Abstract
Methylmercury is an environmental toxicant that biomagnifies and causes severe neurological degeneration in animals. It is produced by bacteria in soils and sediments that have been contaminated with mercury. To explore the potential of plants to extract and detoxify this chemical, we engineered a model plant, Arabidopsis thaliana, to express a modified bacterial gene, merBpe, encoding organomercurial lyase (MerB) under control of a plant promoter. MerB catalyzes the protonolysis of the carbon—mercury bond, removing the organic ligand and releasing Hg(II), a less mobile mercury species. Transgenic plants expressing merBpe grew vigorously on a wide range of concentrations of monomethylmercuric chloride and phenylmercuric acetate. Plants lacking the merBpe gene were severely inhibited or died at the same organomercurial concentrations. Six independently isolated transgenic lines produced merBpe mRNA and MerB protein at levels that varied over a 10- to 15-fold range, and even the lowest levels of merBpe expression conferred resistance to organomercurials. Our work suggests that native macrophytes (e.g., trees, shrubs, grasses) engineered to express merBpe may be used to degrade methylmercury at polluted sites and sequester Hg(II) for later removal.
Keywords: environment, heavy metals, Minamata disease, organic mercury, remediation
Mercury is among the most hazardous of the heavy metals (1), primarily because its charged species have great affinity for the thiol group on cysteine residues of proteins and other important biological molecules (2, 3). Early studies demonstrated that mercury species inactivate metabolic enzymes and structural proteins (4, 5). The strong interaction of mercury species with cellular ligands may also account for its tendency to accumulate in organisms. Organomercurials are 1–2 orders of magnitude more toxic in some eukaryotes and are more likely to biomagnify across trophic levels than ionic mercury [Hg(II)] (1, 6, 7). The biophysical behavior of organic mercury is thought to be due to its hydrophobicity and efficient membrane permeability.
Industrial and agricultural activities have released several hundred thousand tons of mercury into the biosphere during the past century (8). The most notable ecological disasters have been caused by the efflux of mercury-contaminated wastes into semicontained fresh- and saltwater basins. Between 1956 and 1968, widespread poisoning at Minamata Bay, Japan brought international attention to the hazards of mercury pollution. During this period, medical researchers discovered that high levels of monomethylmercury (CH3—Hg+) in seafood were responsible for severe neurological degeneration in birds, cats, and humans (9). This mercury species, traced from Minamata Bay sediment to wastewater from a chemical manufacturing plant, is the principal form of mercury that accumulates in fish and biomagnifies in their predators (10, 11).
Methylmercury has also been detected in lakes and estuaries into which only inorganic forms of mercury have been released. Microbes present in the sediment were found capable of processing Hg(II) to CH3—Hg+ (12, 13). Sulfate-reducing bacteria isolated from the aerobic–anaerobic interface of these sediments were later found to be the principal methylators (14, 15). Using Desulfovibrio desulfuricans LS as a model system, methylcobalamin was identified as the intermediate in transferal of a methyl group from CH3-tetrahydrofolate to Hg(II) (16, 17). Although sulfate-reducing bacteria manage to survive in the presence of methylmercury by converting it to less soluble products (18), they do not carry out these reactions efficiently enough to prevent harmful levels of methylmercury from leaching into the surrounding environment.
Mercury-resistant bacteria eliminate organomercurals by producing an enzyme, organomercurial lyase (MerB), that catalyzes the protonolysis of the carbon—mercury bond (19). The products of this reaction are a less toxic inorganic species, Hg(II), and a reduced carbon compound.
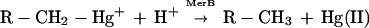 |
These bacteria also synthesize a second enzyme, mercuric ion reductase (MerA), that catalyzes the reduction of the inorganic product, Hg(II), to a volatile and much less reactive elemental form, Hg(0) (20). The enzymes are encoded by the merB and merA genes of the plasmid-borne, broad-spectrum mercury-resistance (mer) operon (21). Both enzymes require a low molecular weight thiol-organic cofactor (e.g., cysteine, 2-mercaptoethanol) for in vitro activity and may require sulfhydryl-bound substrates (19, 20).
