Abstract
We have characterized the Drosophila mitotic checkpoint control protein Bub1 and obtained mutations in the bub1 gene. Drosophila Bub1 localizes strongly to the centromere/kinetochore of mitotic and meiotic chromosomes that have not yet reached the metaphase plate. Animals homozygous for P-element–induced, near-null mutations of bub1 die during late larval/pupal stages due to severe mitotic abnormalities indicative of a bypass of checkpoint function. These abnormalities include accelerated exit from metaphase and chromosome missegregation and fragmentation. Chromosome fragmentation possibly leads to the significantly elevated levels of apoptosis seen in mutants.
We have also investigated the relationship between Bub1 and other kinetochore components. We show that Bub1 kinase activity is not required for phosphorylation of 3F3/2 epitopes at prophase/prometaphase, but is needed for 3F3/2 dephosphorylation at metaphase. Neither 3F3/2 dephosphorylation nor loss of Bub1 from the kinetochore is a prerequisite for anaphase entry. Bub1's localization to the kinetochore does not depend on the products of the genes zw10, rod, polo, or fizzy, indicating that the kinetochore is constructed from several independent subassemblies.
Keywords: checkpoint, kinetochore, chromosome missegregation, apoptosis
The spindle checkpoint is a surveillance mechanism that monitors the attachment of spindle microtubules to the kinetochores, thereby ensuring that the onset of anaphase is dependent on the correct completion of metaphase (reviewed by Elledge 1996; Rudner and Murray 1996; Wells 1996; Nicklas 1997; Wolf and Jackson 1998). To date, genetic studies in yeast have identified seven genes which encode components of the spindle checkpoint: MAD1, MAD2, MAD3, BUB1, BUB2, BUB3, and MPS1. Mutations disrupting these genes bypass the operation of the spindle checkpoint, leading to the initiation of anaphase in the presence of microtubule-depolymerizing drugs (Hoyt et al. 1991; Li and Murray 1991). Homologues of MAD1, MAD2, BUB1, and BUB3 from multicellular eukaryotes have been identified and establish spindle checkpoints in these organisms as well. For example, immunodepletion of Mad1 and Mad2 from Xenopus extracts inactivates the spindle checkpoint (Chen et al. 1996, Chen et al. 1998).
These metazoan spindle checkpoint proteins have been shown to localize most strongly to kinetochores unattached to the spindle apparatus (Chen et al. 1996, Chen et al. 1998; Li and Benezra 1996; Taylor and McKeon 1997; Taylor et al. 1998; Chan et al. 1998; Yu et al. 1999). The differential association of these molecules with attached versus unattached kinetochores is consistent with several observations implying that unattached kinetochores emit an inhibitor that delays anaphase onset (reviewed by Nicklas 1997; Rieder and Salmon 1998). Recent evidence indicates that the checkpoint operates by inhibiting the ability of the anaphase-promoting complex (APC)1 to ubiquitinate substrates whose degradation is a prerequisite for sister chromatid separation and other aspects of the exit from mitosis (Elledge 1998; Hwang et al. 1998; Kim et al. 1998).
Although the function of the Bub and Mad proteins has been well established under conditions in which microtubule depolymerizing reagents or mutations in spindle components were employed, the importance of these proteins for normal cell division is less clear. In Saccharomyces cerevisiae, strains carrying null mutations in most BUB or MAD genes grow somewhat more slowly, accompanied by a weak increase in chromosome missegregation (Hoyt et al. 1991; Li and Murray 1991; Farr and Hoyt 1998). Similarly, Schizosaccharomyces pombe knockouts of bub1 are viable and show modest effects on the fidelity of chromosome segregation during mitosis (Bernard et al. 1998). In higher eukaryotes, tissue culture cells overexpressing presumed dominant negative versions of Bub1 exit from mitosis more quickly than usual (Taylor and McKeon 1997). Microinjection of antibody against Mad2 into tissue culture cells similarly induces premature entry into anaphase (Gorbsky et al. 1998). Interestingly, mutations in a human Bub1–related kinase have been detected in colorectal cancer cell lines showing chromosomal instability (Cahill et al. 1998). These mutations behave neither as null mutations or hypomorphs, but instead generate a version of this protein that also acts in a dominant negative fashion. These results do not provide a clearcut framework for understanding how the checkpoint influences normal cell cycle progression, as we do not yet know the consequences of the absence of any checkpoint component in a developing multicellular eukaryote. To address these issues in more detail, we have begun to characterize the operation of the spindle checkpoint in Drosophila melanogaster, as we believe the combination of genetic and cytological approaches that can be employed in this organism will provide new insights into the mechanisms governing anaphase onset.
In this paper, we present a detailed phenotypic characterization of Drosophila bub1 mutants, the first mutational analysis of any component of the spindle checkpoint in any multicellular organism. We show that loss of function mutations affecting Drosophila bub1 cause severe mitotic abnormalities consistent with accelerated transit through metaphase. In addition, in partial contrast to previous findings indicating that loss of Bub1 function leads to the escape of cells from an apoptotic fate (Taylor and McKeon 1997), we find that mutations in bub1 generate a massive apoptotic response. We have further employed an anti-Drosophila Bub1 antibody to show that the cell cycle distribution of Bub1, including its association with unattached kinetochores, has been conserved between Drosophila and humans. The genetic and immunological reagents we have generated additionally allowed us to examine several other issues, such as the role of Bub1 during meiosis, and the relationship between Bub1 kinase and other kinetochore components. These include 3F3/2 phosphoepitopes and the ZW10 protein, both of which have been suggested to be intimately involved in signaling the metaphase/anaphase transition (Williams et al. 1992; Campbell and Gorbsky 1995). Our results considered together clarify the importance of the spindle checkpoint to normal cell division in higher eukaryotes.
Materials and Methods
Identification of Drosophila Bub1 cDNAs and Drosophila bub1 Mutants
The ESTs LD06986 and LD18419 were identified in the Berkeley Drosophila Genome Project (BDGP) EST database when searched with the amino acid sequence of mouse Bub1 (Taylor and McKeon 1997), and cDNAs containing these ESTs were ordered from Genome Systems Inc. The longest of these cDNA inserts (that containing EST LD06986) was sequenced to completion (Cornell University Sequencing Facility, Ithaca, NY), and was found to contain the entire amino acid coding sequence of Drosophila Bub1.
The lethal P-element insertions l(2)K06109 and l(2)K03113 (gifts of Dr. Todd Laverty, University of California, Berkeley, CA) were identified by searching the BDGP database of sequences adjacent to P-element insertion sites with the sequence of EST06986. We have independently determined the DNA sequence of Drosophila genomic DNA flanking the P-element insertion sites in the l(2)K06109 and l(2)K03113 lines, and our results are in accord with the sequences in the BDGP database.
We determined that the cytological location of the bub1 gene is polytene chromosome interval 42A1-3 by hybridizing a probe made from the LD06986 cDNA clone to larval salivary gland polytene chromosomes as described in Williams et al. 1992. This result confirms BDGP's localization of the P-elements causing the lethal mutations l(2)K06109 and l(2)K03113 to the same polytene chromosome bands. In further support of this position for the bub1 gene, we determined that two deletions uncovering this region of the genome, Df(2R)nap1 (breakpoints 41D2-41E1, 42B1-42B3; obtained from the Drosophila stock center, Bloomington, IN) and Df(2R)nap2 (breakpoints 41F4-41F9, 43A1; the gift of Dr. John Roote, Department of Genetics, University of Cambridge, Cambridge, UK), failed to complement l(2)K06109 or l(2)K03113 for any of the phenotypes we have studied.
To verify that the bub1 mutant phenotype was caused by the l(2)K06109 and l(2)K03113 P-element insertions, we remobilized the P-elements in these lines by introducing P[ry + Δ 2-3] (99B), a source of P-element transposase (Robertson et al. 1988), and selecting for loss of the white + eye color in the next generation (Gatti and Goldberg 1991). Out of 43 white− excision stocks generated from l(2)K06109, 21 showed complete rescue of the lethality and associated mitotic and apoptotic defects of the original bub1 mutants. For l(2)K03113, 19 out of the 37 white− stocks obtained similarly behaved as precise excisions.
Generation of Anti-Bub1 Antibody
To obtain large amounts of Bub1-specific epitopes, a 1219 bp BamH1/Kpn1 fragment from LD06986 was first subcloned into the expression vector pWR590 (Guo et al. 1984). This created an in-frame fusion in which sequences encoding amino acids 54–460 of Drosophila Bub1 were joined to DNA specifying the first 590 amino acids of β-galactosidase (LacZ). When transfected into E. coli XL-1 Blue cells (Stratagene), this construct led to the production of an ∼120-kD LacZ/Bub1 fusion protein. To confirm that this fusion protein indeed contained a LacZ moiety, Western blots of bacterial extracts containing the fusion construct were probed with an antibody against an unrelated LacZ fusion protein (polyclonal anti-LacZ/ZW10; see Williams et al. 1992 for details). Purification of the LacZ/Bub1 fusion protein was carried out by excising the appropriate band from SDS-polyacrylamide gels as described by Basu et al. 1998a.
Antibodies against the LacZ/Bub1 fusion protein were generated in chickens as described in Basu et al. 1998a. The crude IgY fractions containing anti-Bub1 IgY were further purified by affinity chromatography on a column composed of CnBr Sepharose (Sigma Chemical Co.) covalently coupled according to the manufacturer's instructions to the same LacZ/Bub1 fusion protein used as the immunogen. Immunoblotting was performed as previously described (Basu et al. 1998b), except that the secondary antibody employed was peroxidase-conjugated Affinipure donkey anti–chicken IgY (Jackson ImmunoResearch Labs) at a dilution of 1:10,000. Detection of antibody signals were performed with the ECL system (Amersham) according to the manufacturer's instructions. This affinity purified anti-Drosophila Bub1 antibody was used for all Western blotting and immunofluorescence experiments described in this report.
Cytology and Immunofluorescence Observations
To identify embryos homozygous for bub1 mutations, embryos from stocks of genotype bub1/CyO, engrailed-lacZ were collected, dechorionated, and fixed in formaldehyde after the procedure of Karr and Alberts 1986. Embryos were then stained with a rabbit antibody that recognizes β-galactosidase (polyclonal anti–β-galactosidase-ZW10), followed by rhodamine-labeled anti–rabbit IgG, and Hoechst staining to visualize DNA as described by Williams et al. 1992. Embryos homozygous for bub1 mutations were those that did not show the engrailed stripe pattern dictated by the engrailed-lacZ construct on the CyO balancer chromosome. Our cytological analysis focused on metaphase chromosome alignment and anaphase chromosome segregation within the mitotic domains of post-cellularization embryos (Foe 1989); no obvious defects were seen.
