (He et al. 2000; Tanaka et al. 2000). The same cell is imaged in b1–b3 and another cell in c1–c2. Bar, 2 μm.
In conclusion, these data together with earlier results (Osborne et al. 1994; Wigge et al. 1998) show that ts mutants in all four components of the Ndc80p complex show defects in chromosome segregation, and in spc24-1 cells, centromeres fail to split during mitosis and in some cells may detach from the spindle.
Genetic Interactions
We next looked for in vivo evidence for the existence of the Ndc80p complex. Two-hybrid interactions between Ndc80p (also called Tid3 or Yil144w) and Spc25p (Yer018c) and Spc24p (Ymr117c) have been reported (Cho et al. 1998) but numerous other interactions with proteins of apparently diverse biological function have been detected (Fromont-Racine et al. 1997; Cho et al. 1998; Uetz et al. 2000). We looked for genetic interactions between the four components of the complex since ts mutants are now available for all of them. We tested both for suppression by 2μ overexpression and for synthetic lethal effects. We found partial suppression between SPC24 and SPC25, thus SPC24 on a 2μ plasmid suppresses spc25-1 at 34°C and SPC25 on a 2μ plasmid partly suppresses spc24-1 at 34°C. Synthetic lethality at 23°C was found for all six of the double mutant combinations (Table ). The decrease in spore viability was entirely explained by the lethality of the double ts mutants. In addition when the double ts was rescued by one of the wt genes on a CEN-URA plasmid, the cells were unable to grow on fluoroorotic acid plates which selects against the plasmid (data not shown). These suppression and synthetic lethal data support the close association of Ndc80p, Nuf2p, Spc25p, and Spc24p.
Table 1.
Synthetic Lethality between Components of the Ndc80p Complex
| Cross | 4-spore tetrads | 3-spore tetrads | 2-spore tetrads | 1-spore tetrads | Total tetrads | Predicted doubles | Actual doubles | Percent viable spores |
|---|---|---|---|---|---|---|---|---|
| ndc80-1 × nuf2-61 | 3 | 24 | 8 | 0 | 35 | 40 | 0 | 71 |
| nuf2-61 × spc25-1 | 3 | 26 | 7 | 0 | 36 | 40 | 0 | 72 |
| ndc80-1 × spc25-1 | 7 | 19 | 9 | 0 | 35 | 37 | 0 | 74 |
| ndc80-1 × spc24-1 | 4 | 21 | 15 | 0 | 40 | 51 | 0 | 68 |
| spc24-1 × spc25-1 | 8 | 19 | 11 | 0 | 38 | 41 | 0 | 73 |
| nuf2-61 × spc24-1 | 1 | 28 | 10 | 0 | 39 | 48 | 0 | 79 |
Spores were grown at 23°C. In all the 4-spore tetrads, each spore was ts. In the 3-spore tetrads, there was always one wt spore and two ts spores. All 2-spore tetrads were wt. The predicted doubles were calculated from the actual segregation of the markers.
We also looked for genetic interactions with genes encoding some of the centromere-associated components in S. cerevisiae. This is because of the defect in chromosome segregation and because a conditional synthetic lethal effect was detected between ndc80-1 and a deletion of CTF19, which encodes a centromere component (Hyland et al. 1999). Since this deletion of CTF19 shows synthetic lethality (Hyland et al. 1999) with mutants in genes encoding all four components of the centromere–binding factor 3 (CBF3) complex which directly binds CEN DNA (Lechner and Carbon 1991), we checked for similar synthetic lethal effects with the four ts mutants in the Ndc80p complex. We looked for interactions with ts mutants in three of the CBF3 components: ndc10-1 (Goh and Kilmartin 1993), ndc10-2 (Kopski and Huffaker 1997), ctf13-30 (Doheny et al. 1993), and cep3-1 (Strunnikov et al. 1995). No interactions were detected with ndc10-2 (Fig. 5), ctf13-30, or cep3-1 (data not shown). However, a synthetic growth defect was detected between ndc10-1 and ts mutants in all four components of the Ndc80p complex. This varied between the different ts mutants, being strongest with ndc80-1, intermediate with nuf2-61 (Fig. 5) and spc25-1, and weakest with spc24-1 (data not shown). These results suggest an association between the Ndc80p complex and the yeast centromere.
Figure 5.