Although many governments now require companies to recover mercury from their sludge and liquid wastes, they have been less exacting regarding the cleanup of previously polluted landfills and waterways. This lack of resolve is due partly to the fact that the physical and chemical remediation techniques currently used to extract or immobilize mercury are extremely expensive, environmentally disruptive, and sometimes ineffective. A phytoremediation system, in which plants extract, sequester, and/or detoxify mercury pollutants (22), may be a more attractive solution. Besides being cost-effective, phytoremediation offers a promising alternative because plants naturally dominate most ecosystems, use solar energy, have large reservoirs of reducing power from photosystem I, have extensive root systems capable of extracting a variety of metal ions, and can stabilize and rehabilitate damaged environments (23, 24). In a previous paper (7), we communicated that the expression of a modified bacterial merA9pe gene enables Arabidopsis thaliana plants to grow on toxic levels of Hg(II) by converting this species to elemental mercury. We now report that plants transformed with the bacterial merB gene express organomercurial lyase and grow on concentrations of phenylmercuric acetate or methylmercuric chloride that are lethal to their wild-type progenitors.
MATERIALS AND METHODS
Strains and Plasmids.
The Escherichia coli strain SK1592 (F−, gal−, thi−, sup−, tonA−, hsdR4, endA−, SbcB15) was provided by Sidney Kushner (Univ. of Georgia) as a spontaneous T1 phage-resistant derivative of SK1590 (25). The plasmid pDU202, which was used in metal ion-sensitivity disk assays, contains a narrow-spectrum mer operon (26). This operon contains the merA, merD, merP, merR, and merT genes, but not merB. Accordingly, it confers resistance to Hg(II) but not to organomercurials (21). The pBluescript II SK(−) (pBSSKII) vector was obtained from Stratagene. The binary vector pVST I, designed for Agrobacterium-mediated plant transformations, was constructed by Malik and Wahab (27).
Reconstruction of merB for Plant Expression.
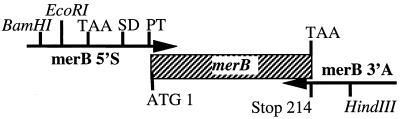
Mercury-resistance plasmids have been isolated from a variety of bacterial species and strains. One well characterized merB gene (GenBank accession no. U77087) is found on the broad-spectrum resistance plasmid R831b (28). This gene was subcloned on a 1.5-kb EcoRI fragment into pBR322 to make pCT12 (29). The 214-codon merB gene was amplified by PCR using long synthetic primers that modified the merB flanking sequences as shown in Fig. 1. The sense primer, merB5′S, consisted of the 57-nt sequence 5′-GCGGTCGGAT CCGAATTCGT CGACTAAGGA GGAGCCACAA TGAAGCTCGC CCCATAT-3′ and contained BamHI, EcoRI, and SacI cloning sites, a TAA stop codon to end the translation of an upstream β-galactosidase fusion protein in E. coli, a GGAGGA bacterial translation signal to assist expression in E. coli, an AGCCACA consensus sequence for plant translation (30), an ATG start codon, and the first 18 nt of the merB coding sequence to prime the forward PCR reaction (7). The antisense primer, merB3′N, had the 43-nt sequence 5′-CGTATCGGAT CCGAATTCAA GCTTATCACG GTGTCCTAGA TGA-3′, with HindIII, EcoRI, and BamHI cloning sites and anticodons to the last seven merB codons to prime the reverse PCR reaction. PCR was carried out for 35 cycles with denaturing, annealing, and extending temperatures/times of 95°C for 1 min, 42°C for 1 min, and 72°C for 1 min. The amplified fragment, merBpe, was cleaved in the flanking BamHI and HindIII sites and ligated into the multilinker of pBSSKII to make pBSmerBpe. The pBSmerBpe construct was electroporated into a strain of SK1592 that had previously been transformed with pDU202. The same E. coli strain was also transformed with an empty pBluescript II SK(−) plasmid to serve as a control in metal ion-sensitivity filter disk assays (Table 1). The merBpe sequence was transferred from pBSmerBpe to the plant vector pVSTI (27) by using the 5′ BamHI site and the 3′ XhoI site in the Bluescript multilinker to create pVSTImerBpe.
Figure 1.
PCR primers and strategy for modification of the merB gene. Two PCR primers were used to add synthetic flanking sequences to the merB gene that have elements necessary for bacterial and plant expression. The sense primer, MerB5′S, contained restriction endonuclease cloning sites, a TAA stop codon, bacterial (SD) and plant (PT) translation signals, an ATG start codon, and the first 18 nt of the merB coding sequence to prime the forward PCR reaction. The antisense primer, merB3′N, contained cloning sites, and 21 nt of anticodon sequence (covering the last seven merB codons) used to prime the reverse PCR reaction.
Table 1.
