Abstract
Cell physiology in the weevil Sitophilus oryzae is coordinated by three integrated genomes: nuclear, mitochondrial, and the “S. oryzae principal endosymbiont” (SOPE). SOPE, a cytoplasmic bacterium (2 × 103 bacteria per specialized bacteriocyte cell and 3 × 106 bacteria per weevil) that belongs to the proteobacteria γ3-subgroup, is present in all weevils studied. We discovered a fourth prokaryotic genome in somatic and germ tissues of 57% of weevil strains of three species, S. oryzae, Sitophilus zeamais, and Sitophilus granarius, distributed worldwide. We assigned this Gram-negative prokaryote to the Wolbachia group (α-proteobacteria), on the basis of 16S rDNA sequence and fluorescence in situ DNA–RNA hybridization (FISH). Both bacteria, SOPE and Wolbachia, were selectively eliminated by combined heat and antibiotic treatments. Study of bacteria involvement in this insect’s genetics and physiology revealed that SOPE, which induces the specific differentiation of the bacteriocytes, increases mitochondrial oxidative phosphorylation through the supply of pantothenic acid and riboflavin. Elimination of this γ3-proteobacterium impairs many physiological traits. By contrast, neither the presence nor the absence of Wolbachia significantly affects the weevil’s physiology. Wolbachia, disseminated throughout the body cells, is in particularly high density in the germ cells, where it causes nucleocytoplasmic incompatibility. The coexistence of two distinct types of intracellular proteobacteria at different levels of symbiont integration in insects illustrates the genetic complexity of animal tissue. Furthermore, evolutionary timing can be inferred: first nucleocytoplasm, then mitochondria, then SOPE, and finally Wolbachia. Symbiogenesis, the genetic integration of long-term associated members of different species, in the weevil appears to be a mechanism of speciation (with Wolbachia) and provides a means for animals to acquire new genes that permit better adaptation to the environment (with SOPE).
Intracellular symbiosis, or endosymbiosis, is the most sophisticated association between eukaryotic cells and intracellular microorganisms. It implies the coordination between associated genomes (1) forming a new entity, the symbiocosm (2). This concept encompasses the endosymbiotic theory dealing with the origin of plastids and mitochondria (3, 4). In insects, endosymbioses often display specialized adaptations such as the differentiation of host cells or bacteriocytes (5). Furthermore, endosymbionts are often intimately involved in several metabolic pathways of the host, and most cannot divide by themselves in vitro (6–8). The association is generally considered as mutualistic. However, the current knowledge on symbiosis shows a continuum between parasitic and mutualistic relationships. A striking example, which rather evokes parasitism, is the Wolbachia symbiosis. This intracellular rickettsia-like organism is widespread in arthropods and is known to alter host reproduction in mainly three ways: cytoplasmic incompatibility (CI) (9, 10), parthenogenesis (11), and feminization of genetic males (12). Moreover, recent work demonstrated that Wolbachia may also be virulent, causing tissue degeneration and early death in Drosophila (13).
The rice weevil Sitophilus oryzae (Coleoptera, Rhynchophoridae) shares an endosymbiotic association with a so-called Sitophilus oryzae principal endosymbiont (SOPE), a Gram-negative bacterium belonging to the Enterobacteriaceae family (14). This symbiosis has been investigated for more than 70 years (2). Investigators have shown that SOPE supplies the weevil with vitamins, such as pantothenic acid, riboflavin, and biotin (15), and interacts with mitochondrial oxidative phosphorylation by increasing mitochondrial enzymatic activity (16), thus enhancing greatly the flight ability of adult insects (17). It also interacts with amino acid metabolism (18, 19). All these features indicate that SOPE is greatly integrated in the host physiology. Moreover, the SOPE genome apparently has undergone genetic changes throughout its intracellular life history (20), and its expression is partly controlled by the host (21). We interpret these observations to indicate a long period of microorganism–insect coadaptation. Here, we show with 16S rDNA sequencing and fluorescence in situ hybridization (FISH) that the weevil symbiocosm is complicated by the coexistence of four genomes: nuclear, mitochondrial, SOPE, and a fourth intracellular genome of the Wolbachia group (α-proteobacteria). Combined heat and antibiotic treatments resulted in a selective elimination of both types of endosymbiotic bacteria, which allowed study of their involvement in the biology of the insect. The comparison between SOPE and Wolbachia leads to the conclusion that they represent two different levels of organism integration in the host biology, and it illustrates the genetic complexity of eukaryotic cells.