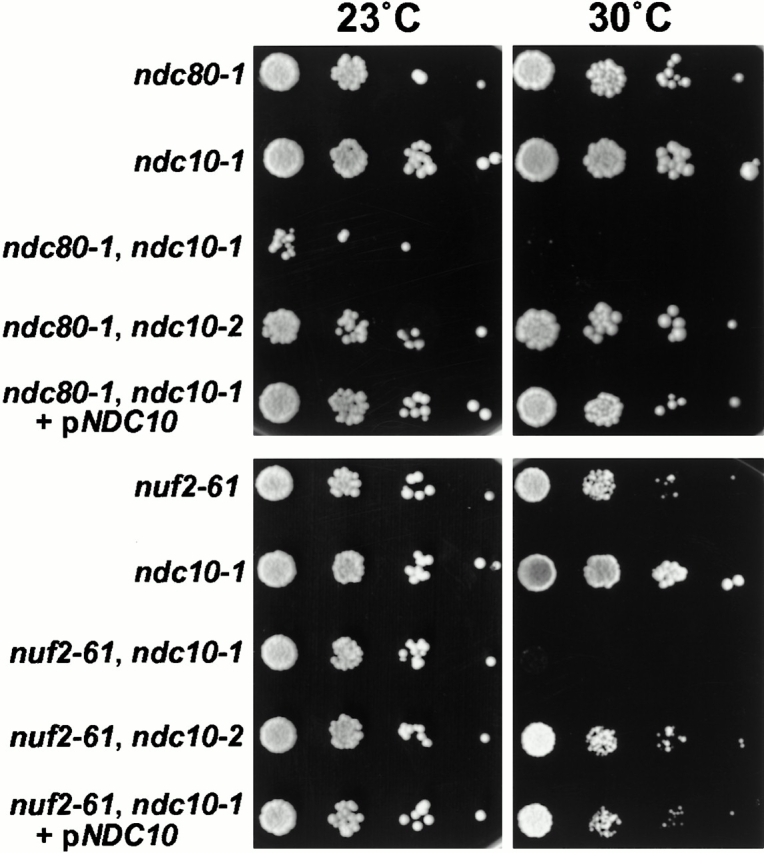
Synthetic growth defect in the ndc80-1, ndc10-1 and nuf2-61, ndc10-1 double mutants when cells were spotted on plates with increasing dilutions at 30°C. This effect was not observed when the ndc10-2 allele was used or the double mutants were transformed with a plasmid containing NDC10.
ChIP Assay
We tested for the presence of the Ndc80p complex at the yeast centromere using the ChIP assay (Meluh and Koshland 1997). Here the cells are cross-linked with formaldehyde then sonicated to shear DNA. The tagged protein is precipitated and any associated centromere DNA detected by PCR. We used the prA-tagged components of the Ndc80p complex because the protein precipitation is particularly clean (Fig. 1). Centromere DNA coprecipitated with all four components of the Ndc80p complex (Fig. 6), while sequences 1 kb to either side or an AT-rich region were not detected. Centromere DNA was not detected with two control proteins (Fig. 6), prA-tagged Spc110p, a component of the SPB (Kilmartin et al. 1993), and prA-tagged Spc34p, a spindle component associated with all of the spindle microtubules (Wigge et al. 1998).
Figure 6.
ChIP assay of strains containing prA-tagged Ndc80p complex components together with controls, prA-tagged Spc110p and Spc34p, and wt cells (K699). Regions of DNA amplified were across CEN3, across 1 kb to either side of CEN3, and across an AT-rich region (see Materials and Methods).
Homologues of Nuf2p and Spc24p
The data presented so far in this paper suggest that components of the Ndc80p complex function at the budding yeast centromere. We wished to confirm this by localization of the components during mitosis. This is a difficult task in budding yeast because of the large number of chromosomes (16) and the finding that association of sister centromeres occurs only on very short spindles (Goshima and Yanagida 2000; He et al. 2000; Tanaka et al. 2000). This makes it very difficult to resolve the 32 individual centromeres during mitosis by immunofluorescence or by GFP tagging of centromere proteins. The immunoEM localizations of Nuf2p (Fig. 2) and other components of the Ndc80p complex (Rout and Kilmartin 1990; Wigge et al. 1998) are consistent with centromere association, but there are other interpretations. For example, the localization could also represent microtubule-associated proteins that are associated only with short spindles. To distinguish between these different interpretations, any localization of centromere proteins during mitosis should be quantized and largely reflect the karyotype. This has been accomplished in budding yeast by analysis of pachytene chromosome spreads where up to 16 kinetochores have been imaged (Hayashi et al. 1998; Klein et al. 1999; Zeng et al. 1999). However, these chromosomes appear before meiosis I and thus do not have microtubules attached (Goetsch and Byers 1982). Therefore, they may lack centromere components dependent on microtubule attachment. To test whether components of the Ndc80p complex localize to the centromere during mitosis, we have turned instead to cytologically more favorable systems and exploited the S. pombe and mammalian genome projects to identify homologues. The advantages of S. pombe are that the sequence of its genome is almost complete, it has only three chromosomes, and recently developed recombinant PCR methods make it easier to tag S. pombe proteins with GFP (Bahler et al. 1998). We also searched within the mammalian genomes because centromere components conserved between yeast and mammals are particularly interesting and the cytology of the mammalian centromere is well characterized.
