Abstract
We have cloned and characterized the troponin C gene, pat-10 of the nematode Caenorhabditis elegans. At the amino acid level nematode troponin C is most similar to troponin C of Drosophila (45% identity) and cardiac troponin C of vertebrates. Expression studies demonstrate that this troponin is expressed in body wall muscle throughout the life of the animal. Later, vulval muscles and anal muscles also express this troponin C isoform. The structural gene for this troponin is pat-10 and mutations in this gene lead to animals that arrest as twofold paralyzed embryos late in development. We have sequenced two of the mutations in pat-10 and both had identical two mutations in the gene; one changes D64 to N and the other changes W153 to a termination site. The missense alteration affects a calcium-binding site and eliminates calcium binding, whereas the second mutation eliminates binding to troponin I. These combined biochemical and in vivo studies of mutant animals demonstrate that this troponin is essential for proper muscle function during development.
Keywords: troponin C, Caenorhabditis elegans, troponin I, muscle gene, gene expression
Troponin C is a component of the troponin complex regulating acto-myosin interaction through calcium concentration in muscle cells in vertebrates. This protein binds four calcium ions and forms a complex with both troponin I and troponin T (Ebashi and Endo 1968; Ohtsuki et al. 1986; Grabarek et al. 1992). Troponin C was first isolated and sequenced from skeletal and cardiac muscles of vertebrates (Collins et al. 1977; Wilkinson 1980; Gahlmann and Kedes 1990), but has also been isolated from invertebrates including crayfish (Kobayashi et al. 1989), scallops (Ojima et al. 1994), nematodes (Kimura et al. 1987), and insects (Bullard et al. 1988). Much is known about the structure and biochemical details of how this protein functions (Herzberg and James 1988; Reinach and Karlsson 1988; Parmacek and Leiden 1989; Fujimori et al. 1990; Parmacek et al. 1990, Parmacek et al. 1992; Schreier et al. 1990; Smith et al. 1994), including its calcium-binding properties (Putkey et al. 1989) and sites for interaction with troponin I (Tripet et al. 1997; Vassylyev et al. 1998), but little is known about the consequence of altering some of these functional properties within a developmental context. Studies on mutated versions of this protein within a well-studied muscle system offer this opportunity.
The nematode Caenorhabditis elegans offers a system within which to study altered troponins. C. elegans has two primary muscle types: body wall muscle for locomotion and pharyngeal muscle for feeding (Brenner 1974). A combination of genetic and molecular approaches has identified >80 genes involved in muscle development and function in this organism (reviewed in Waterston 1988; Anderson 1990; Moerman and Fire 1997). Included in this set of genes are those encoding the structural components of nematode thick filaments, myosin and paramyosin (Kagawa et al. 1989; Gengyo-Ando and Kagawa 1991) and thin filaments, actin and tropomyosin (Kagawa et al. 1995). Regulating the interaction of these filament types is complex and involves both thin and thick filament regulatory networks (Harris et al. 1977). For the thin filaments, regulation is through the troponin/tropomyosin complex, whereas regulation of the thick filaments is mediated by twitchin (Moerman and Fire 1997). Mutations in several contractile regulatory components have been described; those affecting thin filaments invariably lead to late embryonic or early larval lethality (Williams and Waterston 1994), whereas those affecting thick filament regulation lead to unregulated spontaneous contractions and an uncoordinated phenotype (Moerman and Fire 1997).
In this study, we describe the cloning and characterization of a troponin C gene pat-10 in the nematode C. elegans and the identification of pat-10, a previously described locus important for muscle development, as the structural gene of this troponin C. Similar to other thin filament regulatory proteins in the nematode, loss of troponin C leads to lethality. We describe the biochemical defects in these mutations and speculate about how their malfunction may lead to the terminal phenotype observed. This study should help us further understand how troponin C functions in calcium binding, protein–protein interactions, and muscle development.
Materials and Methods
Worm Culture and Molecular Cloning
Worm culture and handling were done by established methods (Brenner 1974; Sulston and Hodgkin 1988). The nematode C. elegans Bristol N2 was used for DNA and protein analysis. The pat-10 mutant strains RW3608: pat-10(st575)/dpy-5(e61)unc-29(e1072), and RW3613: pat-10(st568)/unc-11(e47)dpy-5(e61) were used for analysis of mutation site and muscle structure by using segregated homozygous worms (Williams and Waterston 1994; see Fig. 7 I). Exon expression cloning of the troponin gene was essentially the same as was described (Kagawa et al. 1989) except using a cDNA library. A bacteriophage ZapII library of cDNA provided by R. Barstead (Oklahoma Medical Research Foundation, Oklahoma City, OK; Barstead and Waterston 1989) was screened to obtain positive clones with anti–troponin C against Ascaris protein (Nakae and Obinata 1993). Positive clones were obtained by screening four to eight plates (∼10,000 plaques per plate). Standard DNA recombinant techniques were followed (Sambrook et al. 1989). A 3.5-kb PstI fragment from C15C10 or F54C1 was subcloned into the PstI site of pUC119 to generate pTNC1 (see Fig. 1C and Fig. D). DNA and protein sequence data analyses were done by using the programs of HITACHI DNASIS and GENETYX-MAC. The nucleotide sequence data reported in this paper will appear in the GSDB, DDBJ, EMBL, and NCBI nucleotide sequence databases with the accession numbers D45895 and D45896 for genomic and cDNA sequences, respectively.
Figure 7.