Filter disk assay for mercury sensitivity in Escherichia coli strain SK1592 with plasmid pDU202-merA+
| Metal salt | Concentration, M | Zone diameter, mm
|
|
|---|---|---|---|
| pBSSKI1-merB− | pmerBpe-merB+ | ||
| HgCl2 | 0.1 | 18 | 20 |
| PMA | 0.1 | 22 | 8 |
| MeHgCl | 0.1 | 26 | 10 |
E. coli strain SK1592 containing the various mer gene and control plasmids were assayed for mercury and organomercurial resistance. Each 6-mm filter diameter disk contained 2 μl of freshly prepared metal salt stock solution. The genotype of each strain and sensitivity (s) or resistance (r) phenotype on different forms of mercury is indicated below the strain and plasmid designations.
Disk Assays.
Phenylmercuric acetate (PMA; Sigma) and the chloride salt of methylmercury (CH3HgCl; Aesar) were prepared in DMSO or ethanol. Because of the extreme toxicity and membrane permeability of these chemicals, dry stocks and stock solutions were handled using protective clothing, eye protection, and 4H or Viton gloves (Fisher). All metal ion-sensitivity filter disk assays were performed in the presence of ampicillin to maintain the pBSSKII plasmid and streptomycin to retain pDU202, as described in Rugh et al. (1996). The data reported are the average results of several replicates, which, in all cases, varied by <1 mm.
Construction of Transgenic Plants.
pVSTImerBpe was electroporated into an LBA4404 Agrobacterium tumefaciens strain (GIBCO/BRL). Transformants were verified by using Southern blotting and grown up in YEP medium (10 g/liter Bacto peptone/10 g/liter yeast extract/5 g/liter NaCl) in the presence of streptomycin and kanamycin to maintain the T-DNA and pVSTI plasmids, respectively. Wild-type A. thaliana (ecotype RLD) were transformed with this A. tumefaciens strain using the vacuum infiltration procedure (31).
Germination/Growth Experiments.
Wild-type (RLD), merA9pe (transgenic control) (7), and merBpe (B4 line) Arabidopsis seeds were sterilized, vernalized at 4°C for at least 24 h, and germinated on 1% Phytagar plates (GIBCO/BRL) made with Murashige and Skoog (4.3 g/liter, GIBCO/BRL) medium containing PMA or CH3HgCl. Seedlings were grown at 22°C with a 16 h light/8 h dark regime and photographed at 3–8 weeks.
Quantitative Northern Blot.
Total RNA was prepared from transgenic and control plants (>15 seedlings per line) (32), resolved by electrophoresis on a 1% agarose/formaldehyde gel, and blotted to a Biotrans nylon membrane (ICN) (33). The membrane was probed with the 0.7-kb BamHI–XhoI merBpe fragment. [32P]dATP was incorporated into the probe by using a random primer method to yield a specific activity of approximately 5 × 108 cpm per μg (34).
Quantitative Western Blot/Isolation of MerB-Specific mAbs.
Crude protein was prepared from merBpe and control plants (15 seedlings per line) in a buffer containing 5 mM EDTA/10 mM MgCl2/10 mM NaCl/25 mM Tris⋅HCl, pH 7.5/1 mM PMSF. Extracts were denatured by adding an equal amount of 2× SDS sample buffer and boiling for 5 min and then were separated with SDS/12%PAGE (35). Resolved protein was electroblotted onto an Immobilon-P polyvinylidene fluoride membrane (Millipore).
mAbs were prepared against MerB following the procedure developed by Kohler and Milstein (36). Spleen cells were isolated from mice that had been immunized with a hexa(His)-tagged version of MerB prepared by Qiandong Zeng (Univ. of Georgia) and A.O.S. The hybridoma cell line 2H8 was found to secrete an IgG antibody that strongly recognized the original His-tagged antigen, organomercurial lyase expressed from pBSmerBpe in E. coli, and a protein expressed by Arabidopsis plants resistant to PMA. This antibody, Mab2H8, was purified over a Protein A column (Bio-Rad) and used for quantitative Western blots. Mab2H8 was reacted with membrane-bound plant protein for 90 min after blocking for 1 h with 5% dry milk/10% goat serum (Sigma) TBS-T buffer.
RESULTS
The merBpe construct used to transform A. thaliana was modified from the original bacterial merB sequence by adding flanking regions containing consensus plant and bacterial translation signals as well as convenient restriction sites to facilitate cloning (Fig. 1). Sequencing identified a single silent transversion at base pair 369 in the 642-bp coding region that probably occurred during the PCR amplification. To demonstrate that this clone made an active MerB enzyme, a metal ion-sensitivity filter disk assay was used to compare the growth of E. coli strains with and without the merBpe construct in the presence of organomercurial compounds. A strain carrying a narrow-spectrum mercury-resistance plasmid, pDU202, which confers resistance to Hg(II) but not organomercurials, was used as the recipient for either pBSmerBpe or an unmodified Bluescript plasmid. Bacteria expressing an active MerA enzyme from pDU202 and MerB enzyme from pBSmerBpe should eliminate organomercurials via a coupled reaction that produces Hg(0). As expected, the SK1592/pDU202/pBSmerBpe strain grew much closer to filter disks with PMA or CH3HgCl than did the control strain (Table 1), indicating that the modified merBpe produces an active organomercurial lyase.