Ndc80p has both an S. pombe and a human homologue (Wigge et al. 1998; Zheng et al. 1999), and we also found S. pombe homologues of both Nuf2p and Spc24p (Fig. 7, a and d). Nuf2p and Spc24p are both coiled-coil proteins, and their S. pombe homologues have a similar pattern of coiled-coil domains (Fig. 7c and Fig. e) and in particular homology outside the coiled-coil region (Fig. 7, a and d). We call these S. pombe homologues SpNdc80, SpNuf2, and SpSpc24. We also found a human homologue of Nuf2p. We restricted the BLAST (Altschul et al. 1990) search to the Homo sapiens EST database, increased the E value to 100, and used the most conserved part of the NH2 terminus between S. cerevisiae and S. pombe (residues 79–130 in S. pombe) to avoid matches with other coiled-coil proteins. The top match (Image 1626830) had a low score (E value 13) but did encode some of the residues conserved between the two yeasts. The EST was sequenced and used to match more ESTs (refer to Materials and Methods) to give a sequence encoding a protein (Fig. 7 a) from chromosome I which we call hNuf2R (human Nuf2p-related). A mouse homologue was also identified (Fig. 7 a) which was 73% identical to hNuf2R and one amino acid shorter. When hNuf2R was used to search the nonredundant sequence database using FastA (Pearson and Lipman 1988), the top three matches (Fig. 7 a) were S. pombe Nuf2 (E value 2 × 10−9), S. cerevisiae Nuf2p (1.7 × 10−6), and a Caenorhabditis elegans protein R12B2.4 (7 × 10−6). All of the proteins were similar in length and had a coiled-coil region in the COOH-terminal half (Fig. 7 c), and in addition the S. pombe, C. elegans, mouse and human proteins had a break in the coiled-coil at about residue 250 associated with a SPEKLK motif (Fig. 7 b).
Figure 7.
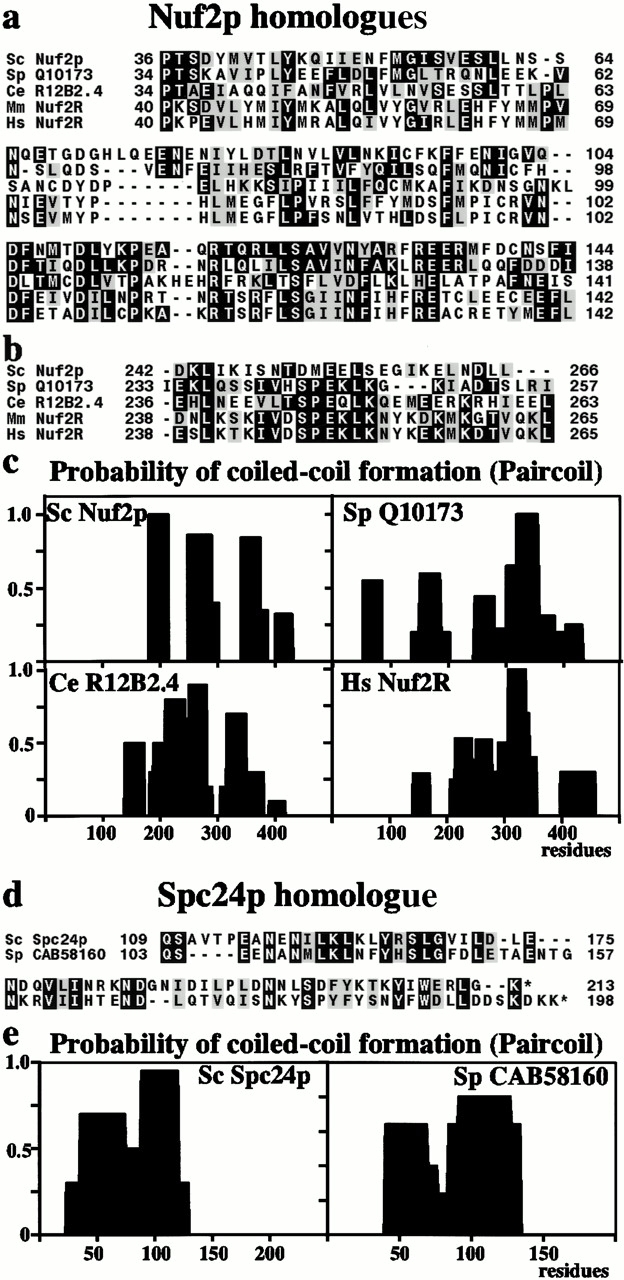
(a) Homologues of S. cerevisiae (Sc) Nuf2p in S. pombe (Sp), C. elegans (Ce), mice (Mm), and humans (Hs), showing homology in the mainly noncoiled coil NH2-terminal region. (b) In four of the homologues there is a break in the coiled coil at around residue 250 associated with an SPEKLK motif. (c) There is a broadly similar distribution of coiled coil domains in the COOH-terminal region of all four homologues. (d) Homologues of Spc24p in S. cerevisiae and S. pombe showing homology in the COOH-terminal region and a similar coiled coil distribution (e).