MATERIALS AND METHODS
Insect Rearing.
Experiments were conducted with two S. oryzae strains: a Chinese strain (Ch) from Guangzhou which was shown to harbor both SOPE and Wolbachia, and a French laboratory strain (SFr) harboring only SOPE. Weevils were reared on wheat at 27.5°C and 70% relative humidity.
16S rDNA Cloning and Sequencing.
Total DNA was isolated from the Chinese strain as described by Heddi et al. (14). Primers used for 16S rDNA amplification were the eubacterial universal primers: 27for 5′-AGAGTTTGATCATGGCTCAG-3′ [nucleotides 8–27, Escherichia coli numbering (GenBank accession no. J01859)] and 1487rev 5′-TACCTTGTTACGACTTCACC-3′ (nucleotides 1487–1506). Reaction mixtures for PCR amplification consisted of 1.5 units of Taq DNA polymerase (Appligene, Illkirch, France), 1.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphate, 0.3 μM primers, and 10 ng of DNA template in a final volume of 50 μl. The PCR parameters were 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 53°C for 45 s, and 72°C for 45 s. PCR products were cloned in a pMOSBlue vector (Amersham), and positive clones were characterized with specific endonuclease digestions. Two panels were identified and two representative clones of each panel were sequenced as described previously (14), to detect possible PCR misincorporations. The clones from each panel were found to be identical.
FISH Procedure.
Insect tissues were fixed in alcoholic Bouin’s solution and embedded in paraffin. Five-micrometer-thick sections were mounted on poly(l-lysine)-coated microscope slides. After toluene dewaxing and rehydration, sections were digested in 100 μg/μl pepsin in 0.01 M HCl for 10 min at 37°C. Specific oligonucleotide probes were designed by sequence alignment of Wolbachia and SOPE 16S rDNA. Two Wolbachia probes 5′ end labeled with rhodamine were used to increase the signals: W1, 5′-AATCCGGCCGARCCGACCC-3′, and W2. 5′-CTTCTGTGAGTACCGTCATTATC-3′. SOPE probe (S) was 5′ end labeled with fluorescein: S, 5′-TACCCCCCTCTACGAGACTC-3′. Hybridization was performed at 45°C in a dark moisture chamber in 0.9 M NaCl/20 mM Tris⋅HCl/5 mM EDTA/0.1% SDS/10× Denhardt’s solution. After a 30 min preincubation period, 50 ng of each probe (W1, W2, and S) was added and incubation was continued for 3 hr. Slides were washed twice in the same buffer at 48°C for 20 min, rinsed with distilled water, and mounted in Vectashield medium (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole (DAPI).
Wolbachia and SOPE Detection in Sitophilus Populations.
Individuals were tested for the presence or the absence of Wolbachia and SOPE by PCR and FISH. For PCR, specific primers were designed on the 16S gene: forward 5′-CGGGGGAAAAATTTATTGCT-3′, reverse 5′-AGCTGTAATACAGAAAGTAAA-3′, generating a-589 bp fragment for Wolbachia; and forward 5′-TAATAGCGCCATCGATTGAC-3′, reverse 5′-CCGAAGGCACCAAGGCAT-3′, generating a 530-bp fragment for SOPE. PCR parameters were described above, except for the hybridization temperature (TH), 50°C (30 s) for Wolbachia and 53°C (30 s) for SOPE.
Insect Treatment and Genetic Analysis.