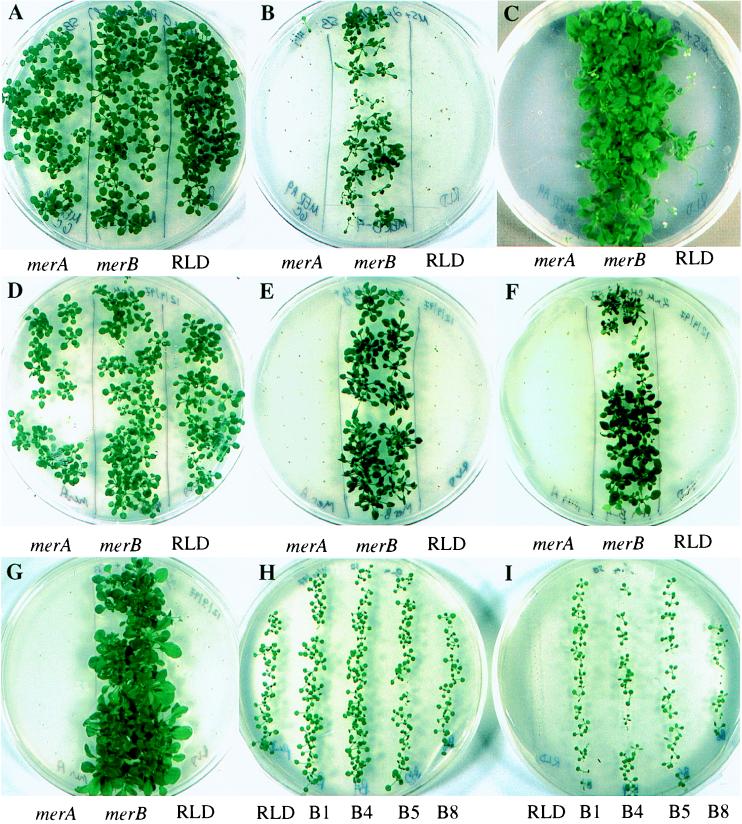
The merBpe sequence was subcloned into the plant transformation vector, pVST I, which provides a constitutive promoter (CaMV 35S), nopaline synthetase (NOS) 3′ termination and polyadenylation signals, and a selectable kanamycin-resistance marker (NPT II). A. thaliana plants were transformed by using vacuum infiltration, and the T1 generation seeds they produced were screened for kanamycin resistance. Seven resistant germinants were selected for further study and used to establish independent merBpe lines. T2 progeny of the B4 line were tested for organomercurial resistance by germinating on medium containing PMA or CH3HgCl. As seen in Fig. 2, the B4 seedlings grew vigorously in the presence of 2 μM PMA (Fig. 2 B and C), producing strong shoots, round, deep-green leaves, and fully branched root systems. By contrast, RLD and merA9pe control seedlings flanking the B4 seedlings either failed to germinate or germinated, bleached white, and died within 2–3 weeks. merBpe Arabidopsis had a clear advantage over the control plants at concentrations between 0.1 and 5 μM PMA or CH3HgCl (0.5 and 2 μM are shown in Fig. 2), flowering and setting seed after 7–8 weeks. However, at 5 μM CH3HgCl (data not shown), the merBpe seedlings and plants were clearly stressed, forming spindly shoots, lanceolate leaves, and relatively few lateral roots.
Figure 2.
Growth of parental and transgenic A. thaliana on organomercurials. (A–C) Growth on PMA. Seeds from the merBpe line B4 (center of each plate) germinated and grew on growth media containing no mercury (A) and 2 μM PMA (B and C). Transgenic merA9pe seeds (Left) and wild-type RLD (Right) seeds used as controls, did not germinate on PMA. Plates in A and B were photographed at 4 weeks. The plate shown in C is the same as in B, photographed 2 weeks later. (D–G) Growth on methylmercury. Seeds from the B4 line (Center) also grew on substrates with no mercury (D), 0.5 μM CH3HgCl (E), and 2 μM CH3HgCl (F and G). Plates were photographed after 5 (D–F) and 8 (G) weeks incubation. Transgenic merA9pe (Left) and RLD (Right) seeds do not grow on these concentrations of CH3HgCl. (H–I) Growth comparisons among independent lines on PMA. Seeds from four independently isolated transgenic lines (B1, B4, B5, B8) representing a range of merBpe expression levels germinated and plants grew on no mercury (H) and 1 μM PMA (I), while the wild-type RLD seeds and plants grew only on the control plate (H). Plates in H and I were photographed at 3 weeks post-germination.