Localization of Homologues of Components of the Ndc80p Complex in S. pombe
We tagged each of the three potential S. pombe homologues with GFP together with a known S. pombe SPB component Cut12 (Bridge et al. 1998) as a control. All the cells containing Cut12-GFP showed only one or two dots of staining (not shown) as was previously found (Bridge et al. 1998). Cells containing SpNdc80-GFP, SpNuf2-GFP, and SpSpc24-GFP also showed mainly one or two dots. However in larger single cells which would be expected to enter mitosis soon, the dots partly separated and moved continuously relative to each other in the center of the cell, eventually forming a procession of five to six dots (Fig. 8 a). Within a minute or slightly longer, these coalesced into two dots at either end of the procession and then moved to the ends of the cell. There were never more than six dots except in a few very large, presumably polyploid, cells. This pattern of staining suggests centromere localization (Funabiki et al. 1993; Ekwall et al. 1995; Saitoh et al. 1997). To confirm this we used immunofluorescence to compare the localization of the GFP in the three strains with an SPB marker, Sad1 (Hagan and Yanagida 1995), tubulin, and a known centromere component, Bub1 (Bernard et al. 1998). Centromeres cluster around the SPB in interphase S. pombe cells (Uzawa and Yanagida 1992) and as expected GFP staining in all three strains colocalized with Sad1 in these cells (Fig. 8 e). In mitotic cells where the centromeres are distributed along the spindle, the GFP staining was also distributed as a series of dots along the spindle (Fig. 8 f), and coincident (Fig. 8 g) with a known centromere marker, Bub1 (Bernard et al. 1998). These results suggest that SpNdc80, SpNuf2, and SpSpc24 are centromere components in S. pombe.
Figure 8.

(a) Live cell imaging of SpNdc80p-GFP (b1 and b2), SpNuf2p-GFP (c1 and c2), and SpSpc24p-GFP (d1 and d2) in S. pombe. Cells about to complete anaphase A (b1, c1, and d1) show between five and six spots which within a few minutes coalesce into two spots during anaphase B (b2, c2, and d2). (e–g) Fluorescence of GFP compared with immunofluorescence with anti-Sad1 (e), antitubulin (f), and anti–HA-tagged Bub1 (g). Bars, 2.5 μm.
A Human Homologue of Nuf2p, hNuf2R, Localizes to Mammalian Centromeres
We prepared antibodies against hNuf2R which in immunoblots of whole HeLa cells (Fig. 9 a) reacted with a band (∼52 kD) close to the expected size for hNuf2R (54.3 kD). No clear immunofluorescent staining pattern was seen in formaldehyde-fixed cells, even for fixation times as short as 1 min. In methanol-acetone–fixed cells, again no clear staining pattern was seen in interphase cells; however, in prophase and metaphase cells dots appeared (Fig. 9b and Fig. d) which had a very similar (Fig. 9c and Fig. f) but not completely identical staining pattern to anti–CENP-B (Earnshaw et al. 1987). Two sets of double-labeled images are shown, one where the CENP-B staining is predominantly dots (Fig. 9 c) and another (Fig. 9 f) where the staining is elongated probably by stretching of the centromere (Shelby et al. 1996). We also stained these cells with antibodies against HEC, the mammalian homologue of Ndc80p (Zheng et al. 1999), and found a very similar staining pattern to anti-hNuf2R (Fig. 9 e). This also coincided in part with anti–CENP-B (Fig. 9 g) and was susceptible to formaldehyde fixation. This susceptibility probably explains why earlier results with anti-HEC (Chen et al. 1997) did not show centromere staining in formaldehyde-fixed cells but did with isolated chromosomes. The extensive cross-linking of cells by formaldehyde can reduce accessibility to buried antigens (Kilmartin et al. 1993), and isolated chromosomes probably have a more open structure. The overall pattern of dots found with both anti-hNuf2R and anti-HEC is similar to the pattern with anti–CENP-B, but when the CENP-B staining was extended, the anti-hNuf2R and HEC staining was associated only with the ends of the extensions (Fig. 9h and Fig. i) and appeared to extend beyond them. These results suggest that the centromeres were stained. By anaphase, the staining in HeLa cells appeared much weaker (data not shown), but we do not know whether this reflects occupancy of the centromere by the antigens or an increase in the accessibility problem. In Indian muntjac cells, which have large centromeres (Brinkley et al. 1984) and thus brighter staining, we saw centromere staining at all stages of mitosis including anaphase (data not shown). In conclusion we have shown that two mammalian homologues of components of the Ndc80p complex, HEC and hNuf2R, are also located at the centromere.