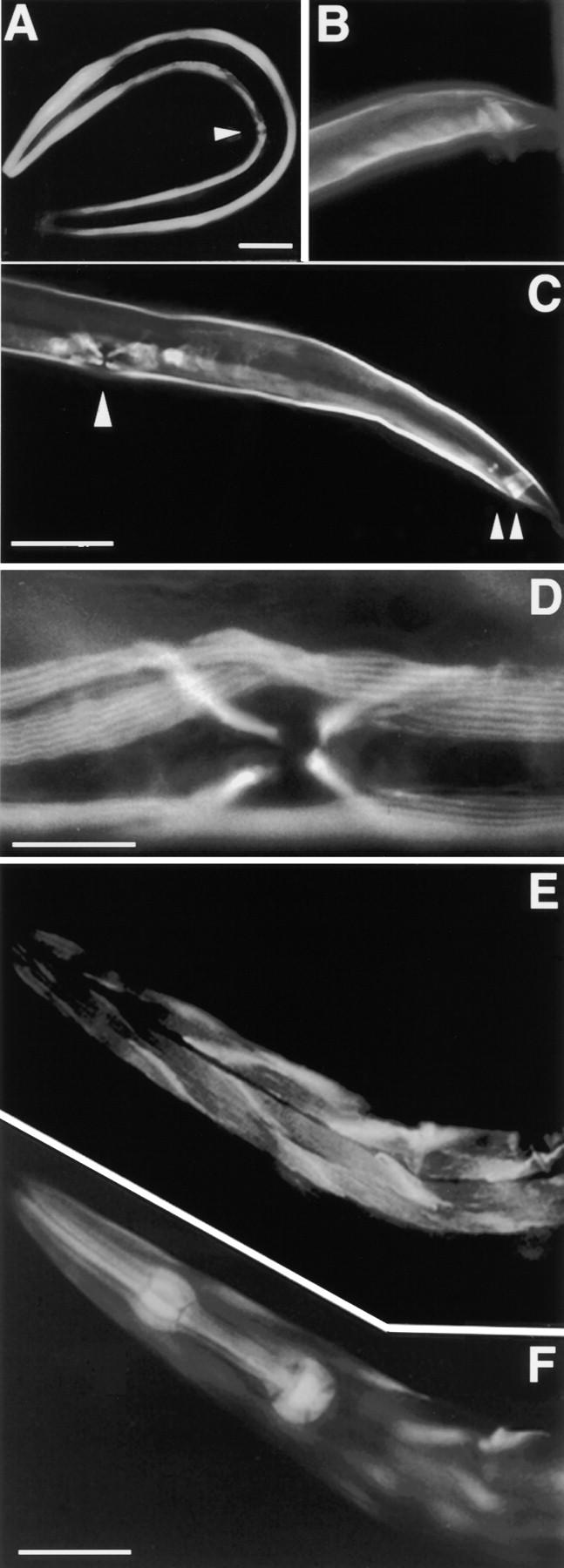
Expression pattern of the pat-10/lacZ gene in early stages of wild-type development and the phenotypes of pat-10 animals. pTNC292 expressions at gastrulation (A), comma bean (B and C), twofold (D), and threefold (B) stages, respectively. pTNC292 expression in adult worm (E). It was noted that pat-10 started expression at comma bean stage (B and C). pat-10 heterozygotes yield both wild-type (I) and pat-10 homozygous animals that stop development at the twofold stage (F–H and I). Bars, 0.1 mm.
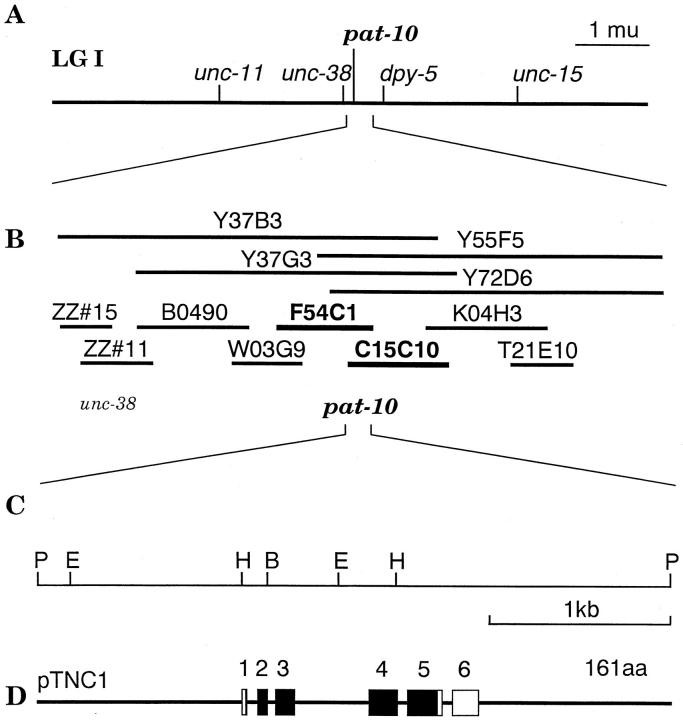
Figure 1.
Map position and genome structure of the troponin C gene, pat-10. (A) Genomic map of the central part of chromosome I of the worm is shown. (B) Contig map of the region surrounding pat-10. Three YAC clones, Y37B3, Y55F5, and Y72D6 hit with cDNA fragment of the troponin C. Cosmid clones of B0490, W03G9, F54C1, C15C10 K04H3, and T21E10 were hybridized with cDNA fragment, and F54C1 and C15C10 contained the genomic fragment of pat-10. (C and D) Restriction map and intron/exon structure from sequence data of pat-10. It should be noted that only one transcript from the plasmid clone pTNC1 was matched to 674 bp of cDNA encoding 161 amino acid residues. H, HindIII; B, BamHI; E, EcoRI; P, PstI. The nucleotide sequence of pat-10 is in Fig. 2.
Other DNA Recombinant Techniques and Construction of Mutant Clones
Placement of tnc-1 on the physical map of the chromosome was essentially the same as described previously (Coulson et al. 1988). PCR products for determining pat-10 mutation sites in tnc-1 used the following oligonucleotides as upstream primers: TNCS1 (AGCCTTGTCTCTCGAATCCTGTGT), TNCS2 (GCTGAGGATATCGAAGAGATTCTTG), TNCS3 (ATCTATGTGGCATCTAACTTCATTC), and the oligonucleotides: TNCA3 (CCTCAATTTGGGATCCGTCGAT), TNCA1 (TGCGGATCAGTTTACGAAGGGTCT), and TNCA2 (GTTGGTGACTGGTCCCCACAGTTGA) as downstream primers, respectively (see Fig. 2), and total DNA from st575 and st568 as templates. Three PCR fragments were cloned into pBluescript SK(−) vectors and were sequenced by designed primers. 30 cycles for reactions were 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min. 5′ RACE1 was done by two steps with the protocol of GIBCO BRL (Gaithersburg, MD). At the first step cDNA was synthesized by using the oligonucleotides TNCA1 and total RNA as a template. The second step PCR was done by using the anchor oligonucleotide as an upstream primer and TNCA3 as a downstream primer and purified cDNA fragment as a template. Forty cycles for 5′ RACE were 95°C for 30 s, 50°C for 1 min, and 72°C for 1 min.
Figure 2.
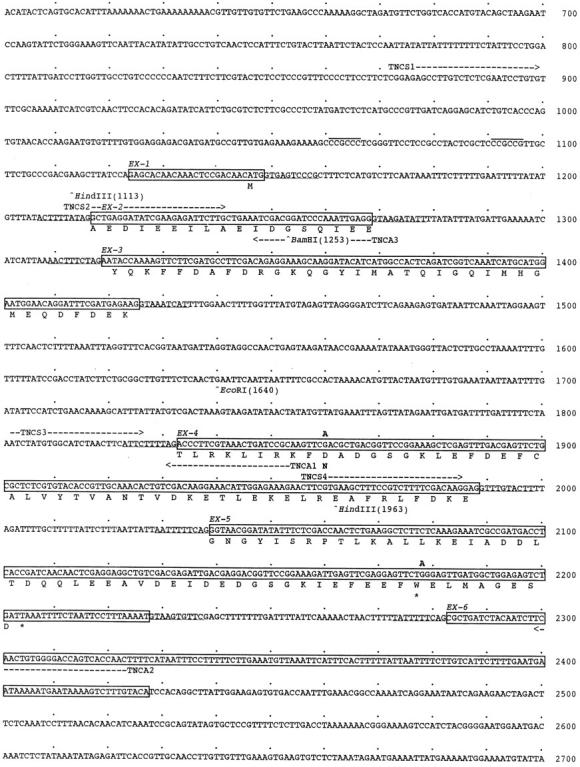
Nucleotide sequence of the troponin C gene pat-10. The exons have been translated using the standard one letter code. The splicing pattern of the gene is given in Fig. 1 D. Mutation sites in pat-10(st575) animal are shown on the top of the sequence at G1860A and G2179A. Restriction sites and the positions of primers for mutation sites determination are indicated. The accession number of pat-10 sequence in the GSDB, DDBJ, EMBL, and NCBI is D45895.