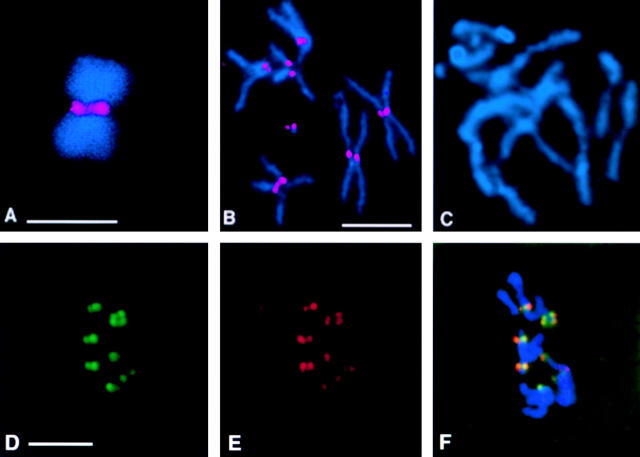
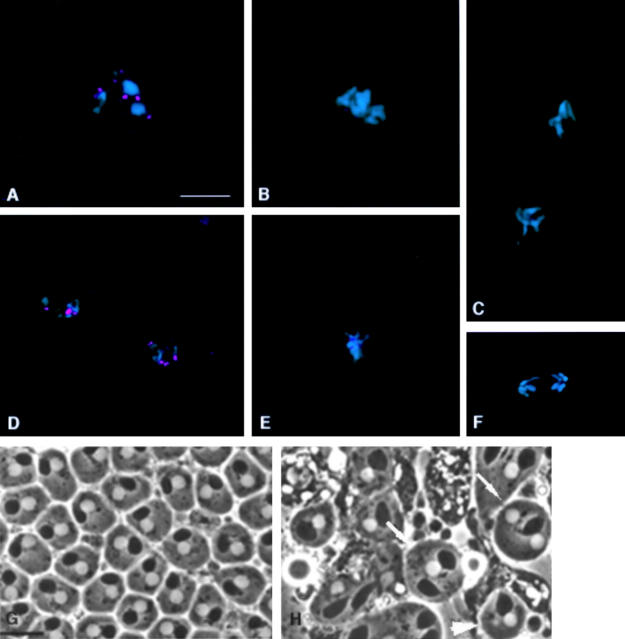
To identify third instar larvae homozygous for bub1 mutations, chromosomes bearing both the l(2)K06109 and l(2)K03113 P-element insertions were rebalanced over T(2;3)SM6a-TM6B, a translocation between the second chromosome balancer SM6a and the third chromosome balancer TM6B synthesized in the laboratory of A. Garcia-Bellido (Universidad Autonoma de Madrid, Madrid, Spain). T(2;3)SM6a-TM6B includes the dominant larval/pupal marker Tubby, so the desired mutant animals were chosen on the basis of their non-Tubby phenotype. Orcein stained preparations of neuroblasts from the brains of third instar larvae were obtained as described by Gatti and Goldberg 1991. Cytological preparation and immunolocalization studies of these larval neuroblasts were performed as described by Williams and Goldberg 1994. Living testes from third instar larvae were observed by the techniques of Cenci et al. 1994, while fixed larval testes were analyzed by immunofluorescence as described by Williams et al. 1996. Preparation and immunolocalization analysis of Drosophila S2 tissue culture cells, and of metaphase-arrested chromosomes isolated from these cells, was as described by Bousbaa et al. 1997; the same reference describes experimental protocols involving the 3F3/2 antibody. Secondary antibodies used for immunofluorescence localization of Bub1 were TRITC or FITC conjugated Affinipure donkey anti–chicken IgY (Jackson ImmunoResearch Labs), both at dilutions of 1:200. In all figures requiring comparisons of Bub1 or 3F3/2 staining between panels (Fig. 2B and Fig. C; 3, A–I; 7, A–D; 9, A–F; and 10, A–D), the gain on the digital camera was held constant, and all images were digitally processed in the same fashion.
Figure 2.
Drosophila Bub1 is recruited to kinetochores. DNA is shown in blue and Bub1 is in red. (A) A chromosome isolated from a Drosophila S2 cell arrested with colchicine, showing strong Bub1 staining at the kinetochores. (B) Bub1 is localized to the kinetochores in a wild-type (Oregon-R) neuroblast arrested with colchicine. (C) A neuroblast from a bub1 mutant (l(2)K06109/l(2)K06109) brain arrested with colchicine and stained with affinity-purified anti-Bub1 antibodies under identical conditions to those used in B. Note the complete absence of Bub1 staining in this figure; complete lack of Bub1 staining is also observed in all other bub1 allelic and deficiency combinations (not shown). D–F show that Bub1 (red) colocalizes with the kinetochore marker ZW10 (green) in wild-type neuroblasts, although the levels of staining of individual kinetochores with the two reagents are not always in concert. Bars, 5 μm. A–C are at the same magnification, as are D–F.
Detection of Markers for Programmed Cell Death
Labeling of apoptotic nuclei with a FITC anti-digoxygenin conjugate was performed using the ApopTag Plus In Situ Apoptosis Detection Kit (Oncor) according to the manufacturer's instructions.
To follow the redistribution of phosphatidylserine (an early apoptotic marker) the Annexin V-FITC kit was used (PharMingen). Brains were dissected in phosphate buffered saline (PBS; 80 mM Na2HPO4, 20 mM NaH2PO4, and 100 mM NaCl, pH 7.5) and incubated for 5–10 min in binding buffer (10 mM Hepes, pH 7.4, 140 mM NaCl, and 2.5 mM CaCl2). FITC-conjugated Annexin V was added at a 1:40 dilution, and the mixture was incubated in the dark for 1 h at 25°C. The tissue was washed at room temperature in PBS and stained with propidium iodide (50 μg/ml in PBS) for 5 min. The sample was washed again in PBS for 5 min, and then fixed in 2% formaldehyde in PBS for 3–5 min. After another 5-min wash in PBS at room temperature, the brains were mounted on a slide in Vectashield (Vector Laboratories) and observed with a BioRad MRC 600 confocal laser microscope.
To follow the expression of the reaper gene (another early apoptosis marker), we used a lacZ reporter for reaper described by Nordstrom et al. 1996. By genetic crosses, we generated a stock of genotype bub1; rpr-lacZ/T(2;3)SM6a-TM6B. Larval brains with their associated imaginal discs were dissected (in PBS) from non-Tubby animals in this stock and from control animals. These tissues were then fixed in PBS and 1.75% glutaraldehyde (Electron Microscopy Services) for 45 min. The tissues were subsequently rinsed for 30 min in 0.1 M phosphate buffer (pH 7.4), followed by incubation in FeCN solution (3 mM potassium ferrocyanide, 3 mM potassium ferricyanide, 0.15 M NaCl, and 1 mM MgCl2 in 0.01 M phosphate buffer, pH 7.2) and 0.2% X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for 3 h at 37°C. Stained imaginal discs were briefly rinsed in phosphate buffer and mounted in glycerol.
Results
A Drosophila Homologue of Bub1
Two Drosophila EST sequences were identified through a BLAST search of the Berkeley Drosophila Genome Project (BDGP) database with the amino acid sequence of mouse Bub1 (Taylor and McKeon 1997). The largest of the two corresponding cDNAs was sequenced in its entirety; this sequence has been deposited in GenBank under accession number AF080399. The cDNA sequence contains an open reading frame predicting a 165-kD protein closely related to Bub1. This protein shows 24.6% identity to human Bub1, 23.8% identity to mouse Bub1, and 14.5% identity to budding yeast Bub1p; it also displays 17.2% amino acid sequence identity to the human Bub1-related protein BubR1. The size of this Drosophila protein is somewhat larger than that of previously characterized human, mouse, and yeast members of the Bub1 family, whose predicted sizes range from 117–122 kD. The COOH-terminal third of the fly protein contains the strongly conserved kinase domain characteristic of Bub1. The NH2-terminal third of the fly protein, in common with the other members of the Bub1 family, shares significant sequence similarity with the yeast checkpoint control component Mad3p (Li and Murray 1991; Chan et al. 1998; Taylor et al. 1998). Most of the additional residues resulting in the relatively larger size of the fly protein are located in its middle third. Evidence presented below on the intracellular distribution of this Drosophila protein further substantiates its assignment as a Bub1 homologue.
Lethal P-Element Insertions into the Drosophila bub1 Gene
A search of the BDGP database of genomic sequences flanking P-element insertion sites with the complete sequence of the Bub1 cDNA identified the lethal P-element insertions l(2)K06109 and l(2)K03113 as mutations that could potentially affect the expression of the bub1 gene. Sequence analysis performed both by ourselves and by BDGP shows that the P-elements in the two separate mutants are inserted in exactly the same position within sequences transcribed into the 5′-untranslated leader of the bub1 mRNA, 48 bp upstream of the initiator ATG.
Several lines of evidence, presented in more detail in Materials and Methods, show that the lethality and associated mitotic phenotypes (see below) of l(2)K06109 and l(2)K03113 homozygotes is due to the P-element insertions into the bub1 gene. In brief, these two independently isolated mutations are allelic to each other, and they do not complement either of two deletions [Df(2R)nap1 and Df(2R)nap2] that remove polytene chromosome region 42A1-3, the location to which the bub1 gene and the l(2)K06109 and l(2)K03113 P-element insertions map by in situ hybridization. In addition, precise excision of the P-element in both mutant stocks by remobilization with a source of P-element transposase resulted in complete rescue of the lethality and associated mitotic defects seen in l(2)K06109 or l(2)K03113 homozygotes, showing that the P-element alone is responsible for the phenotype of these mutants. Importantly, as discussed below, many of the mitotic phenotypes visible in the larval neuroblasts of mutant animals are precisely those that would be expected from mutations affecting the expression of a component of the spindle checkpoint in Drosophila. These observations, taken together with the Western blot and immunofluorescence data described below, argue strongly that these mutant stocks contain P-element–induced hypomorphic mutations specifically affecting the Drosophila bub1 gene.
Drosophila Bub1 Remains at the Kinetochore in Response to Spindle Perturbation
In order to examine Drosophila Bub1 distribution during the cell cycle, affinity-purified antibodies were generated against a LacZ/Bub1 fusion protein as described in Materials and Methods. Affinity-purified IgY identifies two bands of ∼165 kD (the predicted molecular mass for Drosophila Bub1) on Western blots of larval brain extracts. These bands disappear almost completely in brain extracts made from bub1 mutants to levels <2–3% of wild-type (Fig. 1), and are completely absent in identical blots probed with preimmune IgY made from the same chickens (not shown). The antibody preparations also recognize a 100-kD band unaffected by bub1 mutations (Fig. 1); this band is also seen when blots are probed with preimmune IgY (not shown). The two Bub1-specific bands probably represent alternatively spliced or phosphorylated forms of Bub1 as shown previously by Roberts et al. 1994. The near complete absence of these bands in extracts from l(2)K06109 and l(2)K03113 homozygotes indicate that these mutations represent strongly hypomorphic, near null alleles of Drosophila bub1. It is possible that the residual low levels (seen at longer exposures) represent the perdurance of maternally supplied product contributed by the heterozygous mothers of these mutant animals.
Figure 1.