Figure 9.
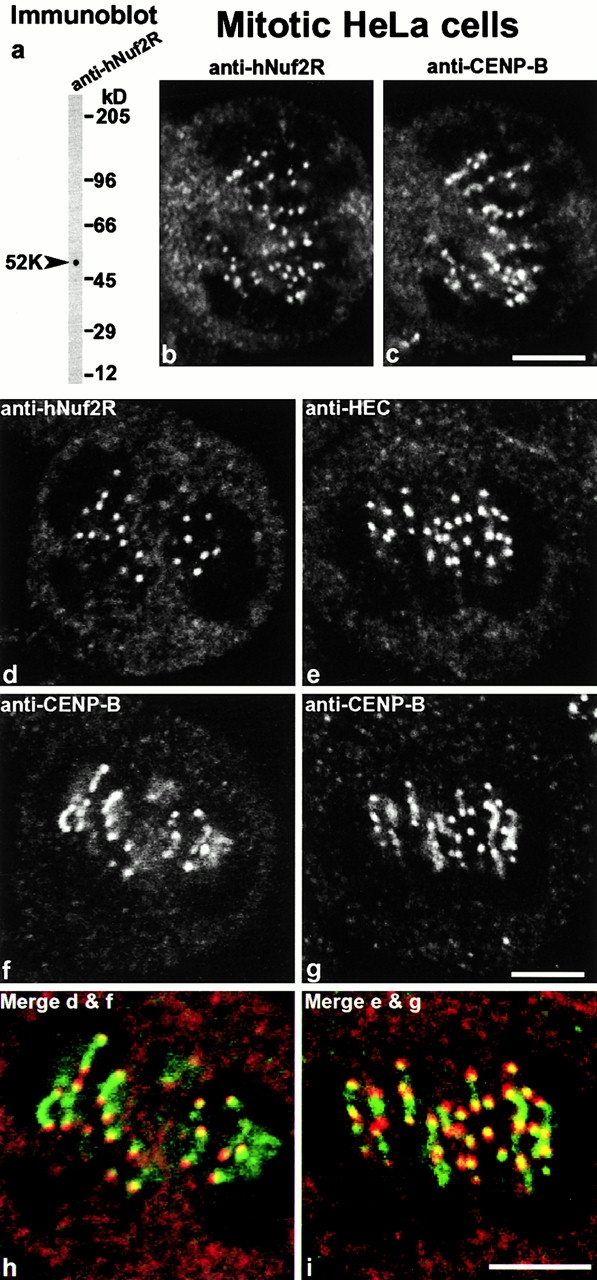
(a) Immunoblot of HeLa cell extract with anti-hNuf2R. Double label immunofluorescent staining by confocal microscopy of mitotic HeLa cells with anti-hNuf2R (b and d), anti-HEC (e), and anti–CENP-B (c, f, and g). Bars, 5 μm.
Discussion
In this paper we describe an initial characterization of an essential complex in budding yeast, the Ndc80p complex, and present evidence suggesting that it is associated with the centromere. In addition, we show that three components of the complex have homologues in S. pombe and two have homologues in mammals, and that all of these are localized to the centromere in these two species. As yet, we do not know whether these components are directly associated in these species; however, given their common location and conservation, it seems likely that the Ndc80p complex also exists in these species. The centromere association or localization is also consistent with the phenotype of ts mutants in yeast and depletion of HEC, the human homologue of Ndc80p, in human cells. Thus in yeast there is chromosome loss in nuf2-61 (Osborne et al. 1994) and an almost complete failure of chromosome segregation in ndc80-1 (Wigge et al. 1998), hshec1-113 (Zheng et al. 1999), spc25-1, and spc24-1 (Fig. 3). Microinjection of an anti-HEC mAb into human T24 cells showed a similar phenotype where the normal relationship between the spindle and the centromeres was disrupted (Chen et al. 1997). This phenotype is very similar to the detachment of the GFP-labeled centromeres from the spindle in some of the spc24-1 cells observed here (Fig. 4).