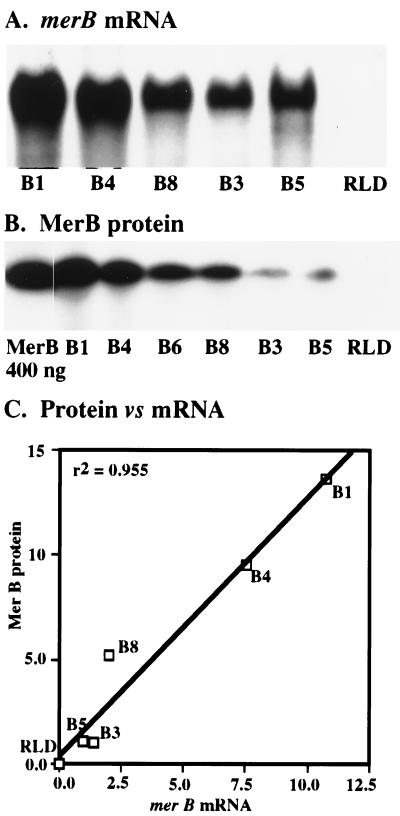
Independent transformants may vary with respect to steady-state RNA and protein levels because of differences in the genomic positioning and copy number of the transgene. Such variation commonly results from A. tumefaciens-mediated transformation (37). Differences in expression levels were confirmed by Northern and Western blots shown in Fig. 3. Total RNA samples from five merBpe lines and the RLD parent line were blotted and hybridized with a 32P-labeled merBpe probe. A strong 700-bp mRNA band was detected in samples from the merBpe lines but not in those from RLD (Fig. 3A). This is the size predicted from the sequence of the merBpe construct. Based on the normalized intensities of these bands, the levels of merBpe mRNA varied over an 11-fold range (Fig. 3C).
Figure 3.
Relative merBpe mRNA and MerB protein levels in independent transgenic plant lines. (A) merBpe mRNA levels. Five plant lines were tested for relative expression of merBpe mRNA by Northern blotting and probing with a 32P-labeled BamHI–XhoI merBpe fragment. Approximately 10–15 μg of total RNA prepared from >30 seedlings (2- to 3-cm plants) was loaded for each sample. merB mRNA was visualized, and the relative band intensities were quantified using a Molecular Dynamics PhosphorImager. To control for variation in loading and transfer, these values were normalized to levels of 18S rRNA (data not shown), determined by reprobing the filter with a oligonucleotide encoding the soybean 18S rRNA gene (42). An 11-fold difference was detected between the highest expressing line, B1, and the lowest expressing line, B5. (B) MerB protein levels. Western blots were performed on total protein extracted from six merBpe plant lines using the MerB specific mAb, Mab2H8 (see Materials and Methods). Fifteen 3- to 4-cm seedlings were harvested for each sample. The protein–antibody complex was visualized on x-ray film using a horseradish peroxidase-based chemoluminescent detection system (Amersham Pharmacia). Relative amounts of MerB were measured by scanning the film with a Molecular Dynamics densitometer, integrating the intensities of the bands over their respective areas, and normalizing to total protein. Relative amounts of total protein were determined by quantifying the band intensities of a parallel gel stained with Coomassie blue (data not shown) on a densitometer. Values recorded for MerB are the averages of three experiments while those for total protein are the averages of two. All data reported for quantitative experiments described here are measurements of band intensities and reflect relative differences in expression between merBpe lines. The B1 sample was shown to have ≈14-fold more steady-state organomercurial lyase than the lowest expressing line, B3. (C) Relative mRNA and protein levels. A linear regression demonstrates that steady-state levels of mRNA and protein are correlated, confirming differences in merBpe gene expression and indicating that gene regulation is straightforward. The relative merBpe mRNA and MerB protein levels in independent transgenic lines (see Materials and Methods) were normalized to the level in the lowest expressing line in each category (B5, mRNA; B3, protein). The relevance of MerB expression levels to organomercurial resistance is discussed in the text.