Female fertility and development time of S. oryzae progeny were measured from crosses between naturally symbiotic Chinese strain (Ch), or its SOPE-aposymbiotic heat-treated relative (ChH), and their tetracycline-treated derivatives (ChT and ChHT). ChH insects were obtained by heat treatment of adults: 35°C and 90% relative humidity for 1 month, followed by female oviposition under normal conditions (22). Tetracycline-treated insects were fed two generations on wheat flour pellets mixed with tetracycline (1 mg/g of flour). To avoid possible direct effects of treatments, virgin pairs of adults were fed for 2 weeks on wheat grains before crossing. Progenies from a 14-day egg-laying were counted to estimate fertility per female and from a 2-day egg-laying to measure the development time from egg-laying to adult emergence. Wolbachia and SOPE were detected by FISH and PCR.
Mitochondrial Enzymatic Assays.
Mitochondria-specific enzymatic activities were assayed by using the procedure of Heddi et al. (16). For vitamin supplementation experiments, insects were reared during one generation on wheat flour artificial pellets supplemented with pantothenic acid (2.38 mg/kg of wheat flour) and with riboflavin (1.88 mg/kg of wheat flour).
RESULTS
Phylogenetic Position of S. oryzae Endosymbionts.
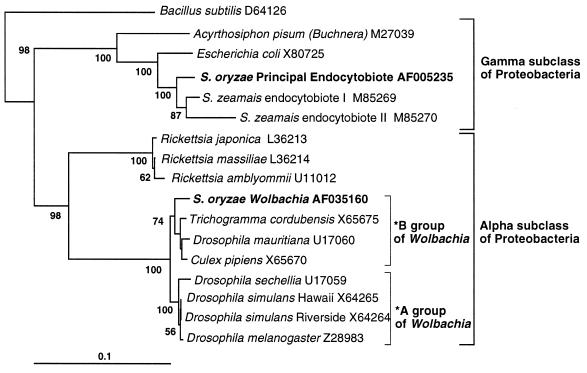
The cloning and sequencing of PCR products from reactions using 16S rDNA universal primers on total DNA from a Chinese strain of S. oryzae (Ch) revealed two different sequences. One sequence was identical to SOPE 16S rDNA (14) and the other belongs to the α-proteobacteria. Fig. 1 positions these sequences relative to several representatives of the α and γ subdivisions of proteobacteria. Whereas SOPE is found within the Enterobacteriaceae, the other sequence (GenBank accession no. AF035160) falls into the B-group of Wolbachia (23), with which it forms a monophyletic group relative to the other α-proteobacteria. The 16S rDNA sequence of S. oryzae Wolbachia is over 97% identical to Wolbachia known from other insects.
Figure 1.
Phylogenetic tree of the SOPE and Wolbachia based on sequences of 16S rDNA genes. The other rDNA sequences were searched at the National Center for Biotechnology Information by using the blast network service. Alignments were performed and the phylogenetic tree was created as in ref. 14. Bacillus subtilis, a Gram-positive bacterium, was used as the out-group. The name of the bacteria or the host insect species is followed by the GenBank accession number. Scale bar, 0.1 substitution per site. *According to the classification of Werren et al. (23).
SOPE and Wolbachia Distribution in Sitophilus Populations.
Twenty-three Sitophilus strains, 10 individuals of each, belonging to three species (S. oryzae, S. zeamais, and S. granarius), were collected from different locations worldwide and tested by using specific PCR primers and FISH experiments for the presence of SOPE and Wolbachia (Table 1). While all strains and individuals were SOPE-symbiotic, only 13 of 23 strains were Wolbachia-symbiotic, constituting 9 strains totally infected and 4 strains partially infected.
Table 1.