Where in the centromere is the Ndc80p complex most likely to function? We believe that it is in the periphery of the centromere close to the microtubules. All the components of this complex were identified from a highly enriched spindle pole preparation which contains SPBs with nuclear microtubules attached (Rout and Kilmartin 1990; Wigge et al. 1998). Centromeres are clustered around the SPB throughout most of the cell cycle (Guacci et al. 1997; Jin et al. 2000), and thus this spindle pole preparation is likely to contain centromere components. We have identified two other centromere components by mass spectrometry in this preparation (Kilmartin, J.V., and S.Y. Peak-Chew, unpublished): Slk19p (Zeng et al. 1999) in band 2a and Mtw1p (Goshima and Yanagida 2000) in band 5e (Wigge et al. 1998). However core centromere components such as Ndc10p (Doheny et al. 1993; Goh and Kilmartin 1993; Jiang et al. 1993) are apparently absent from this preparation (Kilmartin, J.V., unpublished), probably as a result of the DNaseI digestion step. Both Slk19p and Mtw1p are likely to be peripheral centromere components since they or their homologue have genetic interactions with proteins associated with microtubules: Slk19p with the kinesin Kar3p (Zeng et al. 1999) and Mis12, the S. pombe homologue of Mtw1p, with the microtubule-binding protein Dis1 (Goshima et al. 1999). Thus, the presence of Ndc80p complex components in this preparation suggests that they too may be peripheral centromere components. We have not yet been able to establish the more precise localization of HEC and hNuf2R in mammalian cells by immunoEM because formaldehyde fixation blocks antibody access. However, these are probably also peripheral centromere components close to the kinetochore, because the immunofluorescent staining of the human centromere with both anti-HEC and anti-hNuf2R occurs at the ends of the elongated anti–CENP-B staining (Fig. 9). This staining relationship is very similar to that between anti–CENP-B and antibodies to the kinetochore kinesin CENP-E (Shelby et al. 1996).
Given its probable presence in the periphery of the centromere, does the Ndc80p complex have a function in chromosome attachment to the spindle? We have not been able to reach a clear conclusion on this issue. There is an increase in lateral displacement of the centromeres from the presumed spindle axis at later stages of the block in spc24-1 (Fig. 4), and these displaced centromeres do not appear to be connected to the pole by microtubules. However, as discussed earlier, connecting microtubules might be unstable and thus difficult to detect. A role in chromosome attachment might be more likely if the Ndc80p complex bound microtubules. We have tested the binding of the four yeast components, translated together in an in vitro system, to taxol-stabilized brain microtubules. All of the components could be sedimented with microtubules but only partly and only at high tubulin concentration (Wigge, P.A., and J.V. Kilmartin, unpublished), suggesting low affinity for brain microtubules. If the yeast Ndc80p complex does bind to microtubules, then either further components are required for the high affinity binding to brain microtubules, or the low affinity is due to sequence divergence in the microtubule binding site of the Ndc80p complex. Currently, we are testing the binding to yeast microtubules, but these experiments present difficulties because of the very low amounts of yeast tubulin that can be obtained (Kilmartin 1981) and the absence of a reagent such as taxol to stabilize yeast microtubules.
Our finding of a centromeric association for the Ndc80p complex is in agreement with the interaction of Hec1p/Ndc80p with Smc1p in yeast (Zheng et al. 1999), since Smc1p and other cohesins are concentrated in the centromere (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999). At present, it has not been established whether this interaction is direct or indirect, but it may be indirect since suppression of smc1-2 requires high (GAL-driven) overexpression of Hec1p (Zheng et al. 1999). This is in contrast to our genetic interactions within the Ndc80p complex which were all detectable at endogenously expressed levels (Table ), reflecting the direct interactions within the complex. A further interaction is with the complex containing Ctf19p, Mcm21p, and Okp1p (Ortiz et al. 1999) since there is synthetic lethality at 28°C between ndc80-1 and ctf19Δ1 (Hyland et al. 1999). Ctf19p interacts genetically (Hyland et al. 1999), and by two-hybrid and coimmunoprecipitation analysis (Ortiz et al. 1999), with components of the CBF3 complex which directly binds centromere DNA (Lechner and Carbon 1991). We have not detected any of the components of the Ctf19p or CBF3 complexes by mass spectrometric analysis of our spindle pole preparation, suggesting that these complexes are ordered from the microtubules to centromere DNA as Ndc80p, Ctf19p, and CBF3 complexes. The only direct interaction in this chain is between CBF3 and centromere DNA (Lechner and Carbon 1991). It remains to be established whether there are direct interactions between these complexes or whether and more likely other proteins or complexes mediate these interactions.
The finding that our highly enriched yeast spindle pole preparation (Wigge et al. 1998) contains a subset of centromere components, including the Ndc80p complex, now provides an opportunity, possibly with further prA-based purifications, of establishing the macromolecular relationship between this subset of centromeric components and the microtubules of the spindle. In addition, further versions of the spindle pole preparation modified to retain more centromere components could be analyzed similarly to establish the relationship between the Ndc80p complex and adjacent parts of the centromere.