A mutant clone having one of each mutation was constructed by two-step procedures as follows. Two fragments, one having a mutation sequence at the mutation site and another having a mutation at restriction site, were synthesized by PCR. Second PCR was performed by using two annealed fragments as a template. After digestion with restriction enzymes, only a fragment having a mutation site was ligated into vector. Constructed mutant clones were named pat-10-m1 for mutation at the second calcium-binding site and pat-10-m2 for mutation missing COOH-terminal helix, respectively (Fig. 5).
Figure 5.
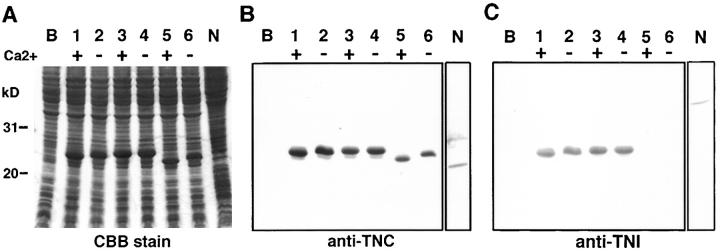
Mobility-shift assay of the wild-type troponin C and characterization of mutant troponin C by using bacterially expressed proteins. (A) SDS-PAGE and Coomassie brilliant blue staining; (B) Western analysis using affinity-purified anti–troponin C antibody. Lane B, Total protein of E. coli JM109; lanes 1 and 2, bacterial protein from cDNA clone, pCTNC-1; lanes 3 and 4, mutant troponin C clone, pCPAT-10-m1; lanes 5 and 6, mutant troponin C clone, pCPAT-10-m2; lane N, total protein of the wild-type N2. Sample solution contained Ca2+ + (1 mM CaCl2, lanes 1, 3, and 5) and Ca2+ − (5 mM EGTA, lanes 2, 4, and 6), respectively. (C) Blotted sheet incubated with bacterially produced troponin I followed by detection with affinity-purified anti–TNI-1. For C, samples and conditions were the same as in A and B. It was noted that PAT-10-m1 did not show a band shift with and without calcium (B, lanes 3 and 4) and PAT-10-m2 did not bind troponin I (C, lanes 5 and 6). For details see the text.
Construction of Plasmids Used in Microinjection
Various upstream and internal regions of the troponin C gene, pat-10 of C. elegans were inserted into pPD transformation vectors (Fire et al. 1990) in-frame with the E. coli lacZ reporter gene. DNA fragments from 7.6 kb of BamHI containing 7,600 bp upstream of the first ATG at 1,146 and to 108 bp of the second exon were cloned into the BamHI site of pPD22.11. Series of another fragments deleting the 5′ upstream end of pat-10; 1,248 bp of PstI-BamHI, 647 bp of ApaLI-BamHI, and 292 bp of EcoRV-BamHI were also ligated to the processed vector. The number of constructed plasmid indicate the numbers of nucleotide from the first ATG at 1,146 (see Fig. 6). Preparation of plasmid DNAs for injection and transformation of C. elegans were carried out as was described (Mello et al. 1991).
Figure 6.

Tissue-specific expression of the pat-10/lacZ genes. A fusion gene containing 5′ UTS of pat-10 and lacZ was used to study cellular expression. (A) Staining of body wall muscles, pTNCZ647; (B) pTNCZ292; (C) Expression of anterior part. It was noted that pharyngeal muscle was not stained. (D and E) pTNCZ647, side and top views of the vulva of young adult, respectively. Bar, 50 μm. (F and G) Summary of 5′ UTS of pat-10 and body wall–specific expressions. The position of regulatory sequences were shown: D, 1330/hlh-1 recognition; G, GC box; M, MEF2-binding sites, respectively.
Transformation Rescue
We generated extra-chromosomal array stEx14 by coinjecting pTNC1 and pRF4 at final concentrations of 2 mg/ml and 200 mg/ml, respectively, into wild-type N2 hermaphrodites using standard methods (Mello et al. 1991). The pRF4 plasmid contains the dominant rol-6(su1006) marker, permitting us to follow extra-chromosomal array segregation. To test for rescue of the pat-10(st568) homozygotes, +/+; stEx14 hermaphrodites were crossed with unc-38(e20) pat-10(st568)/++ males, generating the heterozygous strain unc-38(e20)pat-10(st568)/++; stEx14. This strain was permitted to self-fertilize, and rescued animals with genotype unc-38(e20)pat-10(st568); stEx14 were recognized by their Unc Rol double mutant phenotype. Individual Unc Rol animals were then picked to establish the rescued homozygous strain RW1577 unc-38(e20)pat-10(st568); stEx14, which segregates the Rol Unc parental class and a significant fraction of Pat developmentally arrested progeny due to the sporadic loss of the stEx14 extra-chromosomal array.
It is noteworthy that our initial attempts to generate extra-chromosomal arrays containing pTNC1 may have been complicated by the deleterious effects of troponin C over expression, the consequence of extra-chromosomal arrays that are likely to contain many concatenated copies of the pTNC1 plasmid sequences (Mello et al. 1991). We had difficulty obtaining stable transformed strains in our initial experiments, and the few lines that we did generate were characterized by slow locomotion and slow growth. In subsequent experiments we reduced the ratio of pTNC1 relative to the pRF4 marker plasmid to the levels indicated above. These conditions are likely to generate arrays with a lower pTNC1 copy number, and permitted us to isolate transformed lines that grew well, had no apparent defect in locomotion, and were useful for the rescue experiments.
Fixation and X-Gal Staining
Reagent preparation, fixation, and X-Gal staining were performed as was described (Mello et al. 1991). We used vectors incorporating a nuclear localization peptide at the NH2 terminus of β-galactosidase; this leads to predominant staining in the nuclei of expressing cells, facilitating cell identification (Fire et al. 1990).
Protein Analysis and Immunological Methods
SDS-PAGE.
Troponin C from the recombinant proteins produced in bacterial cells were analyzed by SDS-PAGE under the protocols and analyzed as was described (Kagawa et al. 1989, Kagawa et al. 1995). Total proteins obtained from the nematode or from bacteria cultured in the presence of IPTG and control bacteria were boiled after addition of 2× Laemmli sample buffer. Amino acid sequence analysis was done by GENETYX-MAC (Software Development Co. Ltd.).
Antibody Preparation and Immunostaining.