SOPE and Wolbachia distribution in Sitophilus (weevil) populations
| Species | Strain (collection date) | Source of weevil | Presence of Wolbachia | No. of Wolbachia, positive weevils per 10 tested |
|---|---|---|---|---|
| S. oryzae | GuadI (83) | Guadeloupe | − | 0 |
| GuadII (83) | Guadeloupe | + | 9 | |
| ChI (83) | China | + | 7 | |
| ChII (83) | China | − | 0 | |
| Ind (85) | India | − | 0 | |
| Aust (90) | Australia | + | 10 | |
| TunI (81) | Tunisia | + | 10 | |
| TunII (81) | Tunisia | + | 10 | |
| Ben (92) | Benin | + | 10 | |
| Hai (85) | Haiti | + | 5 | |
| S. zeamais | Syr (85) | Syria | − | 0 |
| AzoI (90) | Azores | − | 0 | |
| AzoII (90) | Azores | − | 0 | |
| AzoIII (90) | Azores | − | 0 | |
| MexI (84) | Mexico | − | 0 | |
| MexII (84) | Mexico | − | 0 | |
| Cong (84) | Congo | + | 10 | |
| Thai (94) | Thailand | + | 10 | |
| Peru (90) | Peru | + | 10 | |
| Reun (79) | Réunion | + | 10 | |
| S. granarius | FrI (82) | France | − | 0 |
| FrII (82) | France | + | 10 | |
| FrIII | France | + | 2 |
SOPE was found in all studied Sitophilus strains. SOPE and Wolbachia were detected by PCR and FISH experiments.
Tissue Distribution of SOPE and Wolbachia.
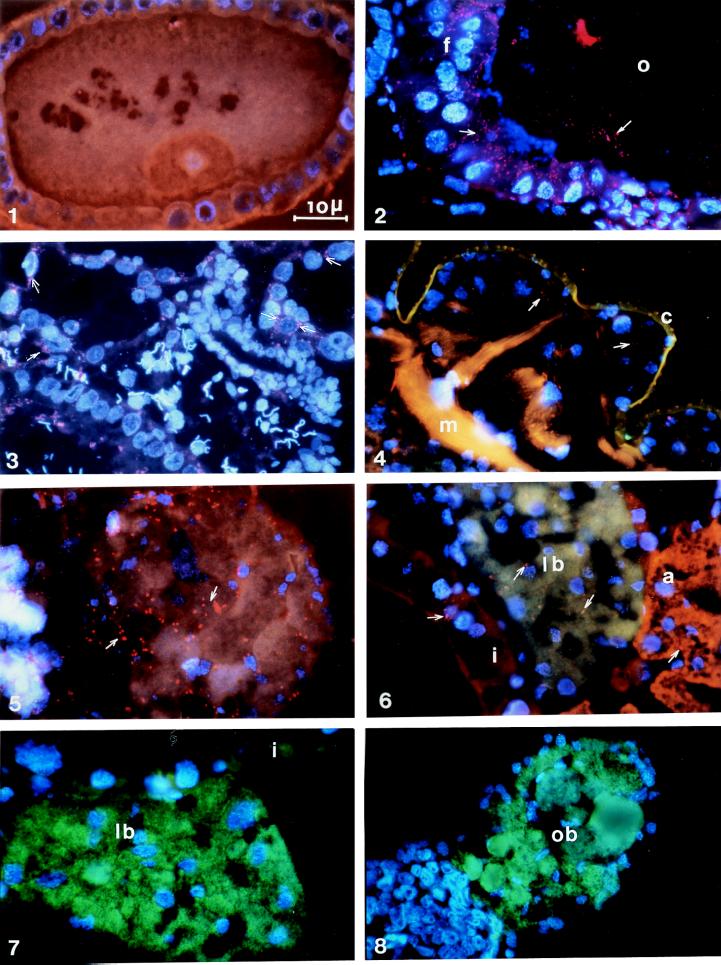
The distribution of Wolbachia and SOPE in weevil tissues was investigated with the FISH approach (24) (see Fig. 2). Oligonucleotide probes (19–23 nucleotides) were designed by sequence alignment of the Wolbachia and the SOPE 16S rDNA sequences. These probes were shown to be highly specific when applied together on sections: (i) devoid of SOPE and Wolbachia (Fig. 2 1, oocyte negative control, no signal), (ii) containing Wolbachia only [2, oocyte (o) and follicular cells (f) of a SOPE-aposymbiotic strain, red spots only], and (iii) containing both Wolbachia and SOPE (6, red spots and green color, respectively). The principal endosymbiont is limited to specialized tissue: the larval bacteriome (specialized bacterial organ, lb) (6 and 7), the ovarian bacteriome (ob) (8), and to female germ cells. It is absent from all other tissue, including male germ cells (3). Bacteriomes fail to differentiate in the absence of the principal endosymbiont even when Wolbachia is present (Fig. 2 5 shows the apex of the ovary lacking the bacteriome). Bacteriome differentiation apparently is induced by the integrated endosymbiotic bacterium of the weevils.