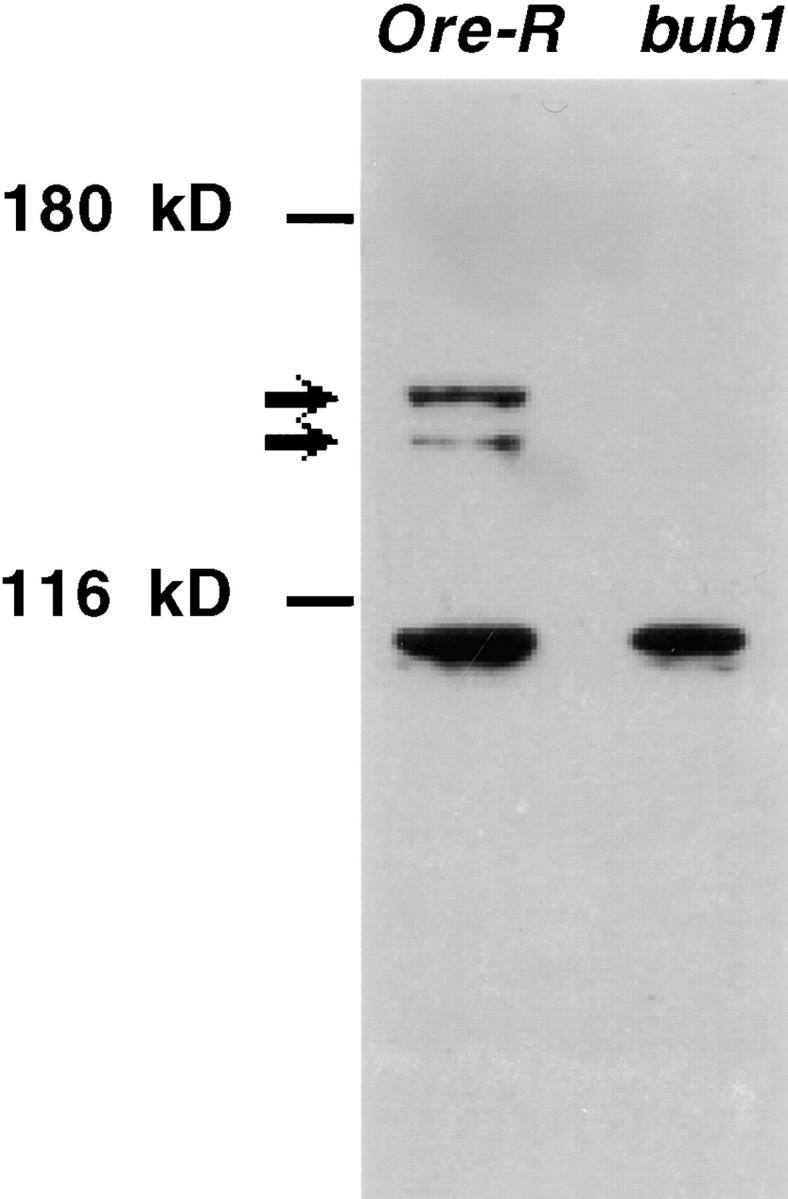
Specificity of affinity-purified anti-Drosophila Bub1 antibodies. Identical amounts of Drosophila third instar larval brain extracts, from either wild-type (Oregon R) or bub1 mutant homozygotes, were loaded onto the indicated lanes of a Western blot probed with affinity-purified anti-Drosophila Bub1 antibodies (see Materials and Methods). A doublet of ∼165 kD (the predicted size for Drosophila Bub1) is recognized in wild-type extracts. These two bands are almost completely absent in brain extracts made from bub1 mutant brains. The antibody also recognizes a band of ∼100 kD that is not Bub1-specific and that is also recognized by pre-immune IgY. This cross-reacting band serves as an internal loading control.
Previous findings have shown that in other organisms, Bub1 and other components of the spindle checkpoint associate with the kinetochore during early prophase and remain until late metaphase, but when mitotic arrest is induced by microtubule depolymerizing agents such as nocodazole or colchicine, they do not leave the kinetochore (Chen et al. 1996; Taylor and McKeon 1997; Taylor et al. 1998; Chan et al. 1998). Treatment of Drosophila cells with colchicine leads to prolonged arrest in a prometaphase-like configuration, demonstrating that the checkpoint responds to this drug in flies as well (Gonzales et al. 1991). Fig. 2 shows that Bub1 is recruited to kinetochores in chromosomes isolated from colchicine-treated Drosophila S2 tissue culture cells (Fig. 2 A) and larval neuroblasts (Fig. 2 B). No Bub1 signal is observed at the kinetochores of larval neuroblasts from prometaphase-arrested bub1 mutants (Fig. 2 C). Preimmune IgY antibodies do not specifically stain any intracellular structure in similarly treated larval neuroblasts (not shown). Thus, our affinity-purified anti-Bub1 antibodies recognize epitopes recruited to the kinetochore when the spindle is perturbed, as would be expected for a Bub1 protein (Taylor and McKeon 1997; Chan et al. 1998). These results additionally confirm the observations gained from the Western blots in Figure 1 that the Bub1 protein recognized by this antibody is nearly completely absent from the larval brains of bub1 mutants.
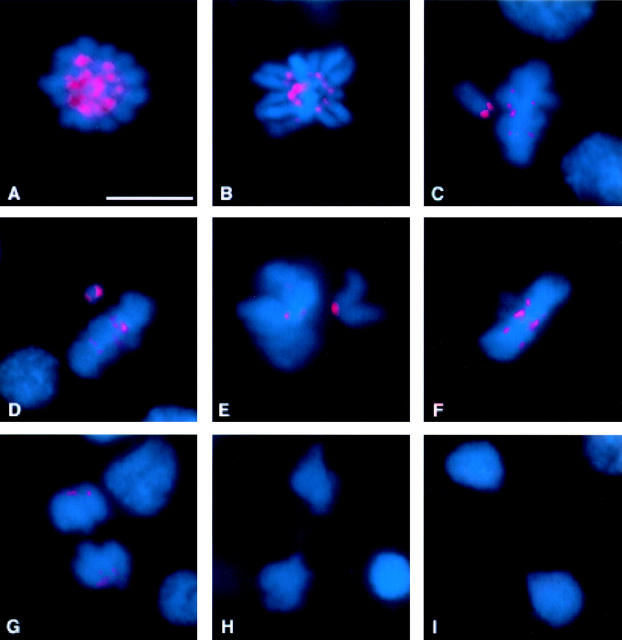
Drosophila Bub1 Shows a Dynamic Cell Cycle–dependent Localization Pattern
Next we used our affinity-purified anti-Bub1 antibodies to examine in detail the distribution of Bub1 during mitosis in cycling Drosophila S2 cells. Interphase cells show a generalized, diffuse nucleoplasmic staining pattern (not shown). At prophase (Fig. 3 A), Bub1 associates strongly with the kinetochore regions of the condensed chromosomes; as shown in Fig. 2D–F, Bub1 indeed substantially colocalizes with the kinetochore marker ZW10 (Williams et al. 1992, 1994, Williams et al. 1996). Kinetochore staining becomes weaker at prometaphase (Fig. 3 B). At metaphase, the Bub1 signal weakens specifically for those chromosomes that have migrated to the metaphase plate (Fig. 3, C–F). Chromosomes in the same cells that have not yet reached the metaphase plate continue to show strong Bub1 staining at their kinetochores (Fig. 3, C–E). Depending on the orientation of the chromosome with respect to the spindle, one kinetochore may stain more strongly for Bub1 than the other (Fig. 3 D). Very weak kinetochore signals continue to be visible into anaphase (Fig. 3 G), but are not observed during late anaphase (Fig. 3 H) or telophase (Fig. 3 I). Some staining of the spindle midzone is detectable at late anaphase (Fig. 3 H).
Figure 3.
Bub1 distribution in cycling Drosophila S2 tissue culture cells. DNA is shown in blue and Bub1 is in red. (A) Prophase. Bub1 is strongly associated with the kinetochores of the condensing chromosomes. (B) Strong kinetochore staining continues to be observed into prometaphase. (C–E) As cells approach metaphase, chromosomes that are aligned along the metaphase plate show only weak Bub1 staining, while chromosomes that have not yet reached the metaphase plate continue to show intense Bub1 staining at one or both kinetochores. Occasionally, as in D, the two kinetochores of the lagging chromosome stain show different intensities of Bub1 signals. (F) At metaphase, all the chromosomes show weak Bub1 staining at the kinetochores, which continues to be detectable into early anaphase (G). (H) Later in anaphase, kinetochore staining is no longer detectable, although some staining of the spindle midzone is visible. In addition to the mitotic figures, interphase nuclei are also visible in G and H. (I) During telophase, no specific Bub1 staining pattern is observed. Bar, 5 μm.
Similar intracellular protein distributions have already been documented by us for the Drosophila mitotic checkpoint control component Bub3 (Basu et al. 1998a), and have also been observed for human Bub1 and BubR1 (Chan et al. 1998; Jablonski et al. 1998). A previous report for mouse Bub1 failed to detect its association with kinetochores during metaphase or subsequent stages of mitosis (Taylor and McKeon 1997); it is not clear whether this represents a true difference between the mouse and the human or Drosophila patterns of Bub1 distribution, or is instead the result of lower signal intensities obtained with the monoclonal anti–mouse Bub1 antibody employed in that study.
Mitotic Defects in bub1 Mutants
To determine the developmental stage at which bub1 mutant homozygotes arrest their development, we rebalanced the l(2)K06109- or l(2)K03113-bearing chromosomes over a balancer chromosome bearing the dominant marker Tubby, whose effects are visible in larvae and pupae. In these rebalanced stocks, ∼30% of the third instar larvae were non-Tubby, in line with Mendelian expectations that bub1 homozygotes would constitute one-third of the animals that hatch from embryos. Many pupae were also non-Tubby, but these constituted a slightly smaller percentage (approximately 22%) of the total pupae. These results indicate that the lethality caused by the two bub1 mutations occurs mainly during the pupal stages, with most mutant homozygotes surviving through the third larval instar. Gatti and Baker 1989 have previously argued that animals homozygous for mutations in genes controlling essential cell cycle functions in Drosophila should survive to third instar larval stages or to the larval–pupal transition, because cell divisions prior to these stages could be supported by maternally supplied components contributed by their heterozygous mothers. This expectation has been borne out by many subsequent investigations of cell cycle mutants in flies (e.g., Williams et al. 1992). Indeed, we have detected no mitotic abnormalities in any of >1,000 post-cellularization divisions from a total of 22 homozygous mutant embryos observed at high resolution (see Materials and Methods and Discussion). Thus, we analyzed squashed preparations of neuroblasts taken from the brains of homozygous bub1 mutant third instar larvae to define the functional role of Bub1 in Drosophila cell divisions.