For antibody preparation bacterially expressed troponin C from cDNA clone pCTNC1 was prepared by 60% of ammonium sulfate fractionation followed by 0.25 M NaCl fraction of ion exchange chromatography. Antiserum was prepared from a rabbit raised with purified troponin C and was affinity-purified as follows. Worms from a full growth plate (9 cm) were treated with 2× Laemmli buffer and run on SDS-PAGE followed by blotting to nitrocellulose membrane (Amersham Corp.). Corresponding band visualized with Ponceau red (in 5% acetic acid; Sigma Chemical Co.) was cut from a membrane and used for specific antibody adsorption. This membrane can be used for preparing antibody several times (Kagawa et al. 1995). Anti–troponin I antibody was also prepared using essentially the same procedure (Kuroda et al., unpublished data). Immunostaining employed freeze-cracking for young larvae and β-mercaptoethanol-collagenase treatment for adults, respectively (Barstead and Waterston 1989; Williams and Waterston 1994).
Results
Identification and Structure of a Troponin C Gene of C. elegans
Initially, we screened a C. elegans cDNA expression library with a polyclonal antiserum raised against purified Ascaris troponin C (Nakae and Obinata 1993), and recovered a 0.7-kb cDNA encoding a 161 residue polypeptide with extensive homology to troponin C from other organisms (see below, Fig. 3). To isolate the corresponding genomic sequence and identify its chromosomal location, we hybridized the cDNA to a filter containing a gridded set of yeast artificial chromosome (YAC) clones that span most of the nearly complete C. elegans genomic physical map (Coulson et al. 1988). We detected strong hybridization to a set of four overlapping YAC clones on chromosome I in the interval defined by the unc-38 and dpy-5 genes (see Fig. 1A and Fig. B). We further mapped the hybridizing region to cosmids F54C1 and C15C10 (Fig. 1 B), and ultimately to a 3.5-kb PstI fragment (Fig. 1 C) which contains the entire troponin C gene, pat-10 (see below, Fig. 2).
Figure 3.

Comparison of amino acid sequences of troponin C-1 of C. elegans with a second troponin C of C. elegans and those of Drosophila, crayfish, and rabbits. Alignment of C. elegans troponin C, CeTNC-1 (pat-10), CeTNC-2 (cosmid ZK673.7), Drosophila troponin C Dm73F (Fyrberg et al. 1994), crayfish type (Kobayashi et al. 1989), rabbits, cardiac (Wilkinson 1980) and skeletal (Zot et al. 1987). Four calcium-binding sites are shown (I–IV). Mutation sites in the pat-10(st575) mutant are shown at 64 D to N and 153 W to termination (*), respectively. Shading indicates the residues that are identical in these six troponin C proteins.
A comparison of the cDNA and genomic sequences for this troponin C revealed a gene with 6 exons (Fig. 1 D and 2). A RACE analysis showed that the cDNA is truncated by only 14 base pairs at the 5′ end, and suggests that troponin C transcripts are not transpliced with either of the leader sequences that are appended to some C. elegans transcripts (Blumenthal and Steward 1997). The 3′ end of the cDNA ends in a short poly A tract that starts just downstream from two consensus polyadenylation signals (Fig. 2). The deduced transcript structure is consistent with the single 0.7-kb band that we detect on Northern blots (data not shown).
A multiple sequence alignment between this nematode troponin C and several other troponins from vertebrate and invertebrate organisms is shown in Fig. 3. We have included a second potential C. elegans troponin C (CeTNC-2), which was identified on cosmid clone ZK673 by the C. elegans genome sequencing consortium. CeTNC-1 is most similar to the other invertebrate proteins, and has overall sequence identities of 48.2, 44.7, and 40.4% to C. elegans CeTNC-2, Drosophila TNC73F (Fyrberg et al. 1994), and the crayfish troponin C alpha isoform (Kobayashi et al. 1989), respectively. Sequence identifies to the rabbit cardiac and skeletal isoforms are 33.9 and 28.6%, respectively. CeTNC-1 contains the four homologous EF-hand motifs that are characteristic of troponin C (Fig. 3), and within these regions sequence identity to the other troponins is ∼60%. The Drosophila Dm73F and the crayfish alpha isoforms have several inappropriately charged residues at the ion coordination sites within EF-hand domains I and III (Fig. 3), suggesting that these domains can not bind calcium ions (Kobayashi et al. 1989; Collins et al. 1991). Site I of the rabbit cardiac muscle protein may also be nonfunctional. All four sites of the rabbit skeletal isoform appear to be functional. The negatively charged residues are a highly conserved feature of these metal binding sites and are likely to be essential for their function (Putkey et al. 1989; Marsden et al. 1990).
Expression Profile of Troponin C in C. elegans
Using both promoter fusion reporter constructs and antibodies to troponin C, we determined that this protein is expressed in body wall, vulval, and anal muscles, but not in the pharynx. Using the 5′ upstream promoter region of pat-10 fused to lacZ we observed staining in the body wall muscles, the vulva muscles, and the minor muscles of the anal region (Fig. 6). From the expression profile of pTNCZ292 it appears that only two hundred base pairs of the 5′ upstream region is necessary for proper expression of this gene (Fig. 6 G). Analysis of this promoter region revealed Sp1 recognition-like sequences (CCCGCCC) at positions 1061 and 1091, corresponding −63 and −33 bp upstream from the transcription start site of the gene (Fig. 2 and Fig. 6 F; Dynan and Tjian, 1985). There is a GC box and a body wall–specific enhancer (1330/hlh-1) recognition sequence at positions 942 and 1055 (Fig. 2 and Fig. 6 F; Krause et al. 1994). Expression of pTNCZ248 which deletes this region at 942 was dramatically decreased in the body wall muscles (Fig. 6 G). This 44-bp region has 66.7% identity to the 1330/hlh-1 enhancer sequence. We also observed CAACTAG sequences within intron 3 which are known to be MyoD-binding sites and may have enhancer activity (Parmacek et al. 1992). These sequences correspond to the M: MEF2-binding sites ATTTTT (Cserjesi and Olson 1991) indicated in Fig. 6, which appear in both intron 3 and in 5′ upstream sequence. Fig. 7A–E, shows that pat-10/lacZ gene expression commences at the comma bean stage of worm development. This time of expression is consistent with what has been observed for other muscle structural proteins (Krause et al. 1994; Kagawa et al. 1995). Interestingly, fusion plasmid pTNC647 expressed also in C. briggsae HK104 and HK105, animals which are isolated in Okayama and Sendai, respectively (data not shown).
Muscle expression of troponin C was confirmed by indirect immunofluorescence staining of wild-type animals (Fig. 8 and Fig. 9). An antibody was generated to nematode troponin C expressed in bacteria (see Materials and Methods), and in nematode protein extracts a band of the expected size (∼18 kD) was detected on SDS-PAGE gels as were larger bands (Fig. 4 B, lanes 1 and 3). These larger bands may be complexes of troponin C with troponin I and troponin I plus troponin T. It is known that troponins strongly bind to each other and can form troponin complexes (Fig. 4 B, lane 1; Grabarek et al. 1990). Using this antisera, we confirmed the expression pattern detected by the reporter constructs. Body wall muscle expression can be seen in embryos and carries on throughout the life of the animal (Fig. 8 and Fig. 9). In older animals vulva and anal muscle expression can also be observed (Fig. 8C and Fig. D). Some staining of pharyngeal muscle may come from cross-reaction with the second troponin C isoform, as suggested by the high degree of sequence homology between these proteins (data not shown).