MerB protein was assayed in crude extracts from six merBpe lines and RLD. Western blots were probed with the Mab 2H8 MerB-specific mAb that was prepared for this study (see Materials and Methods). The organomercurial lyase was detected in all transgenic lines (Fig. 3B) and migrates at 28 kDa relative to standards. MerB protein levels varied over a 14-fold range and correlated linearly with levels of merBpe mRNA (r2 = 0.955; Fig. 3C). Independent repetitions of the Western blot confirmed the rank order of expression levels for the various lines shown in Fig. 3B. The strong correlation between mRNA and protein levels suggests that the transgene is not subject to unusual variations in posttranscriptional regulation.
Given that merBpe confers resistance to organomercurials, we hypothesized that lines expressing more organomercurial lyase should grow faster on various levels of organic mercury (see Discussion). We looked for a relationship between growth rate and expression levels by germinating several merBpe lines on 1 μM PMA (Fig. 2 H and I). However, there were no observable growth differences among the merBpe lines that correlated with expression levels. Similar results were obtained at PMA concentrations between 0.1 and 1 μM. Plants from the B1, B4, and B5 lines all appeared healthy, although they were slightly inhibited compared with unchallenged plants. One other line, B3 (data not shown) gave similar results. Plants from the B8 line with intermediate levels of gene expression grew poorly on PMA, yielding small, slow-growing seedlings with spindly, light green leaves. All lines tested, including a wild-type control, thrived on mercury-free agar plates, sprouting strong, deep green shoots and leaves.
DISCUSSION
A bacterial organomercurial lyase gene, merBpe, reconstructed with an upstream sequence typical of plant genes, enables transgenic plants to germinate and grow well on 0.1–2 μM CH3HgCl, concentrations that retard or kill control plants. A simple interpretation of this result is that merBpe plants efficiently protonolyze organic mercury, producing a more tolerable mercury species, Hg(II). Consistent with this interpretation, we found that 0.5 μM CH3HgCl or PMA was required to kill Arabidopsis seeds and seedlings as efficiently as 50 μM HgCl2 (7). The greater toxicity of organic mercury relative to Hg(II) to eukaryotes is usually attributed to its hydrophobicity and to its tendency to coordinate with a counterion to form a neutral species (6, 38). These characteristics enable organic mercury to pass through cell membranes and reach higher intracellular concentrations than externally applied Hg(II). Furthermore, eukaryotes possess membrane-rich organelles that may form localized sinks for organomecurial deposition (23).
Considering that Hg(II) is generated by MerB within the cytoplasm in the presence of vulnerable cellular proteins, it remains paradoxical that merB plants tolerate more organic mercury than wild-type plants. We suspect that in the intracellular environment, the lipid solubility of organic mercury gives it access to membrane-bound eukaryotic organelles such as mitochondria and chloroplasts, where it may poison essential oxidative and photosynthetic electron transport chains more easily than Hg(II) (7, 23). It is also plausible that cytoplasmic chelators, including phytochelatins and metallothioneins, bind and sequester Hg(II) in preference to the organomercurials. MerB expression would confer an advantage by making mercury available to these protective compounds. A challenging problem to future research on this phytoremediation system would be to quantify the cellular levels of CH3Hg+ and Hg(II) and identify the various compounds to which they are bound.
The Michaelis constants describing the reaction velocities of MerB acting on a variety of organomercurial substrates, including methylmercury and PMA, are very high (Km = 0.5–3.3 mM) (19), suggesting a relatively weak affinity for substrate. However, organomercurial resistance operates in environments and under experimental conditions with low substrate concentrations at which only a small fraction of the enzyme may be bound to organic mercury. Our motivation for generating and maintaining a variety of merBpe lines was to determine whether, at these low substrate concentrations, organomercurial resistance depends on the amount of MerB present in the cytoplasm. Although we detected up to 14-fold differences in MerB expression, we did not observe the direct relationship between enzyme levels and faster growth on medium containing organic mercury that would have been predicted by a simple steady-state model. Specifically, the lines with the most MerB protein were not more resistant to various organic mercury concentrations than those with lower but measurable MerB levels. Apparently, the abundance of MerB enzyme is not the limiting factor in organomercurial degradation. A reasonable explanation for this incongruity is that the reaction is limited by one of several kinetic parameters and is not functioning at steady state (39). For example, in a living cell, the kinetics of the MerB-catalyzed reaction could be constrained by the rate of diffusion of organomercurial substrates from cellular membrane systems to sites of catalysis or by the rate of diffusion of the product, Hg(II), away from the enzyme.