In conclusion, components of the Ndc80p complex join an increasing list of centromere components conserved between budding yeast and mammals, suggesting that despite its comparative simplicity the budding yeast centromere has many features in common with the mammalian centromere.
Acknowledgments
We are particularly grateful to S.Y. Peak-Chew for the mass spectrometry, R. Overman and A. Kaja for technical assistance, D. Kershaw for cutting the serial thin sections and help with the EM, W. Earnshaw for anti–CENP-B, J.-P. Javersat and T. Tanaka for S. pombe and S. cerevisiae strains, and I. Hagan for S. pombe strains, antibodies, and much advice. Thanks are also due to M.S. Robinson for discussion.
Footnotes
Philip A. Wigge's present address is Plant Biology Laboratory, The Salk Institute for Biological Studies, La Jolla, CA 92037.
Abbreviations used in this paper: CBF, centromere-binding factor; ChIP, chromatin immunoprecipitation; GFP, green fluorescent protein; GST, glutathione S-transferase; hNuf2R, human Nuf2p-related; MALDI, matrix-assisted laser desorption/ionization; prA, protein A; SPB, spindle pole body; ts, temperature-sensitive; wt, wild-type.
References
- Adams I.R., Kilmartin J.V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae . J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R.R., Wheatley S.P., Gouldsworthy A.M., Kandels-Lewis S.E., Carmena M., Smythe C., Gerloff D.L., Earnshaw W.C. INCENP binds the aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. Two novel related yeast nucleoporins Nup170p and Nup155pcomplementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe . Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bernard P., Hardwick K., Javerzat J.P. Fission yeast Bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 1998;143:1775–1787. doi: 10.1083/jcb.143.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y., Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Bridge A.J., Morphew M., Bartlett R., Hagan I.M. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 1998;12:927–942. doi: 10.1101/gad.12.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley B.R., Valdivia M.M., Tousson A., Brenner S.L. Compound kinetochores of the Indian muntjac. Chromosoma. 1984;91:1–11. doi: 10.1007/BF00286479. [DOI] [PubMed] [Google Scholar]
- Brown M.T. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 1995;160:111–116. doi: 10.1016/0378-1119(95)00163-z. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Waters J.C., Salmon E.D., Murray A.W. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko A., Mann M., Murray A.W. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Riley D.J., Chen P.L., Lee W.H. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol. Cell. Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R.J., Fromont-Racine M., Wodicka L., Feierbach B., Stearns T., Legrain P., Lockhart D.J., Davis R.W. Parallel analysis of genetic selections using whole genome oligonucleotide arrays. Proc. Natl. Acad. Sci. USA. 1998;95:3752–3757. doi: 10.1073/pnas.95.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny K.F., Sorger P.K., Hyman A.A., Tugendreich S., Spencer F., Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Sullivan K.F., Machlin P.S., Cooke C.A., Kaiser D.A., Pollard T.D., Rothfield N.F., Cleveland D.W. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K., Javerzat J.P., Lorentz A., Schmidt H., Cranston G., Allshire R. The chromodomain protein Swi6a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- Ekwall K., Nimmo E.R., Javerzat J.P., Borgstrom B., Egel R., Cranston G., Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Rain J.C., Legrain P. Towards a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Funabiki H., Hagan I., Uzawa S., Yanagida M. Cell cycle–dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser J.R., Sundberg H.A., Chang B.H., Muller E.G.D., Davis T.N. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae . Mol. Cell. Biol. 1993;13:7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetsch L., Byers B. Meiotic cytology of Saccharomyces cerevisiae in protoplast lysates. Mol. Gen. Genet. 1982;187:54–60. doi: 10.1007/BF00384383. [DOI] [PubMed] [Google Scholar]
- Goh P.Y., Kilmartin J.V. NDC10a gene involved in chromosome segregation in Saccharomyces cerevisiae . J. Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Goshima G., Saitoh S., Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Doye V., Hurt E.C. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. Centromere position in budding yeastevidence for anaphase A. Mol. Biol. Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I.M., Hyams J.S. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe . J. Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hayashi A., Ogawa H., Kohno K., Gasser S.M., Hiraoka Y. Meiotic behaviours of chromosomes and microtubules in budding yeastrelocalization of centromeres and telomeres during meiotic prophase. Genes Cells. 1998;3:587–601. doi: 10.1046/j.1365-2443.1998.00215.x. [DOI] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P.K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Hyland K.W., Kingsbury J., Koshland D., Hieter P. Ctf19pa novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and the mitotic spindle. J. Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Lechner J., Carbon J. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J. Cell Biol. 1993;121:513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Fuchs J., Loidl J. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 2000;113:1903–1912. doi: 10.1242/jcs.113.11.1903. [DOI] [PubMed] [Google Scholar]