Figure 8.
Immunostaing of the worm with affinity-purified anti-TnC. Staining of whole worm (A). The arrowhead indicates vulva muscles. Mail tail (B). High magnification of vulva muscles (arrowhead) and anus muscles (double arrowhead) (C). Vulva and body wall muscles are shown higher magnification (D). Immuno- and phalloidin-stained animals of the head region, respectively (E and F). These results indicate that troponin C protein locates in body wall and other minor muscles except pharyngeal muscles. Bars: (A–C and E) 0.1 mm; (D) 0.02 mm, respectively.
Figure 9.
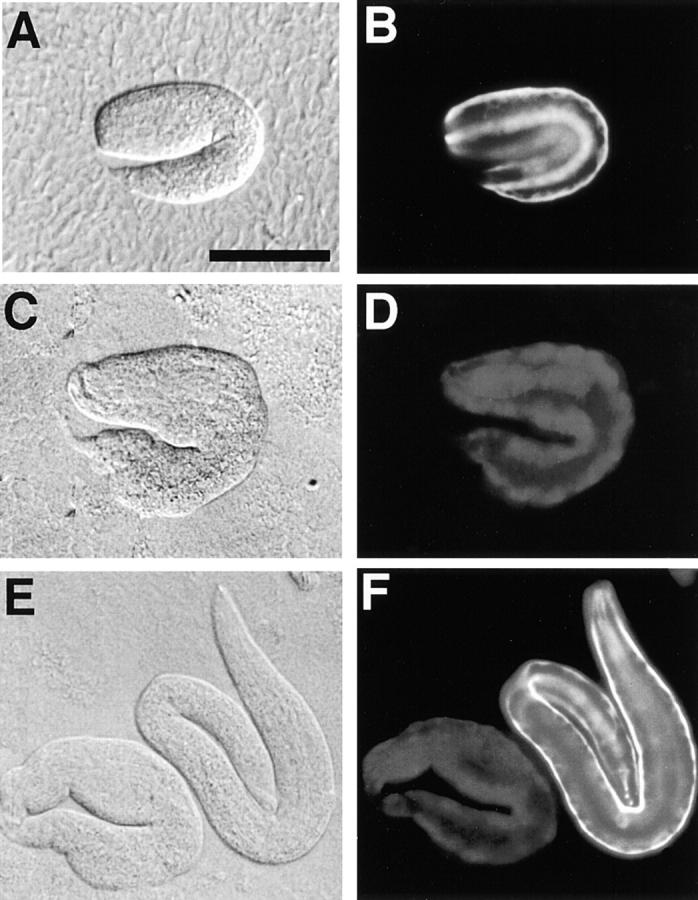
Nomarsky and anti–troponin C immunostaining images of wild-type and pat-10 animals. Staining of the wild-type (B) and pat-10 (st575) animals at the two hold stage (D and F), Nomarsky images of both (A, C, and E). The muscle filaments of the wild-type embryo were visible in a regular array (B and F). In contrast to wild-type, the tail portion of a mutant larva at the threefold stage has an irregular morphology (C and E). Bar, 50 μm.
Figure 4.
Western blot identification of troponin C from a worm extract and bacterial expression. Protein fractions were separated on 10–20% gels and either stained with Coomassie brilliant blue (A) or transferred to nitrocellulose for indirect probing with anti–Ascaris-troponin C antiserum (B). Protein fractions are the immunotransfers contained in a total protein extract from the wild-type N2 (lane 1), bacterial protein from the troponin I clone, pCTNI-1 (lane 2), bacterial protein from the troponin C clone, pCTNC-1 (lane 3), bacterial protein from bluescript (lane 4), respectively. Molecular size marker (M).
The Structural Gene of Troponin C Is pat-10
The genetic map position and the muscle-affecting phenotype of pat-10 suggested that it may be the structural gene encoding the troponin C described here. This previously defined genetic locus maps to the interval between unc-38 and dpy-5 (see Fig. 1 A) and was identified through mutations that cause paralysis of embryonic body wall muscles, and ultimately the characteristic Pat (paralyzed, arrested elongation at twofold) phenotype (Williams and Waterston 1994). This developmental arrest phenotype has been associated with several muscle-affecting genes (Waterston 1988; Williams and Waterston 1994; Kagawa et al. 1997; Moerman and Fire 1997) and thus made this a likely candidate gene for the troponin C. As a first step to confirm this possibility, we showed that pat-10 mutants can be rescued by a transgenic extra-chromosomal array containing clone Y55F5 which we have shown by hybridization includes the troponin C gene (Fig. 1 B). To test directly whether pat-10 mutants can be rescued by a troponin C transgene, we generated an extra-chromosomal array carrying the pTNC1 subclone (Fig. 1 D) and found that it rescues the developmental arrest phenotype of pat-10(st568) mutant homozygotes. These rescuing plasmids contain complete exon and intron fragments, together with 1,145 bp upstream of the 5′ noncoding region (Fig. 6 G). These results are consistent with the premise that pat-10 is the structural gene for troponin C.
To confirm that pat-10 is the structural gene for troponin C, we sequenced two mutant alleles of pat-10 and found corresponding dramatic changes in the troponin C gene. The results confirm that pat-10 is the troponin C gene. In pat-10(st575) we found changes at position 1890, changing D65 to N in the second calcium-binding site, and at position 2179, altering W153 to a stop codon thus deleting the COOH-terminal H-helix of troponin C. In the other mutation, pat-10(st568), we also found same alterations (Fig. 2 and Fig. 3). One would expect both of these changes in the troponin C to be severe alterations. Staining of the putative pat-10(st575) animals with an antibody to troponin C shows that body wall muscle staining is no longer present (Fig. 9D and Fig. F). This experiment both serves to demonstrate that mutant troponin C did not assemble to filaments by the reason of functional defects of the molecule (see later). These combined studies confirm that pat-10 is the structural gene for troponin C.
A Mutation in Troponin C Affects Calcium and Troponin I Binding
In the process of making an antibody to troponin C we noted that bacterially expressed wild-type nematode troponin C appeared to have intact calcium-binding capability. We were able to detect mobility shifts of troponin C produced in bacteria in the presence or absence of calcium on SDS-PAGE followed by Western blot analysis (Fig. 5A and Fig. B, lanes 1 and 2). We took advantage of this observation to analyze the character of mutationally altered troponin C (Fig. 5A and Fig. B, lanes 3–6 and C, lanes 3–6). We analyzed two constructs, PAT-10-m1 with a D64N alteration and PAT-10-m2 with a W153 to nonsense change.