The remediation of mercury-polluted sites has been slow because the chemical engineering technologies currently used to remove mercurials are expensive and disruptive. Excavation and roasting of soil is considered the “best demonstrated treatment technology” (40), but it is impractical for very large sites. Vitrification and concrete capping, which aim to stabilize mercury, render the site uninhabitable for plants, insects, and other organisms. An ideal treatment would degrade methylmercury, sequester other mercury forms, and help restore biological productivity. Our proposed strategy is to introduce engineered native plants that readily assimilate methylmercury and, by virtue of the merBpe gene, convert it into Hg(II). The plants would act as a biological filter, absorbing methylmercury from the sediment and water, and MerB would catalyze the protonolysis of the methylmercury as it enters plant cells. Hg(II) is 50 times less toxic to plants (this study and ref. 7) and lower animals (6, 38) and much less prone to biomagnification than methylmercury (1). Once formed, Hg(II) should accumulate in plant tissues. Plants could be harvested before Hg(II) reached toxic levels, and the mercury could be extracted or disposed of in a greatly reduced volume (41).
The phytoremediation system we have proposed requires the eventual removal of plant material that has accumulated high levels of inorganic mercury. Some benefits to this approach are the ability to immobilize mercury without disrupting the environment and the opportunity to recycle mercury by extracting it from the harvested plants. The size and topography of many contaminated sites, however, may make harvesting economically unfeasible. We have previously reported on merA9pe transgenic plants that electrochemically reduce Hg(II) to Hg(0), a volatile form that escapes from plant tissues (7). Judging by the phenotypes of independently transformed merA9pe and merBpe plants, plants made to express both genes simultaneously will be capable of converting methylmercury into Hg(0), thereby volatilizing it from their local environment. These plants would require very little maintenance and could theoretically be engineered to volatilize mercury at a rate that is safe and consistent with governmental regulations.
Acknowledgments
We thank Margaret Wallace for her technical assistance in cloning the merBpe gene. M. K. Kandasamy, Qiandong Zeng, Elizabeth Lytle, and Lorraine Aron helped with production of MerB-specific mAbs. John Wampler, Gay Gragson, and Bruce Haines made constructive suggestions on the manuscript. This work has been supported by grants from the U.S. National Science Foundation, the Department of Energy Environmental Management Sciences Program, and the National Institute of Health (GM28211).
ABBREVIATION
- PMA
phenylmercuric acetate
References
- 1.Keating, M. H., Mahaffey, K. R., Schoeny, R., Rice, G. E., Bullock, O. R., Ambrose, R. B., Swartout, J. & Nichols, J. W. (1997) (U.S. Environmental Protection Agency, Washington, DC) 2, 1–2:9.
- 2.Liu Y, Cotgreave I, Atzori L, Grafstrom R C. Chem Biol Interact. 1992;85:69–78. doi: 10.1016/0009-2797(92)90053-n. [DOI] [PubMed] [Google Scholar]
- 3.Carty A J, Malone S F. In: The Chemistry of Mercury in Biological Systems. Nriagu J O, editor. Amsterdam: Elsevier Biomedical; 1979. pp. 433–479. [Google Scholar]
- 4.Boyer P D. J Am Chem Soc. 1954;76:4331–4337. [Google Scholar]
- 5.Falchuk K H, Goldwater L J, Vallee B L. In: The Biochemistry and Toxicology of Mercury. McAuliffe C A, editor. New York: Macmillan; 1977. pp. 261–284. [Google Scholar]
- 6.Mason R P, Reinfelder J R, Morel F M M. Environ Sci Technol. 1996;30:1835–1845. [Google Scholar]
- 7.Rugh C L, Wilde D, Stack N M, Thompson D M, Summers A O, Meagher R B. Proc Natl Acad Sci USA. 1996;93:3182–3187. doi: 10.1073/pnas.93.8.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andren A W, Nriagu J O. In: The Global Cycle of Mercury. Nriagu J O, editor. Amsterdam: Elsevier Biomedical; 1979. pp. 1–21. [Google Scholar]
- 9.Minamata Disease Research Group. Minamata Disease. Kumamoto, Japan: Medical School of Kumamoto University; 1968. [Google Scholar]