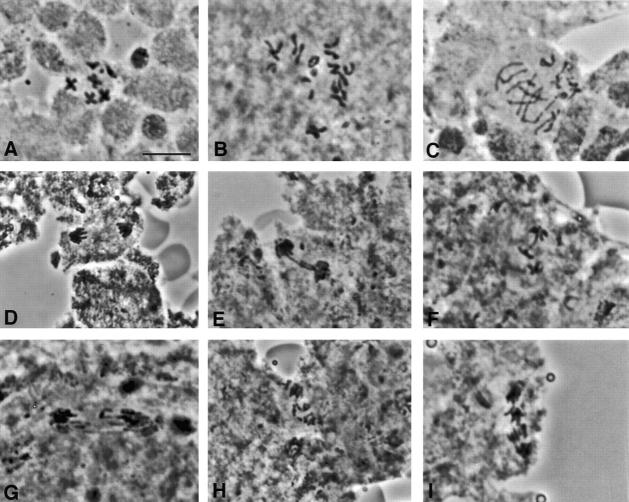
We initially observed that bub1 mutants possess the hallmark of a defect in the spindle checkpoint: that is, a failure to maintain sister chromatid cohesion when the spindle is disrupted. In wild-type brains incubated with colchicine for 1 h, sister chromatids remain attached in 98% of all mitotic figures (Fig. 4 A and Table ), revealing activity of the spindle checkpoint. Similar values have been obtained in previous experiments by our laboratory (Williams et al. 1992) and by others (Gonzales et al. 1991). Under identical conditions, sister chromatids remain attached in only 32% of mitotic figures in bub1 brains, indicating that the spindle checkpoint has often been bypassed (Fig. 4B and Fig. C, and Table ). In fact, the frequency of neuroblasts with separated sister chromatids in bub1 mutant brains is essentially unaffected by colchicine treatment, in stark contrast with wild-type.
Figure 4.
Phenotype of bub1 mutants in Drosophila neuroblasts. (A–C) Orcein-stained mitotic figures from third instar larval brains treated with colchicine and hypotonic solution to perturb spindle assembly. Wild-type neuroblasts arrest in a prometaphase like configuration with attached sister chromatids as in A. In bub1 mutant neuroblasts treated in the same fashion (B and C), sister chromatids separate, and some evidence for aneuploidy or chromosome fragmentation is observed (diploid cells should contain 12 large chromatids and four dot-like 4th chromosome chromatids). (D–I) Orcein-stained anaphase figures from untreated brains. In contrast with a wild-type (D), bub1 mutant anaphases show several abnormalities including extensive chromatin bridging (E and F), chromosomes that lag in the vicinity of the metaphase plate (G), and apparent widespread chromosome fragmentation (H and I). Bar, 5 μm.
Table 1.
Quantification of Mitotic Parameters of Ore-R and bub1 Mutant Neuroblasts after Colchicine Treatment
| Genotype | Time in colchicine | Number of optic fields | Number of cells in mitosis | Number of prophase + prometaphase | Frequency of prophase + prometaphase | Number of cells with SCS‡ | Frequency of mitotic cells with SCS | Mitotic index | Ratio of prophase + prometaphase/ anaphase |
|---|---|---|---|---|---|---|---|---|---|
| min | |||||||||
| Wild-type | 0 | 992 | 1,766 | 1,279 | 0.72 | 487 | 0.28 | 1.78 | 2.63 |
| 30 | 754 | 2,029 | 1,754 | 0.86 | 275 | 0.14 | 2.70 | 6.38 | |
| 60 | 554 | 3,988 | 3,892 | 0.98 | 96 | 0.02 | 7.20 | 40.54 | |
| 1(2)K02113 | 0 | 1,325 | 802 | 169 | 0.21 | 633 | 0.79 | 0.61 | 0.27 |
| 1(2)K03113 | 30 | 787 | 437 | 142 | 0.32 | 295 | 0.68 | 0.56 | 0.48 |
| 60 | 639 | 339 | 109 | 0.32 | 230 | 0.68 | 0.53 | 0.47 | |
| 1(2)K02113 | 0 | 952 | 305 | 154 | 0.50 | 160 | 0.52 | 0.32 | 0.96 |
| Df(2R)nap2 | 30 | 383 | 136 | 56 | 0.41 | 80 | 0.59 | 0.36 | 0.70 |
| 60 | 436 | 113 | 50 | 0.44 | 63 | 0.56 | 0.26 | 0.79 | |
| 1(2)K02113 | 0 | 614 | 131 | 38 | 0.29 | 93 | 0.71 | 0.21 | 0.41 |
| Df(2R)nap1 | 30 | 352 | 117 | 48 | 0.41 | 69 | 0.59 | 0.33 | 0.69 |
| 60 | 575 | 142 | 61 | 0.43 | 81 | 0.57 | 0.25 | 0.75 |
Table details quantitative measurements of various mitotic parameters in squashed preparations of third instar larval brains that provide an overview of the phenotype associated with the l(2)K03113 mutation. In brains untreated with colchicine, the percentage of bub1 mutant mitotic cells with separated sister chromatids is much higher, and the percentage of mitotic cells in prophase or prometaphase is much lower, than in wild-type. Relative to wild-type controls, the brains of bub1 homozygotes show a threefold reduction in the mitotic index, operationally defined as the number of mitotic figures per optic field, with every optic field in the brain being scored. More limited data sets obtained through observations of l(2)K06109 homozygotes or of l(2)K06109/l(2)K03113 trans-heterozygotes yielded almost identical results (not shown).
In all particulars, the brains of larvae heterozygous for either bub1 mutation with either of two deletions removing the bub1 locus displayed phenotypes qualitatively identical to those seen in bub1 homozygotes. However, there is some slight quantitative variation in mitotic parameters between these deletion heterozygotes and the mutation homozygotes (Table ); we do not know whether these effects are due to the activity of the bub1 gene or due to background effects. Based on these genetic criteria, the two bub1 mutations behave as very strong hypomorphs that are probably nearly but not completely null alleles.
A significant proportion of the anaphase figures in bub1 mutant brains are aberrant (61–84%, depending upon genotype; Fig. 4, D–I). Three major kinds of abnormalities are seen at high frequency. First, in many neuroblast anaphases, chromatin bridges extend between the two separating groups of chromosomes (Fig. 4, E–F). In other anaphase figures, lagging chromatids remain at the position of the metaphase plate while the other chromosomes have migrated to positions near the poles (Fig. 4 G). Finally, we observe extensive chromosome fragmentation in many mutant anaphases (Fig. 4, H–I). We believe that these anaphase aberrations explain the observation that many of the cells in colchicine-treated mutant brains appear to be aneuploid (Fig. 4B and Fig. C). These aneuploid cells could be produced by the maldistribution of intact chromosomes during anaphase of a previous cell generation. However, we suspect that many of the chromatids seen in mitotic cells like those depicted in Fig. 4B and Fig. C may actually be chromosome fragments, resulting in an overestimate of the degree of aneuploidy.
Absence of Bub1 Leads to Apoptosis in Drosophila Larval Brains
A striking feature of bub1 mutant brains examined with DNA staining is the occurrence of extremely high frequencies of pycnotic nuclei with highly condensed chromatin. These nuclei are strongly positive when labeled by Tdt-mediated dUTP-biotin nick end labeling (TUNEL)-based techniques (Fig. 5, A–F; see Materials and Methods). Because the TUNEL procedure detects chromosome damage (normally induced in the pathway for apoptosis), the TUNEL signals could reflect either the occurrence of bona fide programmed cell death, or alternatively simply the chromosome fragmentation that occurs during anaphase in bub1 mutant cells. To discriminate between these possibilities, we asked whether mutant nuclei showed elevated expression of two apoptotic events independent of chromosome breakage. The first of these markers was the redistribution of phosphatidylserine, which early in apoptosis rapidly moves from the internal face of the plasma membrane to the outside of the membrane; this redistribution was detected by use of FITC-conjugated Annexin V, a protein with very strong affinity for the serine in phosphatidylserine (Martin et al. 1995; see Materials and Methods). The second marker was a β-galactosidase reporter for reaper, a gene whose expression is needed to activate programmed cell death in Drosophila (White et al. 1994). Use of both markers verifies that mitotic cells in bub1 mutants undergo vastly elevated levels of apoptosis (Fig. 5, G–I, and 6). Levels of apoptotic nuclei are similar in l(2)K06109 or l(2)K03113 homozygotes as well as in trans-heterozygotes for either of the two alleles with deletions of the region (not shown).
Figure 5.
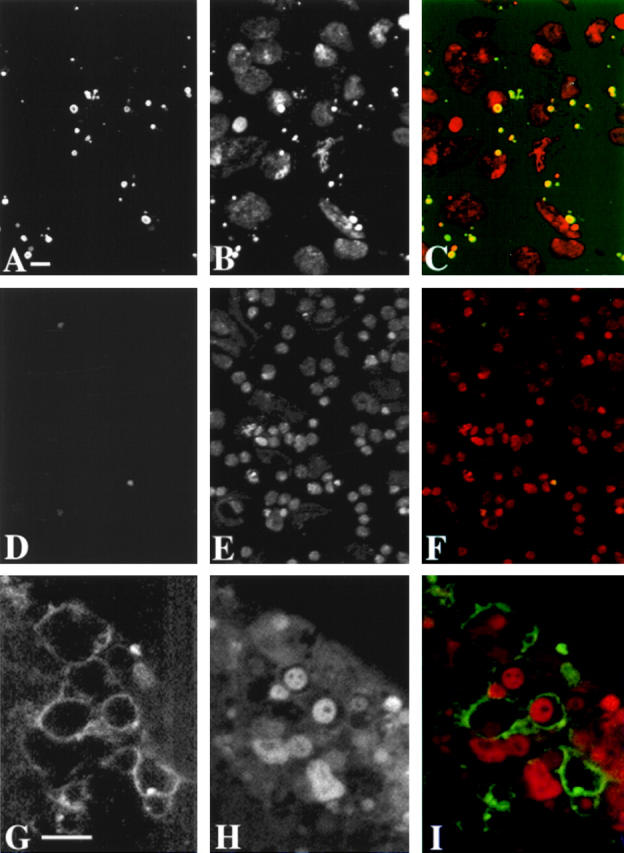
Larval brains of bub1 mutants contain many apoptotic nuclei. bub1 mutant (A–C) and wild-type (Oregon-R; D–F) brains were labeled by a TUNEL-based assay (A and D) for the presence of apoptotic nuclei, and stained with propidium iodide for DNA (B and E); a merged view with DNA in red and the TUNEL signal in green is shown in C and F. Many apoptotic nuclei are seen in bub1 mutant brains but not in wild-type; the majority of these TUNEL-positive nuclei are also pycnotic as seen by the abnormally condensed DNA signal. G–I show bub1 mutant larval brains stained with annexin V to reveal phosphatidylserine on the outside of the cell membrane (G), propidium iodide (H), and a merged view (I) with DNA staining in red and the annexin V signal in green. No annexin V staining is observed within wild-type brains (not shown). Bars: (A) 10 μm; (G) 5 μm. A–F are at the same magnification, as are G–I.