In the first instance, PAT-10-m1 with a substitution within a calcium-binding site appeared to lose the ability to band shift based on the presence or absence of calcium ions (Fig. 5A and Fig. B, lanes 3 and 4). Interestingly, PAT-10-m2 could still band shift but had a faster mobility due to its smaller size (Fig. 5A and Fig. B, lanes 5 and 6).
We also analyzed whether or not these altered molecules could bind troponin I in a protein overlay assay (Fig. 5 C). In this case PAT-10-m1 still retained the ability to bind troponin I in a pattern similar to wild-type troponin C (Fig. 5 C, lanes 3 and 4). In contrast PAT-10-m2 had lost troponin I–binding capacity (Fig. 5 C, lanes 5 and 6). These in vitro observations suggest what the functional consequences of these single base alterations may be, and why these alterations may lead to lethality, i.e., a Pat terminal phenotype.
Discussion
We have determined the genome position, structure, and sequence of a troponin C gene pat-10 of C. elegans and have shown that it corresponds to the pat-10 locus that was previously identified as a gene affecting muscle development and morphogenesis (Williams and Waterston 1994; Kagawa et al. 1997). The troponin C gene, pat-10, maps to the center of chromosome I (Fig. 1) and encodes a protein of 161 amino acid residues (Fig. 2) and a molecular mass of 18 kD (Fig. 4). We have sequenced two mutations in this gene and these occur at the second and near the fourth calcium-binding site in both. Both appear to be functional alleles since they lead to the embryonic terminal phenotype. Our biochemical experiments show that the missense allele has problems binding calcium and thus regulating contraction. On the other hand, the truncated product in vitro, while able to bind calcium, is unable to bind troponin I effectively (Fig. 5 C). We cannot detect the truncated protein in our in situ studies using antibodies (Fig. 9D and Fig. F). The truncated protein may simply be turned over too rapidly for detection by the reason of its functional defects.
The terminal phenotype of pat-10 animals was previously described in detail (Williams and Waterston 1994). These animals reach the one- and half-fold stage of embryonic development and then become paralyzed. Shortly thereafter they also cease elongation at the twofold stage of embryonic development (Fig. 7, F–I, and 9, C and E). This terminal phenotype corresponds quite well with the earliest expression pattern we see for troponin C (Fig. 7, A–D). Similar to other filament associated proteins we first detect it at ∼350–400 min in body wall muscle and later in development we see it in other muscle types except for the pharynx (Fig. 6 C). In agreement with this PAT-10 expression pattern, pharyngeal pumping is observed in homozygous pat-10 animals. Genes with Pat alleles are a rather large class and contain genes whose products affect several different steps in myofilament assembly, stability, and contractile regulation (reviewed in Moerman and Fire 1997). Pat-10 would appear to fall in the latter class since it does not affect myofilament initiation (Williams and Waterston 1994). Included in this group are mup-2, which encodes troponin T (Myers et al. 1996) and tmy-1/lev-11, which encodes tropomyosin (Kagawa et al. 1995, Kagawa et al. 1997). Mutations in none of these three genes affect sarcomere assembly, but all act later, after muscle contraction has begun. At this time, they become essential for proper sarcomere function which is reflected in lack of movement of the late embryo and lack of continued morphogenesis in the form of embryonic elongation (Fig. 9).
In mammals, troponin C is encoded by a multi-gene family with both cardiac- and skeletal-specific isoforms. Troponin C from pat-10 is most similar to that of cardiac types (Fig. 3; Gahlmann and Kedes 1990; Kagawa et al. 1997). Whether one can make much of this is not clear since nematode muscle has several features in common with either skeletal or cardiac muscle. The pat-10 locus encodes only a single transcript and the tnc-1/lacZ fusion gene is expressed in a limited set of muscle tissues within the nematode including the body wall muscles, the vulval muscles and the anal muscles (Fig. 6; Kagawa et al. 1995). One muscle tissue that we did not detect expression in was the pharynx which also was unaffected in pat-10 mutants. There is presumably a separate troponin C for this tissue. We have found a second nematode troponin C in the EST database of Y. Kohara (National Institute Genetics, Mishima, Japan). This troponin C (designated CeTNC-2 in Fig. 3) is a candidate for the pharyngeal isoform. Location of TNC-2 within the animal is in progress in our laboratory (unpublished observation). Only two members of troponin C were found in a search of complete genome sequence. Compared with vertebrates and even many invertebrates, for example, Drosophila, which has three genes encoding troponin C (Fyrberg et al. 1994), the nematode has few copies of this regulatory gene. Troponin C is viewed as a specialized calmodulin that has added sites for troponin T and I binding and in this context it is useful to note that several calmodulin related genes have been described by Salvato et al. 1986 and noted by the nematode genome consortium. A sequenced genome and comparative analysis between species may help us discern the evolution of troponin C from calmodulin and the specialization of this molecule for different muscle types. What is clear from this study is that a model system with few troponin C isoforms is more amenable to a combined genetic and molecular analysis.
This is the first report on the effects of troponin C mutations in vivo. However, there are reports of in vitro mutagenesis-generated alterations in troponin C and there effects on conformational changes between EF-hand structures (Xu et al., 1988), the NH2-terminal helix (Reinach et al., 1988) and calcium-binding sites (Putkey et al. 1989). It has long been known that troponin C has an EF-hand structure and changes conformation during calcium binding (Herzberg and James 1988; Strynadka et al. 1997). The observation that a D64N alteration in a single EF-hand may lead to lack of contraction and embryonic lethality both affirms and extends these earlier observations on the importance of calcium binding for proper functional regulation. This interpretation also supports the observations that showed that troponin C was not stained with anti–troponin C in pat-10(st575) mutant (Fig. 9D and Fig. F). It is known from biochemical studies of other troponin Cs that only EF-hand II known as the so-called low-affinity calcium-binding site responds to conformation change (Putkey et al. 1989; Marsden et al. 1990; Grabarek et al. 1992). Three negative charges in each of EF-hands are essential for calcium binding (Marsden et al. 1990). According to this rule, troponin C of the nematode could bind two calcium at EF-hand II and IV (Fig. 3). Substitution from D64 to N in PAT-10-m1 is the reason of the lost of a band shift on SDS-PAGE (Fig. 5 B, lanes 3 and 4). The major effect of this mutation is therefore on regulating contraction by calcium since the troponin complex, as monitored by the ability of troponin I to bind troponin C, appears intact (Fig. 5B and Fig. C, lanes 3 and 4).