- Kahana J.A., Schnapp B.J., Silver P.A. Kinetics of spindle pole body separation in budding yeast. Proc. Natl. Acad. Sci. USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J.V. Purification of yeast tubulin by self-assembly in vitro. Biochemistry. 1981;20:3629–3633. doi: 10.1021/bi00515a050. [DOI] [PubMed] [Google Scholar]
- Kilmartin J.V., Wright B., Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J.V., Dyos S.L., Kershaw D., Finch J.T. A spacer protein in the Saccharomyces cerevisiae spindle pole body whose transcript is cell cycle-regulated. J. Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Kang J.S., Chan C.S. Sli15 associates with the Ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae . J. Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S.B., Michaelis C., Nairz K., Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Knop M., Schiebel E. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopski K., Huffaker T.C. Suppressors of the ndc10-2 mutationa role for the ubiquitin system in Saccharomyces cerevisiae kinetochore function. Genetics. 1997;147:409–420. doi: 10.1093/genetics/147.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J., Carbon J. A 240kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Li C.J., Heim R., Lu P., Pu Y., Tsien R.Y., Chang D.C. Dynamic redistribution of calmodulin in HeLa cells during cell division as revealed by a GFP-calmodulin fusion protein technique. J. Cell Sci. 1999;112:1567–1577. doi: 10.1242/jcs.112.10.1567. [DOI] [PubMed] [Google Scholar]
- Li Y., Benezra R. Identification of a human mitotic checkpoint genehsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Megee P.C., Mistrot C., Guacci V., Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Meluh P.B., Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Yang P., Glowczewski L., Koshland D., Smith M.M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae . Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe . Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Oegema K., Wiese C., Martin O.C., Milligan R.A., Iwamatsu A., Mitchison T.J., Zheng Y. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J., Stemmann O., Rank S., Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne M.A., Schlenstedt G., Jinks T., Silver P.A. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J. Cell Biol. 1994;125:853–866. doi: 10.1083/jcb.125.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappin D.J.C., Hojrup P., Bleasby A.J. Protein identification by peptide mass fingerprinting. Curr. Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- Pearson W.R., Lipman D.J. Improved tools for biological data comparison. Proc. Natl. Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Kilmartin J.V. Components of the yeast spindle and spindle pole body. J. Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C., Ratrie H.D., Maurer M., Rothfield N.F., Earnshaw W.C. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Saitoh S., Takahashi K., Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Saunders W.S. Action at the ends of microtubules. Curr. Opin. Cell Biol. 1999;11:129–133. doi: 10.1016/s0955-0674(99)80016-7. [DOI] [PubMed] [Google Scholar]
- Schiebel E. Gamma-tubulin complexesbinding to the centrosome, regulation and microtubule nucleation. Curr. Opin. Cell Biol. 2000;12:113–118. doi: 10.1016/s0955-0674(99)00064-2. [DOI] [PubMed] [Google Scholar]
- Schiebel E., Bornens M. In search of a function for centrins. Trends Cell Biol. 1995;5:197–201. doi: 10.1016/s0962-8924(00)88999-0. [DOI] [PubMed] [Google Scholar]
- Shelby R.D., Hahn K.M., Sullivan K.F. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V., Hieter P. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Ann. Rev. Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- Stoler S., Keith K.C., Curnick K.E., Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Strunnikov A.V., Kingsbury J., Koshland D. CEP3 encodes a centromere protein of Saccharomyces cerevisiae . J. Cell Biol. 1995;128:749–760. doi: 10.1083/jcb.128.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K.F., Hechenberger M., Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Cosma M.P., Wirth K., Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Fuchs J., Loidl J., Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha E., McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M., Tokiwa G., Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclinsCln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Uzawa S., Yanagida M. Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci. 1992;101:267–275. doi: 10.1242/jcs.101.2.267. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Alberti-Segui C., Rebischung C., Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae . Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. γ-Tubulin complexes and their interaction with microtubule organizing centers. Curr. Opin. Struct. Biol. 1999;9:250–259. doi: 10.1016/S0959-440X(99)80035-9. [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Jensen O.N., Holmes S., Souès S., Mann M., Kilmartin J.V. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Kahana J.A., Silver P.A., Morphew M.K., McIntosh J.R., Fitch I.T., Carbon J., Saunders W.S. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Chen Y., Lee W.H. Hec1p, an evolutionarily conserved coiled-coil protein, modulates chromosome segregation through interaction with SMC proteins. Mol. Cell. Biol. 1999;19:5417–5428. doi: 10.1128/mcb.19.8.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]