Figure 2.
FISH of Wolbachia (red) and SOPE (green) in different weevil tissues. (1 and 2) Comparison between the Wolbachia-negative control (1) [oocyte from the SOPE-aposymbiotic Chinese strain (ChH) treated with tetracycline (ChHT)] and the Wolbachia-positive control (2) [oocyte (o) and follicular cells (f) of the SOPE-aposymbiotic strain (ChH)]. (3) Testes of the SOPE- and Wolbachia-symbiotic strain (Ch) (see arrows, Wolbachia near spermatid nuclei). (4) Cuticle (c) and muscle (m) of the SOPE- and Wolbachia-symbiotic strain (Ch). (5) SOPE-aposymbiotic apex of ovary (ChH). (6) Intestine (i), the larval bacteriome (lb) in green and the adipocytes (a) of the Ch strain: both SOPE (green) and Wolbachia (red spots, arrows) are visible in the bacteriocytes. (7 and 8) Larval bacteriome (lb) and the ovarian bacteriome (ob) of the French Wolbachia-free strain (SFr). All slides were hybridized with the three probes (W1, W2, and S; see Materials and Methods). (Scale bar, 10 μm.)
Wolbachia (red spots of about 0.5 μm), in contrast, is disseminated throughout the whole body of the insect and particularly is in high density in the male and female germ cells. It is located around the periplasmic membrane of the oocyte and in the surrounding follicular cells (Fig. 2 2, follicular cells are packed full of Wolbachia). In testis, Wolbachia is near many spermatid nuclei (3, see arrows; 4′,6-diamidino-2-phenylindole blue dye indicates the spermatid nuclei), suggesting its interaction with the male chromosomes. Everywhere else, Wolbachia is present only at low density and exhibits a weak signal intensity. It can be seen in the cuticle (c) and muscles (m) (4), in adipocytes (a) and the intestine (i) (6), and in the bacteriocytes, coexisting with SOPE (6) and representing less than 5% of the total bacterial population. In the FISH experiment conditions, the fluorescent signal is proportional to the concentration of 16S rRNA in cells. These transcripts represent the FISH probe targets, and their quantity may indicate the level of the bacterial cell metabolic activity. We interpret high levels of fluorescence of Wolbachia within germ cells (2, 3, and 5), relative to soma line (4 and 6), to suggest a high metabolic activity of this bacterium in the male and female germ line.
Differential Influence of the Two Symbionts.
To investigate separately the influence of both SOPE and Wolbachia, genetic analysis was conducted with different crosses between individuals treated or not with heat and/or tetracycline (Table 2). Heat treatment disrupted the SOPE symbiosis in S. oryzae. Tetracycline treatment for two generations removes both Wolbachia and SOPE. Relative to other insects such as Drosophila or Trichogramma, Wolbachia of weevils are resistant to the heat treatment (evidence given by PCR and FISH experiments). Table 2 displays four salient results. First, Wolbachia seems not to be much involved in weevil physiology: development times in crosses 6 to 8 differ only slightly from the development time of cross 5 (perhaps because of a secondary effect of the antibiotic). Second, unidirectional cytoplasmic incompatibility is seen clearly when females but not males are treated with tetracycline, whatever the SOPE status of the strain (crosses 3 and 7). Third, tetracycline treatment of males reestablishes high levels of fertility, which are almost restored to normal in ChHT × ChHT (as compared with the corresponding untreated cross, ChH × ChH), but which are only partly restored in the cross ChT × ChT, because of the concomitant lack of SOPE in the ChT strain. Finally, heat treatment decreases female fertility and increases the insect development time (compare crosses 1 and 5). The most harmful cross is when Ch females are tetracycline treated and males are not. This generates the weakest fertility. In the experiment to measure the development time only one weevil with a development time of 48 days has emerged, indicating the action of both Wolbachia (cytoplasmic incompatibility) and SOPE (decreased fertility).