Bub1 Is Not Required for Phosphorylation of 3F3/2 Kinetochore Epitopes
Dephosphorylation of 3F3/2 epitopes is associated with the metaphase–anaphase transition (Campbell and Gorbsky 1995). Microinjection of anti-3F3/2 antibodies into cultured cells blocks 3F3/2 dephosphorylation and delays anaphase onset, implying that dephosphorylation of 3F3/2 epitopes may be a prerequisite for entry into anaphase (Campbell and Gorbsky 1995). The Bub1 kinase has been suggested as a candidate 3F3/2 kinase, both because of its function in the spindle checkpoint and because its intracellular distribution shows similarities with that of 3F3/2 epitopes (Chan et al. 1998). In order to examine these questions in more detail, we asked whether bub1 mutations would affect the distribution of 3F3/2 epitopes. As shown in Fig. 7A and Fig. B, Fig. 3F3/2 signals are present at the kinetochores in bub1 prophase/prometaphase and metaphase figures at levels comparable to that of wild-type brains (see Bousbaa et al. 1997 for a description of 3F3/2 staining in wild-type Drosophila neuroblasts). This result demonstrates that Bub1 kinase does not contribute significantly to 3F3/2 kinase activity in vivo.
Figure 7.
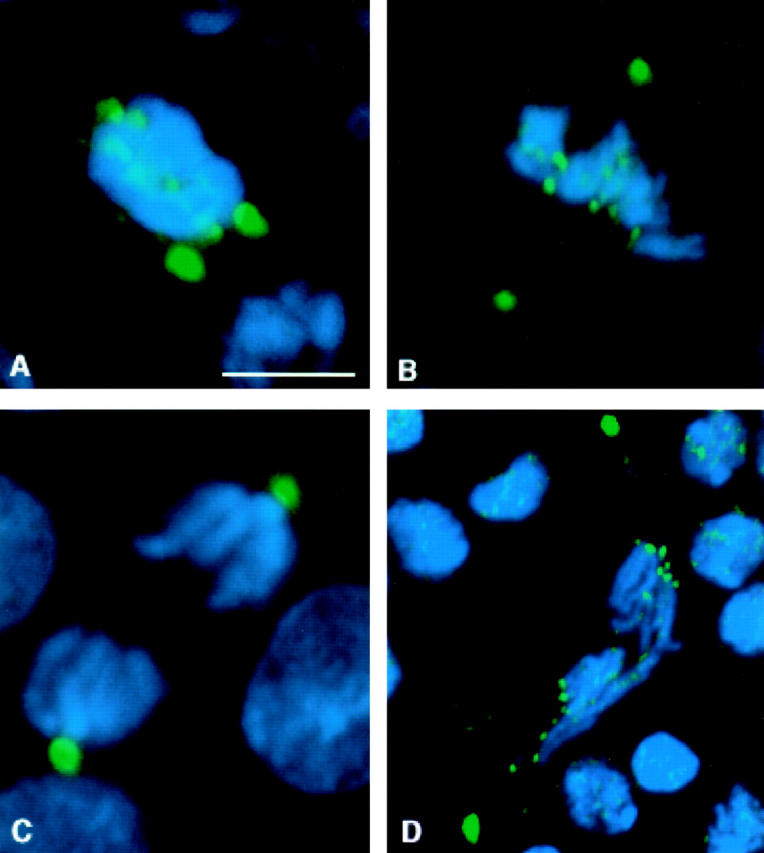
Distribution of 3F3/2 phosphoepitopes in bub1 mutant neuroblasts. DNA is in blue, and 3F3/2 phosphoepitopes are in green. In all panels, the two strongest sites of 3F3/2 staining are the centrosomes. 3F3/2 distribution in prophase (A) and metaphase (B) neuroblasts from bub1 mutants. 3F3/2 epitopes at the centrosomes and kinetochores are strongly recognized, demonstrating that the Bub1 kinase is not a significant source of 3F3/2 phosphorylation activity in vivo. (C) 3F3/2 epitopes are completely dephosphorylated during anaphase in wild-type neuroblasts (see Bousbaa et al. 1997 for a detailed description of 3F3/2 distribution in wild-type Drosophila neuroblasts). (D) 3F3/2 distribution in an anaphase figure from a bub1 mutant neuroblast. 3F3/2 epitopes continue to remain phosphorylated in bub1 anaphases, though at reduced levels relative to those seen during prophase/prometaphase. Thus, total dephosphorylation of 3F3/2 phosphoepitopes cannot be a prerequisite for entry into anaphase. Bar, 5 μm.
Interestingly, 3F3/2 staining continues to be detectable at the kinetochore at significant levels in many anaphase figures from bub1 mutant brains (Fig. 7 D). In wild-type Drosophila neuroblasts, 3F3/2 phosphoepitopes at the kinetochore are completely lost by the start of anaphase (Fig. 7 C; Bousbaa et al. 1997). This observation indicates that dephosphorylation of kinetochore-associated 3F3/2 phosphoepitopes is not essential for entry into anaphase, at least in a bub1 mutant background.
Mutations in Genes Encoding Several Other Kinetochore Components Do Not Disrupt the Association of Bub1 with the Kinetochore
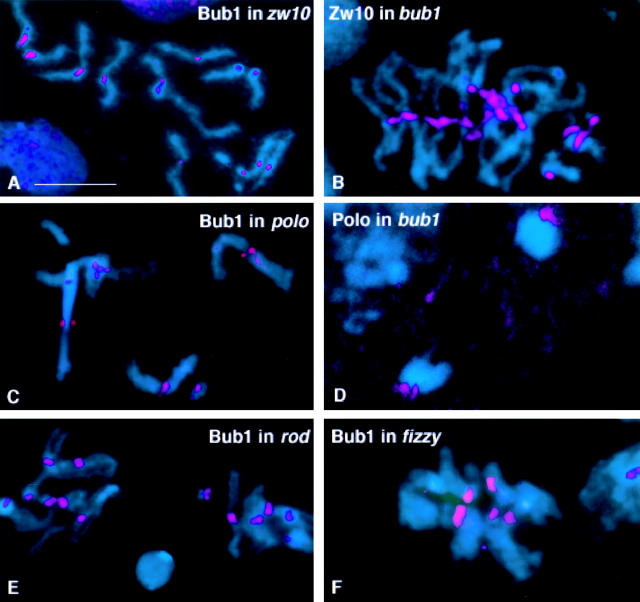
To establish the possible relationship between bub1 and other genes known to influence the fidelity of cell division in Drosophila, we explored the effects of mutations in these genes on the intracellular distribution of Bub1. We have focused on genes encoding other proteins that localize to the kinetochore, as the results of this analysis would further our understanding of kinetochore assembly.
Mutations in zw10 and rough deal disrupt the segregation of chromosomes during anaphase of mitosis and meiosis. Intriguingly, mutations in both genes cause precocious sister chromatid separation in colchicine treated larval neuroblasts, indicating a bypass of the spindle checkpoint (Smith et al. 1985; Karess and Glover 1989; Williams et al. 1992). Both the ZW10 and Rod proteins are associated with the kinetochore during prophase/prometaphase of mitosis and both meiotic divisions (Williams et al. 1992; Williams and Goldberg 1994; Scaerou, F., and R. Karess, personal communication). We found that mutations in zw10 or rod do not affect the localization of Bub1 to the kinetochore (Fig. 8A and Fig. E). Interestingly, in these mutant cells Bub1 continues to be associated with the kinetochores of precociously separated sister chromatids (Fig. 8 A), indicating that sister chromatid separation does not require the loss of Bub1 from the kinetochore. Similar results were observed when precocious sister chromatid separation was induced in wild-type colchicine-arrested neuroblasts subjected to prolonged hypotonic shock (data not shown). Conversely, bub1 mutations do not block the association of ZW10 with the kinetochore (Fig. 8 B).
Figure 8.
Relationship of Bub1 to other kinetochore components. In all panels, DNA is in blue. Neuroblasts were treated with colchicine in all panels except D. (A) Bub1 (red) in a zw10S1/Y mutant neuroblast. Note the Bub1 staining at the kinetochores of separated sister chromatids. (B) ZW10 (red) stains the kinetochores in bub1 mutant neuroblasts. (C) In neuroblasts homozygous for the mutation polo1, Bub1 associates with the kinetochores. (D) In bub1 mutant neuroblasts (here shown at anaphase), Polo protein is bound to the kinetochores of the separating chromosomes, as in wild-type. (E) Null mutations in rough deal (rod) do not prevent the association of Bub1 with kinetochores. (F) Bub1 also binds to kinetochores in the neuroblasts of animals homozygous for the mutation fizzy6. Bar, 5 μm.
The mitotic mutation polo also affects mitotic fidelity and leads to chromosome missegregation and spindle abnormalities (Sunkel and Glover 1988). The polo gene product is a protein kinase which shows a dynamic, cell cycle–dependent localization with several components of the mitotic apparatus, including the kinetochores (Logarinho and Sunkel 1998). However, mutations in polo do not affect the distribution of Bub1 (Fig. 8 C), and the Polo protein kinase is localized normally to the kinetochores in a bub1 mutant background (Fig. 8 D).
Mutations in the Drosophila gene fizzy lead to metaphase arrest (Sigrist et al., 1995), and Fizzy/Cdc20/Slp1/p55CDC has been shown to be required to mediate the Bub/Mad-dependent inactivation of the APC (for review see Townsley and Ruderman 1998). p55CDC, the mammalian homolog of Fizzy, has recently been shown to be concentrated at kinetochores from late prophase to telophase (Kallio et al. 1998). Because the action of the Fizzy protein is thought to be downstream of Bub1, we predicted that mutations in fizzy would not affect the ability of Bub1 to localize to the kinetochores. Fig. 8 F shows that this is indeed the case.
Role of Bub1 in Drosophila Spermatogenesis
The existence of a spindle checkpoint in meiotically dividing Drosophila spermatocytes is currently uncertain. The presence of univalents (chromosomes without pairing partners) does not prevent primary spermatocytes from entering anaphase. Furthermore, although mei-S332 or ord mutations cause sister chromatids to separate during the first meiotic division, chromosomes in mutant secondary spermatocytes still undergo obvious anaphase pole-ward movements (Goldstein 1980; Lin and Church 1982; Kerrebrock et al. 1992; Miyazaki and Orr-Weaver 1992). If a spindle checkpoint were active, it should have prevented anaphase onset under either of these conditions because chromosomes could not be subject to the bipolar tension needed to deactivate the checkpoint (Nicklas et al. 1995; Nicklas 1997). Finally, testes treated with colchicine contain many spermatids with polyploid nuclei, showing that spermatocytes with aberrant spindles do not arrest in metaphase and instead progress through meiosis and differentiate into spermatids (our unpublished results).
To explore the apparent absence or weakness of the spindle checkpoint in meiotic Drosophila spermatocytes, we examined the distribution of Bub1 during spermatogenesis using techniques we have previously developed (Williams et al. 1996). Bub1 localizes to the kinetochores of bivalents in primary spermatocytes during prometaphase I, as shown in Fig. 9 A. The kinetochore association of Bub1 decreases significantly as the bivalents align at the metaphase plate (Fig. 9 B) and becomes undetectable at anaphase (Fig. 9 C), although some nuclear and spindle staining above background is visible during these cell cycle stages. This dynamic localization pattern is repeated during the second meiotic division (Fig. 9, D–F). Thus, the pattern of Bub1 distribution during both meiotic divisions parallels that seen during mitosis.
Figure 9.