Whereas much is known about troponin I and C interactions (Tripet et al. 1997), the precise regions on troponin C important for these interactions is unclear. Here we show that troponin I binding is independent of the second calcium-binding site in troponin C but dependent on the COOH-terminal region of troponin C (Fig. 5 C, lanes 3–6). Within the last H helix region of troponin C methionine 156 and glycine 158 are conserved in all reported sequences (Fig. 3) which might be a clue to help define the region important for troponin I binding. This result is consistent with the recent structural study that shows that this region makes contact with troponin I (Vassylyev et al. 1998). The troponin complex, consisting of troponins C, T, and I binds to actin filaments in muscle and regulates contraction via calcium released from the sarcoplasmic reticulum. Curiously isolated, both pat-10(st568) and pat-10(st575) alleles had two mutations at the same position. It is interesting to know which mutation affects the terminal phenotype. Analysis on a transgenic animal having one of each mutations give us the answer. In vivo analysis on mutants having a defect of calcium binding or troponin I binding allows us to solve the question how in vitro result affects filament assembly and contraction. In the nematode, mutations in troponin T (Myers et al. 1996), tropomyosin (Kagawa et al. 1995, Kagawa et al. 1997) and now troponin C (this study) have been described. In our laboratory we have cloned two troponin I genes and these are currently under study (unpublished observations). Therefore, we now have in hand the entire tropomyosin/troponin thin filament regulatory complex for contraction. A careful genetic dissection of this complex with a judicious choice of mutations could lead to further functional details of thin filament regulated contraction.
Acknowledgments
The authors are grateful to R.H. Waterston for his encouragement, stimulation, and generous help and to D. Moerman for his correction of English usage and encouragement. Genome mapping with a YAC library screening and cosmid clones was mainly done by A. Coulson and J. Sulston. We also thank to N. Toyota for his comment on cloning, Y. Tanaka for his suggestion on mobility-shift assay for calcium binding, A. Fire for lacZ reporter injection vectors, and T. Iio for a comment of calcium-binding sites of the troponin C.
Wild-type and mutant worms were provided by the Caenorhabditis Genetic Center, which is funded by the National Institutes of Health National Center for Research Resources (NCRR). This work was supported by grants to H. Kagawa from The Ministry of Education, Science, Sports and Culture of Japan and Japan Society for the Promotion of Science (Research for the Future 97L00401).
Footnotes
1.used in this paper: 5′ RACE, 5′ rapid amplification of cDNA ends; UTS, untranslated sequence; YAC, yeast artificial chromosome
References
- Anderson P. Molecular genetics of nematode muscle. Annu. Rev. Genet. 1990;23:507–525. doi: 10.1146/annurev.ge.23.120189.002451. [DOI] [PubMed] [Google Scholar]
- Barstead R.J., Waterston R.H. The Basal component of the nematode dense-body is binculin. J. Biol. Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- Blumenthal T., Steward K. RNA processing and gene structure. In: Riddle D.L., editor. C. elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 117–145. [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard B., Kevin L., Larkins A., Butcher G., Karlik C., Fyrberg E. Troponin of asynchronous flight muscle. J. Mol. Biol. 1988;204:621–637. doi: 10.1016/0022-2836(88)90360-9. [DOI] [PubMed] [Google Scholar]
- Collins J.H., Greaser M.L., Potter J.D., Horn M.J. Determination of the amino acid sequence of troponin C from rabbit skeletal muscle. J. Biol. Chem. 1977;252:6456–6462. [PubMed] [Google Scholar]
- Collins J.H., Theibert J.L., Francois J.-M., Ashley C.C., Potter J.D. Amino acid sequences and Ca2+-binding properties of two isoforms of barnacle troponin C. Biochemistry. 1991;30:702–707. doi: 10.1021/bi00217a017. [DOI] [PubMed] [Google Scholar]
- Coulson A., Waterston R.H., Kiff J., Sulston J., Kohara Y. Genome linking with yeast artificial chromosomes. Nature. 1988;335:184–186. doi: 10.1038/335184a0. [DOI] [PubMed] [Google Scholar]
- Cserjesi P., Olson E.N. Myogenin induces the myocyte-specific enhancer binding factor MEF-2 independently of other muscle specific gene products. Mol. Cell. Biol. 1991;11:4854–4862. doi: 10.1128/mcb.11.10.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W.S., Tijan R. Control of eukaryotic messenger RNA synthesis by sequence specific DNA-binding proteins. Nature. 1985;316:774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Ebashi S., Endo M. Calcium ion and muscle contraction. Prog. Biophys. Mol. Biol. 1968;18:123–183. doi: 10.1016/0079-6107(68)90023-0. [DOI] [PubMed] [Google Scholar]
- Fire A., Harrison S.W., Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans . Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Fujimori K., Sorenson M., Herzberg O., Moult J., Reinach F.C. Probing the calcium-induced conformational transition of troponin C with site-directed mutants. Nature. 1990;345:182–184. doi: 10.1038/345182a0. [DOI] [PubMed] [Google Scholar]
- Fyrberg C., Parker H., Hutchison B., Fyrberg E.A. Drosophila melanogaster genes encoding three troponin-C isoforms and a calmodulin-related protein. Biochem. Genet. 1994;32:119–135. doi: 10.1007/BF00554420. [DOI] [PubMed] [Google Scholar]
- Gahlmann R., Kedes L. Cloning, structural analysis, and expression of the human fast twitch skeletal muscle troponin C gene. J. Biol. Chem. 1990;265:12520–12528. [PubMed] [Google Scholar]
- Gengyo-Ando K., Kagawa H. Single charge change on the helical surface of the paramyosin rod dramatically disrupts thick filament assembly in Caenorhabditis elegans . J. Mol. Biol. 1991;219:429–441. doi: 10.1016/0022-2836(91)90184-8. [DOI] [PubMed] [Google Scholar]
- Grabarek Z., Wang T.R.-Y., Tao J.T., Gergely J. Inhibition of mutant troponin C activity by an intra-domain disulphide bond. Nature. 1990;345:132–135. doi: 10.1038/345132a0. [DOI] [PubMed] [Google Scholar]
- Grabarek Z., Tao T., Gergely J. Molecular mechanism of troponin-C function. J. Muscle Res. Cell Mot. 1992;13:383–393. doi: 10.1007/BF01738034. [DOI] [PubMed] [Google Scholar]
- Harris H.E., Tso M.-Y.W., Epstein H.F. Actin and myosin-linked calcium regulation in the nematode Caenorhabditis elegans. Biochemical and structural properties of native filaments and purified proteins. Biochemistry. 1977;16:859–917. doi: 10.1021/bi00624a008. [DOI] [PubMed] [Google Scholar]
- Herzberg O., James M.N.G. Refined crystal structure of troponin C from turkey skeletal muscle at 2.0 A resolution. J. Mol. Biol. 1988;203:761–779. doi: 10.1016/0022-2836(88)90208-2. [DOI] [PubMed] [Google Scholar]