- 10.Lockhart W L, Uthe J F, Kenney A R, Mehrle P M. J Fish Res Board Can. 1972;29:1519–1523. [Google Scholar]
- 11.Kamps L R, Carr R, Miller H. Bull Environ Contam Toxicol. 1972;8:273–279. doi: 10.1007/BF01684556. [DOI] [PubMed] [Google Scholar]
- 12.Jensen S, Jernelov A. Nordforsk Biocidinformation. 1967;10:4. [Google Scholar]
- 13.Jensen S, Jernelov A. Nature (London) 1969;223:753–754. doi: 10.1038/223753a0. [DOI] [PubMed] [Google Scholar]
- 14.Compeau G C, Bartha R. Appl Environ Microbiol. 1985;50:498–502. doi: 10.1128/aem.50.2.498-502.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilmour C C, Henry E A, Mitchell R. Environ Sci Technol. 1992;26:2281–2287. [Google Scholar]
- 16.Choi S C, Chase T, Jr, Bartha R. Appl Environ Microbiol. 1994a;60:4072–4077. doi: 10.1128/aem.60.11.4072-4077.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi S-C, Chase T, Jr, Bartha R. Appl Environ Microbiol. 1994b;60:1342–1346. doi: 10.1128/aem.60.4.1342-1346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldi F, Pepi M, Filippelli M. Appl Environ Microbiol. 1993;59:2479–2485. doi: 10.1128/aem.59.8.2479-2485.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Begley T P, Walts A E, Walsh C T. Biochemistry. 1986;25:7192–7200. doi: 10.1021/bi00370a064. [DOI] [PubMed] [Google Scholar]
- 20.Fox B, Walsh C T. J Biol Chem. 1982;257:2498–2503. [PubMed] [Google Scholar]
- 21.Summers A O. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- 22.Raskin I. Proc Natl Acad Sci USA. 1996;93:3164–3166. doi: 10.1073/pnas.93.8.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meagher R B, Rugh C L, Kandasamy M K, Gragson G, Wang N J. In: Fourth International Conference on the Biogeochemistry of Trace Elements. Ishndar I K, Hardy S E, Chang A C, Pierzynski G M, editors. Berkeley, CA: Ann Arbor Press; 1998. [Google Scholar]
- 24.Salt D E, Blaylock M, Kumar N P B A, Dushenkov V, Ensley B D, Chet I, Raskin I. Biotechnology. 1995;13:468–474. doi: 10.1038/nbt0595-468. [DOI] [PubMed] [Google Scholar]
- 25.Kushner S R. In: An Improved Method for Transformation of Escherichia coli with ColE1 Derived Plasmids. Boyer H W, Nicosia S, editors. New York: Elsevier/North–Holland; 1978. pp. 17–23. [Google Scholar]
- 26.Hamlett N V, Landale E C, Davis B H, Summers A O. J Bacteriol. 1992;174:6377–6385. doi: 10.1128/jb.174.20.6377-6385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik V S, Wahab S Z. J Plant Biochem Biotechnol. 1993;2:69–70. [Google Scholar]
- 28.Ogawa H I, Tolle C L, Summers A O. Gene. 1984;32:311–320. doi: 10.1016/0378-1119(84)90006-4. [DOI] [PubMed] [Google Scholar]
- 29.Tolle C T. M.S. thesis. Athens, GA: Univ. of Georgia; 1985. [Google Scholar]
- 30.Heidecker G, Menning J. Annu Rev Plant Physiol. 1986;37:451–462. [Google Scholar]
- 31.Bent A, Kunkel B N, Dahlbeck D, Brown K L, Schimdt R, Giraudat J, Leung J, Staskawicz B J. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 32.Ausubel F, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1988. [Google Scholar]
- 33.Hightower R C, Meagher R B. EMBO J. 1985;4:1–8. doi: 10.1002/j.1460-2075.1985.tb02309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 35.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Kohler G, Milstein C. Nature (London) 1975;256:495. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 37.Hooykaas P J J, Schilperoort R A. Plant Mol Biol. 1992;19:15–38. doi: 10.1007/BF00015604. [DOI] [PubMed] [Google Scholar]
- 38.Clarkson T W. In: The Toxicology of Mercury and Its Compounds. Watras C J, Huckabee J W, editors. Ann Arbor, MI: Lewis; 1994. pp. 631–642. [Google Scholar]
- 39.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. Boston: Butterworth; 1979. [Google Scholar]
- 40.Smith L A, Means J L, Chen A, Alleman B, Chapman C C, Tixier J S, Brauning S E, Gavaskar A R, Royer M D. Remedial Options for Contaminated Sites. Boca Raton, FL: CRC; 1995. [Google Scholar]
- 41.Cunningham S D, Ow D W. Plant Physiol. 1996;110:715–719. doi: 10.1104/pp.110.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckenrode V K, Arnold J, Meagher R B. J Mol Evol. 1985;21:259–269. doi: 10.1007/BF02102358. [DOI] [PubMed] [Google Scholar]