Role of Bub1 during Drosophila spermatogenesis. DNA is shown in blue and Bub1 is in red. (A–C) Localization of Bub1 during the first meiotic division. (A) Bub1 is strongly associated with the kinetochores at prometaphase I. (B) Kinetochore staining is significantly reduced by metaphase I and lost completely by anaphase I (C). (D–F) Localization of Bub1 during the second meiotic division parallels the behaviour of Bub1 during the first meiotic division. (G and H) Living spermatids from third instar larval testes viewed by phase contrast optics. A field of wild-type “onion stage” spermatids is shown in (G). Note that each spermatid contains a single phase light nucleus and a single phase dark Nebenkern (mitochondrial derivative), and that the volume of all nuclei are the same, indicating that chromosome segregation has occurred correctly. In contrast, a field of spermatids from a bub1 mutant testis (H) displays evidence of chromosome missegregation, in the form of spermatids with abnormal numbers of nuclei (arrows) or with micronuclei (arrowheads). Bars, 5 μm. A–F appear at the same magnification, as do G and H.
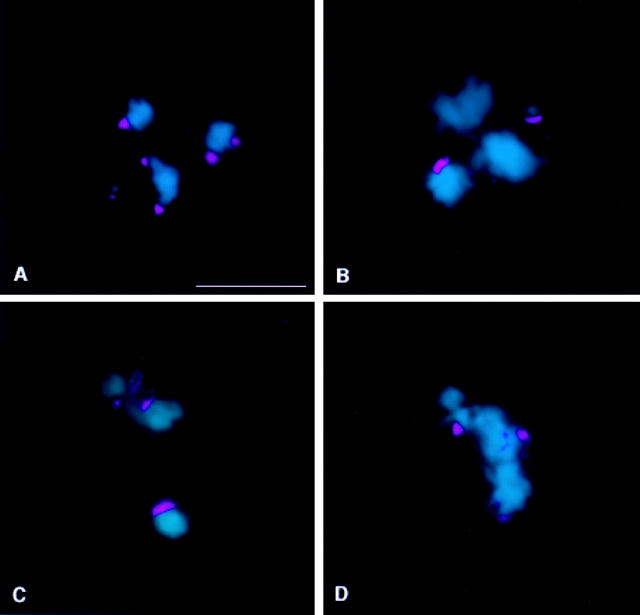
Is the association of Bub1 with the kinetochores during male meiosis in Drosophila responsive to tension? To answer this question, we analyzed the distribution of Bub1 in primary spermatocytes containing univalents: the attached XY (X^Y) and the compound 4th [C(4)RM], which are never subject to bipolar tension as they can attach only to a single pole (Ault and Lin 1984; Ault and Nicklas 1989). Fig. 10 A shows that the intensity of Bub1 staining is comparable between univalents and bivalents at prometaphase I. However, once the bivalents align at the metaphase plate, the intensity of Bub1 staining on their kinetochores decreases drastically, while the univalents in the same cell retain strong Bub1 signals (Fig. 10, B–D). Thus, the spindle checkpoint component Bub1 is not only properly localized during male meiosis, but it is also capable of discriminating between the presence or absence of bipolar tension at kinetochores. Tension has also been recently implicated in regulating the localization of Mad2 at the kinetochores in maize spermatocytes (Yu et al. 1999).
Figure 10.
Bub1 responds to tension. DNA is shown in blue and Bub1 is in red. (A) Prometaphase I spermatocyte from a X^Y; C(4)RM stock, showing strong kinetochore staining of comparable intensity on both bivalents and univalents. (B–D) Once the bivalents align at the metaphase plate, the intensity of kinetochore staining is sharply decreased. Univalents continue to show strong kinetochore association with Bub1, indicating that Bub1 can discriminate between the presence and absence of bipolar tension at the kinetochores. Bar, 5 μm.
Finally, we have examined larval testes from bub1 mutants for evidence of mitotic and meiotic defects. These testes are significantly smaller than wild-type testes, suggesting that mitotic proliferation of the germline has been substantially suppressed or that many mutant germline cells are directed into an apoptotic fate as seen in neuroblasts. Although it is as a result difficult to find meiotic or post-meiotic figures in mutant larval testes, the limited observations we have been able to make indicate that bub1 mutations strongly affect meiosis as well. Living testes from bub1 mutants examined by phase contrast optics show meiotic figures with severe spindle abnormalities at metaphase and anaphase, and multiple nuclei of variable volume at telophase (not shown). Onion stage spermatids from bub1 mutant testes contain abnormal numbers of nuclei of variable size (including micronuclei) associated with a single Nebenkern of normal size (Fig. 9G and Fig. H). As described by Fuller 1993, this phenotype results from chromosomal missegregation not accompanied by cytokinesis defects.
Discussion
A Drosophila Homologue of Bub1
We report in this paper the identification and molecular analysis of a Drosophila protein closely related to the spindle checkpoint component Bub1. Several lines of evidence support the assignment of this protein as the Drosophila Bub1 homologue. First, its primary sequence has been conserved across the phylogenetic spectrum, and is more similar to human and mouse Bub1 than to the related human BubR1 protein kinase (Taylor and McKeon 1997; Chan et al. 1998; Taylor et al. 1998). Second, we show that the cell cycle distribution of the fly protein is essentially the same as that previously reported for human Bub1. Both proteins associate strongly with the kinetochores of chromosomes unattached to the spindle prior to anaphase onset of normal mitosis, and with all the kinetochores in cells treated with microtubule depolymerizing drugs. Reduced amounts of both proteins are also found at the kinetochores of chromosomes either at the metaphase plate or being pulled toward the poles at anaphase (Fig. 2 and Fig. 3; Chan et al. 1998; Jablonski et al. 1998). Third, near null mutations in the gene encoding this Drosophila protein cause phenotypes indicating an abrogation of the spindle checkpoint. Finally, we have previously shown that these same mutations abolish the ability of another checkpoint component, Drosophila Bub3, to localize to the kinetochores (Basu et al. 1998a). This latter finding fits well with a wealth of data substantiating an intimate relationship between Bub1 and Bub3 (Roberts et al. 1994; Taylor et al. 1998; Farr and Hoyt 1998). Taken together, we believe that these observations in Drosophila provide strong evidence for the conservation of Bub1 function throughout evolution.
Bub1 Is an Essential Checkpoint Component Required for Proper Cell Cycle Progression and Chromosome Segregation
This paper describes the first genetic dissection of the function of a spindle checkpoint protein in a multicellular eukaryote. In S. cerevisiae, bub and mad genes are nonessential in the absence of microtubule depolymerizing agents, though the growth of mutant cells is slowed (Hoyt et al. 1991; Li and Murray 1991; Roberts et al. 1994). In S. pombe, bub1 null mutants are viable, though some abnormalities in chromosome segregation are observable during mitosis (Bernard et al. 1998). In marked contrast, loss of bub1 function in Drosophila leads to lethality at the larval/pupal transition. Lethality at this stage has been observed for many mutations affecting essential cell cycle components, presumably because maternally supplied stores of protein obtained from a nonmutant mother are exhausted by this point in development (Gatti and Baker 1989). Examination of neuroblasts dissected from dying third instar bub1 homozygous mutant larvae has thus allowed us to define how loss of checkpoint function affects cell division in a multicellular organism. In the description below, we cannot exclude the possibility that some aspects of the phenotype we report are indirect consequences of problems encountered in earlier cell divisions. However, it should be noted that all embryonic divisions appear to be normal, and essentially all bub1 mutant animals hatch into larvae that survive until the third instar. As there is very little cell division in the larval brain before the third instar (Ito and Hotta, 1992), the number of cell divisions that could take place between the exhaustion of maternal stores of Bub1 protein and the time of analysis is limited. Moreover, we note that these phenotypes are quite specific to bub1 mutants, and have not been observed in our analysis of many other mitotic mutants in Drosophila.
As shown in Fig. 4B and Fig. C, treatment of bub1 mutant neuroblasts with colchicine causes precocious sister chromatid separation, instead of the prometaphase arrest with attached sister chromatids typical of wild-type neuroblasts (Fig. 4 A; Gonzalez et al., 1991). This phenotype is a predictable property of mutations affecting the operation of the spindle checkpoint, as it indicates that bub1 mutant neuroblasts attempt to enter anaphase despite the absence of a functional spindle.
More interesting are the effects of bub1 mutations on normal cell division in neuroblasts that have not been treated with microtubule depolymerizing drugs. Our observations suggest that bub1 mutant neuroblasts enter anaphase prematurely even in these untreated cells. In particular, the ratio of metaphase figures to anaphase figures is decreased 5–10-fold in bub1 brains relative to wild-type brains (Table ). This result is consistent with studies showing that microinjection of Mad2 antibodies into mammalian cells causes premature sister chromatid separation and entry into anaphase (Gorbsky et al. 1998). Interestingly, loss of Bub1 in Drosophila generates a sharp decrease in mitotic index (Table ). This finding could be explained by an accelerated transit through mitosis as has been suggested for mammalian cell cultures expressing dominant negative forms of mouse Bub1 (Taylor and McKeon 1997). However, it is also possible that the lowered mitotic index reflects the assumption of an apoptotic fate by many neuroblasts in the brain (see below).
A high proportion of anaphases in untreated bub1 mutant brains show a variety of aberrations, including extensive chromatin bridging (Fig. 4E and Fig. F), lagging chromosomes (Fig. 4 G, most likely leading to aneuploidy as in Fig. 4B and Fig. C), and chromosome fragmentation (Fig. 4H and Fig. I). We interpret these aberrations as further evidence for the precocious entry into anaphase. In this view, the proper synchronization of different aspects of sister chromatid separation at the metaphase/anaphase transition has not occurred. It is well known that the forces holding sister chromatids together along their arms are separable from the forces joining sister chromatids at their centromeres (for review see Bickel and Orr-Weaver 1996). For example, acentric chromosome fragments in irradiated grasshopper neuroblasts remain associated until the onset of anaphase (Carlson 1938). In addition, sister chromatid cohesion along the arms can also be disrupted independently of centromeric cohesion through treatment with hypotonic solutions (Gatti and Baker 1989; Gonzales et al. 1991). We hypothesize that absence of bub1 function leads to loss of cohesive forces at the centromere before the separation of sister chromatids along their arms is completed. Thus, the chromatin bridging and fragmentation seen in bub1 mutant anaphases most likely reflect a failure to resolve concatenated sister chromatid DNAs along the arms at a time at which the centromeres have already separated and are being pulled toward the poles. In support of this interpretation, mutations in the Drosophila gene barren, which encodes a chromosome-associated protein that interacts with topoisomerase II, cause substantial chromatin bridging during anaphase of late embryonic divisions (Bhat et al. 1996).
Loss of Bub1 Leads to Apoptosis in Drosophila
A striking feature of Drosophila bub1 mutants is the occurrence of significantly elevated frequencies of apoptotic nuclei in larval brains (Fig. 5 and Fig. 6). This result was unexpected, as expression of a dominant negative form of mouse Bub1 has been reported to reduce the frequency of apoptotic nuclei in nocodazole-treated cells (Taylor and McKeon 1997), implying that loss of checkpoint function prevents the apoptotic response. The reasons for the apparent dichotomy between our results in Drosophila and those from mouse tissue culture cells are not clear. These effects could be organism or cell type–specific, or the differences could reflect unusual consequences of the dominant negative forms of Bub1 utilized in the mouse study.