- Kagawa H., Gengyo K., McLachlan A.D., Brenner S., Karn J. Paramyosin gene (unc-15) of Caenorhabditis elegans; molecular cloning nucleotide sequence and models for thick filament structure. J. Mol. Biol. 1989;207:311–333. doi: 10.1016/0022-2836(89)90257-x. [DOI] [PubMed] [Google Scholar]
- Kagawa H., Sugimoto K., Matsumoto H., Inoue T., Takuwa K., Sakube Y. Genome structure, mapping and expression of the tropomyosin gene tmy-1 of Caenorhabditis elegans . J. Mol. Biol. 1995;251:603–613. doi: 10.1006/jmbi.1995.0459. [DOI] [PubMed] [Google Scholar]
- Kagawa H., Takuwa K., Sakube Y. Mutations and expressions of the tropomyosin gene and the troponin C gene of Caenorhabditis elegans . Cell Struct. Funct. 1997;22:213–218. doi: 10.1247/csf.22.213. [DOI] [PubMed] [Google Scholar]
- Kimura K., Tanaka T., Nakae H., Obinata T. Troponin from nematodepurification and characterization of troponin from Ascaris body wall muscle. Comp. Biochem. Physiol. 1987;88B:399–407. [Google Scholar]
- Kobayashi T., Takagi T., Konishi K., Wnuk W. Amino acid sequences of the two major isoforms of troponin C from Crayfish. J. Biol. Chem. 1989;264:18247–18259. [PubMed] [Google Scholar]
- Krause M., Harrison S.W., Xu S.-Q., Chen L., Fire A. Elements regulating cell- and stage-specific expression of the C. elegans MyoD family homolog hlh-1 . Dev. Biol. 1994;166:133–148. doi: 10.1006/dbio.1994.1302. [DOI] [PubMed] [Google Scholar]
- Marsden B.J., Shaw G.S., Sykes B.D. Calcium binding proteins. Elucidating the contributions to calcium affinity from an analysis of species variants and peptide fragments. Biochem. Cell Biol. 1990;68:587–601. doi: 10.1139/o90-084. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. Efficient gene transfer in C. elegans extra chromosomal maintenance and integration of transforming sequence. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman, D.G., and A. Fire. 1997. Muscle: structure and function, development. In C. elegans II. D.L. Riddle, T. Blumenthal, B.J. Meyer, and J. Priess, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 417–470. [PubMed]
- Myers C.D., Goh P.-Y., Allen T.St.C., Bucher E.A., Bogaert T. Developmental genetic analysis of troponin T mutations in striated and nonstriated muscle cells of Caenorhabditis elegans . J. Cell Biol. 1996;132:1061–1077. doi: 10.1083/jcb.132.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae H., Obinata T. Immunocytochemical localization of troponin I and C in the muscles of Caenorhabditis elegans . Zool. Sci. 1993;10:375–379. [Google Scholar]
- Ohtsuki I., Maruyama K., Ebashi S. Regulatory and cytoskeletal proteins of vertebrate skeletal muscle. Adv. Prot. Chem. 1986;38:1–67. doi: 10.1016/s0065-3233(08)60525-2. [DOI] [PubMed] [Google Scholar]
- Ojima T., Tanaka H., Nishita K. Cloning and sequence of a cDNA encoding Akazara scallop troponin C. Arch. Biochem. Biophys. 1994;311:272–276. doi: 10.1006/abbi.1994.1237. [DOI] [PubMed] [Google Scholar]
- Parmacek M.S., Leiden J.M. Structure and expression of the murine slow/cardiac troponin C gene. J. Biol. Chem. 1989;264:13217–13225. [PubMed] [Google Scholar]
- Parmacek M.S., Bengur A.R., Vora A., Leiden J.M. The structure and regulation of expression of the murine fast skeletal troponin C gene. J. Biol. Chem. 1990;265:15970–15976. [PubMed] [Google Scholar]
- Parmacek M.S., Vora A.J., Shen T., Barr E., Jung F., Leiden J.M. Identification and characterization of a cardiac-specific transcriptional regulatory element in the slow/cardiac troponin C gene. Mol. Cell. Biol. 1992;12:1967–1976. doi: 10.1128/mcb.12.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkey J.A., Sweeney H.L., Campbell S.T. Site-directed mutation of the trigger calcium-binding sites in cardiac troponin C. J. Biol. Chem. 1989;264:12370–12378. [PubMed] [Google Scholar]
- Reinach F.C., Karlsson R. Cloning, expression, and site-directed mutagenesis of chicken skeletal muscle troponin C. J. Biol. Chem. 1988;263:2371–2376. [PubMed] [Google Scholar]
- Salvato M., Sulston J., Albatross D., Brenner S. A novel calmodulin-like gene from the nematode Caenorhabditis elegans . J. Mol. Biol. 1986;190:281–290. doi: 10.1016/0022-2836(86)90002-1. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch F.E., Maniatis T. Molecular Cloning. 2nd edition. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schreier T., Kedes L., Gahlmann R. Cloning, structural analysis, and expression of the human slow twitch skeletal muscle/cardiac troponin C gene. J. Biol. Chem. 1990;265:21247–21253. [PubMed] [Google Scholar]
- Smith L., Greenfield N.J., Hitchcock-DeGregori S.E. The effects of deletion of the amino-terminal helix on troponin C function and stability. J. Biol. Chem. 1994;269:9857–9863. [PubMed] [Google Scholar]
- Strynadka N.C., Cherney M., Sielecki A.R., Li M.X., Smillie L.B., James M.N.J. Structural details of a calcium-induced molecular switch; X-ray crystallographic analysis of the calcium-saturated N-terminal domain of troponin C at 1.75 A resolution. J. Mol. Biol. 1997;273:238–255. doi: 10.1006/jmbi.1997.1257. [DOI] [PubMed] [Google Scholar]
- Sulston, J., and J. Hodgkin. 1988. Methods. In The Nematode Caenorhabditis elegans. W.B. Wood, editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 578–686.
- Tripet B., Van Eyk J.E., Hodges R.S. Mapping of a second Actin-tropomyosin and a second troponin C binding site with the C-terminus of troponin I, and their importance in the Ca2+-dependent regulation of muscle contraction. J. Mol. Biol. 1997;271:728–750. doi: 10.1006/jmbi.1997.1200. [DOI] [PubMed] [Google Scholar]
- Vassylyev D.G., Takeda S., Wakatsuki S., Maeda K., Maeda Y. Crystal structure of troponin C in complex with troponin I fragment at 2.3-A resolution. Proc. Natl. Acad. Sci. USA. 1998;95:4847–4852. doi: 10.1073/pnas.95.9.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H. 1988. Muscle. In The Nematode Caenorhabditis elegans. W.B. Wood, editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 281–335.
- Wilkinson J.M. Troponin C from rabbit slow skeletal and cardiac muscle is the product of a single gene. Eur. J. Biochem. 1980;103:179–188. doi: 10.1111/j.1432-1033.1980.tb04302.x. [DOI] [PubMed] [Google Scholar]
- Williams B.D., Waterston R.H. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.-Q., Hitchcock-DeGregori S.E. Synthesis of a troponin C cDNA and expression of wild-type and mutant proteins in Escherichia coli . J. Biol. Chem. 1988;263:13962–13969. [PubMed] [Google Scholar]
- Zot A.S., Potter J.D., Strauss W.L. Isolation and sequence of a cDNA clone for rabbit fast skeletal muscle troponin C. J. Biol. Chem. 1987;262:15418–15421. [PubMed] [Google Scholar]