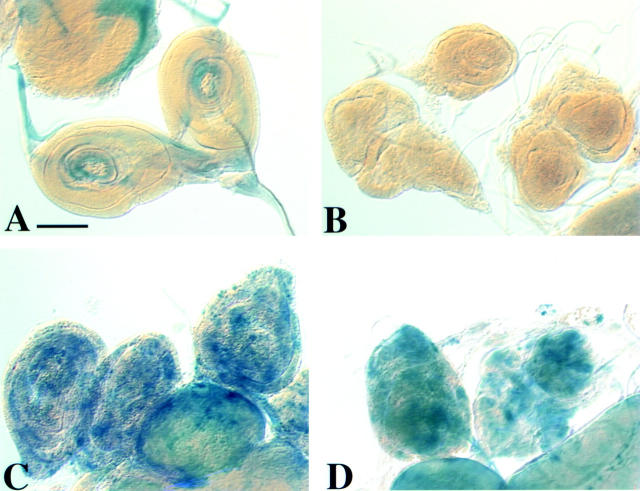
Figure 6.
Expression of reaper in bub1 mutants. Imaginal discs were stained with X-gal to follow expression of a reaper-lacZ reporter construct (rpr-lacZ) in larvae of various genotypes. (A) rpr-lacZ/rpr-lacZ. (B) bub1/bub1 without the rpr reporter gene. (This control was necessitated by the fact that the P elements causing the bub1 mutations contain a lacZ gene.) (C and D) bub1/bub1; rpr-lacZ/rpr-lacZ imaginal discs showing very high levels of β-galactosidase expression, indicating that the cell death program is activated in many cells. Similar results were observed in the brains of these same animals (not shown). Bar, 100 μm.
A strong possibility for the high level of apoptotic cells seen in bub1 mutant brains emerges from our findings that the chromosomes in many mutant anaphase figures are extensively fragmented (Fig. 4H and Fig. I). It has been well documented that chromosome breakage in Drosophila is normally a cell lethal event preventing entry into the next round of mitosis (Gatti 1979; Baker et al. 1982). We have examined the larval brains of a number of new, relatively uncharacterized mitotic mutants that cause massive chromosome fragmentation, and these uniformly have high levels of apoptotic cells (our unpublished results). Moreover, Ahmad and Golic 1999 have recently demonstrated that the induction of chromosome breakage with the FLP/FRT system is also associated with apoptosis. Apoptosis (or in fact any aspect of the bub1 mutant phenotype) cannot be an indirect consequence of aneuploidy, because brains from zw10 and rod mutants, which have many aneuploid cells (Karess and Glover 1989; Williams et al. 1992), do not show the massive apoptotic response (or any of the cell cycle defects) generated by bub1 mutants (data not shown). Regardless of the mechanism underlying the induction of apoptosis in bub1 mutant brains, it is clear that loss of spindle checkpoint function does not prevent a cell's entry into the apoptotic pathway.
Bub1 Is Not a Significant Source of the Kinase Activity Responsible for Phosphorylating 3F3/2 Epitopes
In yeast, Bub1 acts as a kinase that can phosphorylate both itself and Bub3 (Roberts et al. 1994). Because Bub1's cell cycle distribution parallels that of 3F3/2 phosphoepitopes that appear to be intimately involved in the metaphase–anaphase transition (Campbell and Gorbsky 1995), Bub1 has been suggested as a possible source of the kinase activity that generates these phosphoepitopes (Nicklas 1997; Chan et al. 1998). Our results show that this is not the case. As shown in Fig. 7, Fig. 3F3/2 epitopes are strongly phosphorylated in a bub1 mutant, showing that Bub1 cannot be a significant source of 3F3/2 kinase activity in vivo. In addition, we have previously demonstrated that Bub3 fails to associate with the kinetochore in bub1 mutants (Basu et al. 1998a), ruling out Bub3 as a major 3F3/2 phosphoepitope.
If Bub1 does not phosphorylate 3F3/2 phosphoepitopes, what kinase(s) can supply such an activity? A recent report indicates that ERK and MKK (extracellular signal-regulated protein kinase and mitogen-activated protein kinase kinase) localize to the kinetochore and can phosphorylate 3F3/2 phosphoepitopes (Shapiro et al. 1998). It is not clear whether this activity is direct or indirect; in any event, our results indicate that Bub1 does not participate in the same 3F3/2 phosphorylation pathway.
We were surprised to find that 3F3/2 epitopes at the kinetochores remain phosphorylated in anaphase figures from bub1 mutants (Fig. 7 D). In contrast, 3F3/2 phosphoepitopes at the kinetochores are normally lost completely at the start of anaphase (Fig. 7 C; Bousbaa et al. 1997). The implications of this result are twofold. First, dephosphorylation of kinetochore-associated 3F3/2 epitopes is not required for the metaphase/anaphase transition, at least in a bub1 mutant background. One possibility is that 3F3/2 dephosphorylation is not as commonly suggested (Campbell and Gorbsky 1995; Nicklas et al. 1995) as part of the signaling pathway for anaphase onset, but is instead a downstream response to the signal. Alternatively, Bub1 may function downstream of 3F3/2 dephosphorylation in the pathway governing the metaphase–anaphase transition. A second implication of our observation is that Bub1 kinase activity is required, presumably indirectly, for the dephosphorylation of 3F3/2 epitopes at the metaphase-anaphase transition. A possible explanation for the continued phosphorylation of kinetochore-based 3F3/2 epitopes is that the accelerated transit through mitosis in bub1 mutants may not allow enough time for action of the relevant phosphatase(s).
Separable Pathways in the Construction of Kinetochores
We show in this paper that the localization of Bub1 to the kinetochore is not abolished by mutations in several genes encoding other kinetochore components, nor do mutations in bub1 affect the association of ZW10 or Polo proteins with the kinetochore. Combined with previous observations from our laboratories, these findings suggest that the kinetochore may be assembled in at least two independent pathways. In one pathway, interaction between Bub1 and Bub3 is required for the kinetochore targeting of either protein (Roberts et al. 1994; Basu et al., 1998; Taylor et al. 1998). In a second subassembly, ZW10 and Rod proteins form a complex needed for the recruitment of the microtubule motor dynein to the kinetochore (Starr et al. 1998). The fact that polo mutations do not disrupt the kinetochore localization of Bub1, Bub3, or ZW10 (Fig. 8 C and our unpublished observations) suggests either a third independent pathway or that the kinetochore binding of Polo protein is subsequent to the recruitment of one of the two subassemblies.
In colchicine-treated larval neuroblasts from zw10 mutants where the sister chromatids have separated prematurely, high levels of Bub1 protein remain at the kinetochores (Fig. 8 A). This phenomenon is not restricted to a zw10 mutant background, as prolonged treatment of wild-type larval neuroblasts with hypotonic solution after colchicine incubation also generates precocious sister chromatid separation with continued strong Bub1 staining at the kinetochores (not shown). These observations indicate that it is possible to initiate anaphase despite the presence of the Bub1 “wait-anaphase” signal at the kinetochores. It is thus conceivable that the relative loss of Bub1 from kinetochores at metaphase and anaphase (Fig. 3, C–E, and 9, B and C) is not normally a prerequisite for anaphase onset.
Does the Spindle Checkpoint Function in Drosophila Meiosis?
Although the existence of a tension-dependent “wait-anaphase” checkpoint in meiotic grasshopper spermatocytes has been well established (Nicklas et al. 1995; Nicklas 1997), several observations suggest that such a checkpoint may not play a major role in Drosophila spermatogenesis. The presence of univalents (chromosomes without a pairing partner) does not obviously affect meiotic progression (Church and Lin 1988). Mutations in mei-S322 and ord, which lead to sister chromatid separation before the start of the second meiotic division, do not affect entry into anaphase II (Goldstein 1980; Lin and Church 1982; Kerrebrock et al. 1992; Miyazaki and Orr-Weaver 1992). Finally, colchicine-treated spermatocytes that cannot segregate their chromosome still exit meiosis and differentiate into spermatids (our unpublished results).
Nevertheless, Drosophila Bub1 and Bub3 both associate strongly with the kinetochores of primary spermatocytes before metaphase of both meiotic divisions (Fig. 9 A and 10 A; Basu et al. 1998a), and we have recently been able to observe kinetochore staining with antibodies against Xenopus Mad2 (Chen et al. 1996) in prometaphase primary spermatocytes (our unpublished observations). Bub1 responds differentially to the presence and absence of tension across chromosomes during meiosis (Fig. 10, B–D) exactly as would be predicted were it acting as part of a functional spindle checkpoint. In addition, bub1 mutations have a dramatic effect on Drosophila spermatogenesis. Though it is difficult to distinguish aberrations introduced during mitotic germ line cell proliferation from those occurring during meiosis, the appearance of disrupted meiotic spindles (not shown) and of multiple nuclei of uneven volume within “onion-stage” spermatids (Fig. 9G and Fig. H) are suggestive of defects specifically affecting meiosis.
On the basis of these observations, we believe that a spindle checkpoint does exist in Drosophila meiotic spermatocytes, but that it operates with significantly reduced efficiency or according to different signals. The reasons underlying this apparent inefficiency remain unclear, but very likely involve part of the checkpoint pathway downstream of Bub1. One prediction of this viewpoint is that conditions that should enable the checkpoint would delay, but not completely block, cell cycle progression past the metaphase of either meiotic division. It will thus be of importance in the near future to verify this prediction through real-time observations of male meiosis in cultured spermatocytes.
Acknowledgments
We thank Kathy Matthews (Indiana University, Bloomington, IN), John Roote (University of Cambridge, UK), and Todd Laverty (University of California, Berkeley, CA) for the Drosophila stocks described in this paper.
This work was funded by grant no. GM48430 from the National Institutes of Health to M.L. Goldberg and by funding from the Junta Nacional de Investigação Científica of Portugal and the European Union to C.E. Sunkel.
Footnotes
1.used in this paper: APC, anaphase-promoting complex; TUNEL, Tdt-mediated dUTP-biotin nick end labeling
References
- Ahmad K., Golic K.G. Telomere loss in somatic cells of Drosophila causes cell cycle arrest and apoptosis. Genetics. 1999;151:1041–1051. doi: 10.1093/genetics/151.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ault J.G., Lin H.P. Bivalent behaviour in Drosophila melanogaster males containing the In(1)sc4Lsc8RXchromosome. Chromosoma. 1984;90:222–228. doi: 10.1007/BF00292400. [DOI] [PubMed] [Google Scholar]
- Ault J.G., Nicklas R.B. Tension, microtubule rearrangements, and the proper distribution of chromosomes in mitosis. Chromosoma. 1989;98:33–39. doi: 10.1007/BF00293332. [DOI] [PubMed] [Google Scholar]
- Baker B.S., Smith D.A., Gatti M. Region specific effects on chromosome integrity of mutations at essential loci in Drosophila melanogaster . Proc. Natl. Acad. Sci. USA. 1982;79:1205–1209. doi: 10.1073/pnas.79.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, J. 1999. The spindle assembly checkpoint in Drosophila. Ph.D. thesis. Cornell University, Ithaca, NY.
- Basu J., Logarinho E., Herrmann S., Bousbaa H., Li Z., Chan G.K.T., Yen T.J., Sunkel C.E., Goldberg M.L. Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod Chromosoma. 107 1998. 376 385a [DOI] [PubMed] [Google Scholar]
- Basu J., Williams B.C., Li Z., Williams E.V., Goldberg M.L. Depletion of a Drosophila homolog of yeast Sup35p disrupts spindle assembly, chromosome segregation and cytokinesis during male meiosis Cell Motil. Cytoskelet. 39 1998. 286 302b [DOI] [PubMed] [Google Scholar]
- Bernard P., Hardwick K., Javerzat J.P. Fission yeast Bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 1998;143:1775–1787. doi: 10.1083/jcb.143.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M.A., Philp A.V., Glover D.M., Bellen H.J. Chromatid segregation at anaphase requires the barren product, a novel chromosome associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- Bickel S., Orr-Weaver T.L. Holding chromatids together to ensure they go their separate ways. Bioessays. 1996;18:293–300. doi: 10.1002/bies.950180407. [DOI] [PubMed] [Google Scholar]
- Bousbaa H., Correira L., Gorbsky G.J., Sunkel C.E. Mitotic phosphoepitopes are expressed in Kc cells, neuroblasts and isolated chromosomes of Drosophila melanogaster . J. Cell Sci. 1997;110:1979–1988. doi: 10.1242/jcs.110.17.1979. [DOI] [PubMed] [Google Scholar]
- Brinkley B.R., Cox S.M., Pepper D.A. Structure of the mitotic apparatus and chromosomes after hypotonic treatment of mammalian cells in vitro. Cytogenet. Cell Genet. 1980;26:165–174. doi: 10.1159/000131438. [DOI] [PubMed] [Google Scholar]
- Cahill D.P., Lengauer C., Yu J., Riggins G.J., Willson J.K.V., Markowitz S.D., Kinzler K.W., Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Campbell M.S., Gorbsky G.J. Microinjection of mitotic cells with the 3F3/2 anti-phosphoepitope antibody delays the onset of anaphase. J. Cell Biol. 1995;129:1195–1204. doi: 10.1083/jcb.129.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J. Mitotic behaviour of induced chromosomal fragments lacking spindle attachments in the neuroblasts of the grasshopper. Proc. Natl. Acad. Sci. USA. 1938;24:500–507. doi: 10.1073/pnas.24.11.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G., Bonaccorsi S., Pisano C., Verni F., Gatti M. Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J. Cell Sci. 1994;107:3521–3534. doi: 10.1242/jcs.107.12.3521. [DOI] [PubMed] [Google Scholar]
- Chan G.K.T., Scharr B.T., Yen T.J. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and human BUBR1 kinase. J. Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Waters J.C., Salmon E.D., Murray A.W. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko A., Mann M., Murray A.W. Spindle checkpoint protein Xmad1 recruits Xmad2 to the kinetochore. J. Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church K., Lin H.P.P. Drosophila: A model for the study of aneuploidy. In: Vig B.K., Sandberg A.A., editors. Aneuploidy Part BInduction and Test Systems. Alan R. Liss Inc; New York, NY: 1988. pp. 227–255. [Google Scholar]
- Elledge S.J. Cell cycle checkpointspreventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Elledge S.J. Mitotic arrestMad2 prevents Sleepy from waking up the APC. Science. 1998;279:999–1000. doi: 10.1126/science.279.5353.999. [DOI] [PubMed] [Google Scholar]
- Farr K.A., Hoyt M.A. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol. Cell. Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V.E. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989;107:1–22. [PubMed] [Google Scholar]
- Fuller M.T. Spermatogenesis. In: Martinez-Arias A., Bate M., editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 71–147. [Google Scholar]
- Gatti M. Genetic control of chromosome breakage and rejoining in Drosophila melanogaster: spontaneous chromosome aberrations in X-linked mutants defective in DNA metabolism. Proc. Natl. Acad. Sci. USA. 1979;76:1377–1381. doi: 10.1073/pnas.76.3.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Baker B.S. Genes controlling essential cell cycle functions in Drosophila melanogaster . Genes Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- Gatti M., Goldberg M.L. Mutations affecting cell division in Drosophila . Methods Cell Biol. 1991;35:543–586. doi: 10.1016/s0091-679x(08)60587-7. [DOI] [PubMed] [Google Scholar]
- Goldstein L. Mechanisms of chromosome orientation revealed by two meiotic mutants in Drosophila melanogaster . Chromosoma. 1980;78:79–111. doi: 10.1007/BF00291909. [DOI] [PubMed] [Google Scholar]
- Gonzales C., Jimenez C.J., Ripoll P., Sunkel C.E. The spindle is required for the process of sister chromatid separation in Drosophila neuroblasts. Exp. Cell. Res. 1991;192:10–15. doi: 10.1016/0014-4827(91)90150-s. [DOI] [PubMed] [Google Scholar]
- Gorbsky G.J., Chen R.H., Murray A.W. Microinjection of antibody to Mad2 protein in mammalian cells in mitosis induces premature anaphase. J. Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L.H., Stepien P.P., Tso J.Y., Brousseau R., Narang S., Thomas D.Y., Wu R. Synthesis of human insulin gene. Construction of expression vectors for fused proinsulin production in Escherichia coli . Gene. 1984;29:251–254. doi: 10.1016/0378-1119(84)90186-0. [DOI] [PubMed] [Google Scholar]
- Hoyt M.A., Totis L., Roberts B.T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau L.F., Smith D.L., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W. Budding yeast cdc20a target of the spindle assembly checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Jablonski S.A., Chan G.K.T., Cooke C.A., Earnshaw W.C., Yen T.J. hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- Kallio M., Weinstein J., Daum J.R., Burke D.J., Gorbsky G.J. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R.E., Glover D.M. rough deal, a gene required for proper mitotic segregation in Drosophila . J. Cell Biol. 1989;109:2951–2961. doi: 10.1083/jcb.109.6.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr T.L., Alberts B.M. Organization of the cytoskeleton in early Drosophila embryos. J. Cell Biol. 1986;102:1494–1509. doi: 10.1083/jcb.102.4.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrebrock A.W., Miyazaki W.Y., Birnby D., Orr-Weaver T.L. The Drosophila mei-S322 gene promotes sister chromatid cohesion in meiosis following kinetochore differentiation. Genetics. 1992;130:827–841. doi: 10.1093/genetics/130.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lin D.P., Matsumoto S., Kitazono A., Matsumoto T. Fission yeast slp1an effector of the Mad2 dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- Li R., Murray A.W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li Y., Benezra R. Identification of a human mitotic checkpoint genehsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Lin H.P., Church K. Meiosis in Drosophila melanogaster. III. The effect of orientation disrupter (ord) on gonial mitotic and meiotic divisions in males. Genetics. 1982;102:751–770. doi: 10.1093/genetics/102.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E., Sunkel C.E. The Drosophila kinase POLO localizes to multiple compartments of the mitotic apparatus and is required for the phosphorylation of MPM2 reactive epitopes. J. Cell Sci. 1998;111:2897–2909. doi: 10.1242/jcs.111.19.2897. [DOI] [PubMed] [Google Scholar]
- Martin J.S., Reutelingsperger C.P.M., McGahon A.J., Rader J.A., van Schie R.C.A.A., LaFace D.M., Green D.R. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulusInhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki W.Y., Orr-Weaver T.L. Sister chromatid misbehaviour in Drosophila ord mutants. Genetics. 1992;132:1047–1061. doi: 10.1093/genetics/132.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B., Ward S.C., Gorbsky G.J. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J. Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Nordstrom W., Chen P., Steller H., Abrams J.M. Activation of the reaper gene during ectopic cell killing in Drosophila . Dev. Biol. 1996;180:213–226. doi: 10.1006/dbio.1996.0296. [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B.T., Farr K.A., Hoyt M.A. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H.M., Preston C.R., Phillis R.W., Johnson-Schlitz D.M., Benz W.K., Engels W.R. A stable genomic source of P element transposase in Drosophila melanogaster . Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A.D., Murray A.W. The spindle assembly checkpoint. Curr. Opin. Cell. Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Shapiro P.S., Vaisberg E., Hunt A.J., Tolwinski N.S., Whalen A.M., McIntosh J.R., Ahn N.G. Activation of the MKK/ERK pathway during somatic cell mitosisdirect interactions of active ERK with kinetochores and regulation of the mitotic 3F3/2 phosphoantigen. J. Cell Biol. 1998;142:1533–1545. doi: 10.1083/jcb.142.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.A., Baker B.S., Gatti M. Mutations in genes controlling essential mitotic functions in Drosophila melanogaster . Genetics. 1985;110:647–670. doi: 10.1093/genetics/110.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Williams B.C., Li Z., Etemad-Moghadam B., Dawe R.K., Goldberg M.L. Conservation of the centromere/kinetochore protein ZW10. J. Cell Biol. 1997;138:1289–1301. doi: 10.1083/jcb.138.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Williams B.C., Hays T.S., Goldberg M.L. ZW10 helps recruit dynein and dynactin to the kinetochore. J. Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel C.E., Glover D.M. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 1988;89:24–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha E., McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1 related protein kinase. J. Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley F.M., Ruderman J.V. Proteolytic ratchets that control progression through mitosis. Trends Cell Biol. 1998;8:238–244. doi: 10.1016/s0962-8924(98)01268-9. [DOI] [PubMed] [Google Scholar]
- Wells W.A.E. The spindle assembly checkpointaiming for a perfect mitosis, every time. Trends Cell Biol. 1996;6:228–234. doi: 10.1016/0962-8924(96)10018-0. [DOI] [PubMed] [Google Scholar]
- White K., Grether M.E., Abrams J.M., Young L., Farrell K., Steller H. Genetic control of programmed cell death in Drosophila . Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Williams B.C., Karr T.L., Montgomery J.M., Goldberg M.L. The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J. Cell Biol. 1992;118:759–773. doi: 10.1083/jcb.118.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.C., Goldberg M.L. Determinants of Drosophila zw10 protein localization and function. J. Cell Sci. 1994;107:785–798. doi: 10.1242/jcs.107.4.785. [DOI] [PubMed] [Google Scholar]
- Williams B.C., Gatti M., Goldberg M.L. Bipolar spindle attachments affect redistributions of ZW10, a Drosophila centromere/kinetochore component required for accurate chromosome segregation. J. Cell Biol. 1996;134:1127–1140. doi: 10.1083/jcb.134.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D.A., Jackson P.K. Cell cycleoiling the gears of anaphase. Curr. Biol. 1998;8:636–639. doi: 10.1016/s0960-9822(07)00410-1. [DOI] [PubMed] [Google Scholar]
- Yu H.G., Muszynski M.G., Dawe R.K. The maize homolog of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J. Cell Biol. 1999;145:425–435. doi: 10.1083/jcb.145.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]