Table 2.
Female fertility and development time of S. oryzae progeny from crosses between naturally symbiotic Chinese strain (Ch), or its SOPE-aposymbiotic heat-treated relative (ChH), and their tetracycline-treated derivatives (ChT and ChHT)
| Parents
|
Progeny
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cross
|
Female
|
Male
|
Mean fertility, 14-d progeny | nb1 | Mean development time, days | nb2 | |||
| No. | F × M | W | S | W | S | ||||
| 1 | Ch × Ch | + | + | + | + | 57.60 ± 6.37 | 10 | 34.81 ± 0.27 | 159 |
| 2 | Ch × ChT | + | + | − | − | 51.71 ± 4.35 | 7 | 33.99 ± 0.35 | 46 |
| 3 | ChT × Ch | − | − | + | + | 4.80 ± 2.29* | 5 | 48.00∗ | 1 |
| 4 | ChT × ChT | − | − | − | − | 18.40 ± 4.37* | 5 | 35.66 ± 0.26 | 92 |
| 5 | ChH × ChH | + | − | + | − | 28.85 ± 1.05 | 20 | 38.17 ± 0.43 | 78 |
| 6 | ChH × ChHT | + | − | − | − | 25.51 ± 1.47 | 21 | 41.25 ± 0.49∗ | 51 |
| 7 | ChHT × ChH | − | − | + | − | 6.84 ± 1.15* | 17 | 40.04 ± 0.76∗ | 26 |
| 8 | ChHT × ChHT | − | − | − | − | 21.15 ± 2.32* | 20 | 44.41 ± 0.30∗ | 138 |
Wolbachia (W) and SOPE (S) were detected by FISH and PCR using respective specific 16S probes and primers. 14-d progeny, mean progeny per female from a 14-day egg-laying of 2- to 3-week-old females (results are mean ± SE); nb1, pair number; nb2, progeny number from a 2-day egg-laying; +, presence; −, absence. ∗, Significant differences compared with the control crosses of each group—i.e., 1 for 2, 3, 4, and 5 for 6, 7, 8 (bilateral Dunnett test, α = 0.05).
Metabolic Interaction Between Nucleus, SOPE, and Mitochondria.
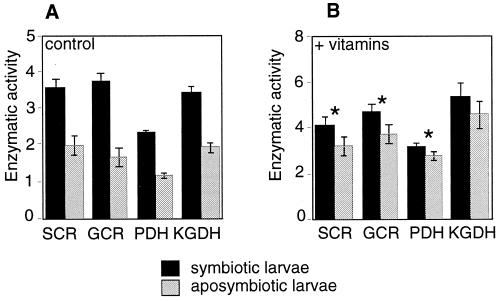
In a previous experiment (16), we have shown that SOPE improves mitochondrial enzymatic activity. Since SOPE supplies the host with vitamins (pantothenic acid and riboflavin), we have investigated in the present study the effect of vitamin supplementation on mitochondrial enzymatic activities of the weevils. Thus, vitamin supplementation experiments were conducted on wheat flour artificial pellets. Fig. 3 shows clearly that mitochondrial enzymatic activities of SOPE-aposymbiotic individuals are much more affected than SOPE-symbiotic ones, when reared on vitamin-supplemented pellets. As a result, differences between the strains are significantly attenuated for three enzymes of four (ANOVA2 test, P > 0.05), suggesting that SOPE influences the weevil energy metabolism principally through nutritional supply. However, we do not exclude that vitamin supplementation could influence other metabolic pathways of the host.
Figure 3.
Specific enzymatic activities of mitochondrial suspensions isolated from SOPE-symbiotic and SOPE-aposymbiotic larvae of a Wolbachia-free French strain (SFr). (A) Control wheat flour artificial pellets, not supplemented with vitamins. (B) Pellets supplemented with pantothenic acid and riboflavin. Data are expressed as the mean of at least 10 repeats ±SD. SCR, succinate cytochrome c reductase (y unit = 1 μmol⋅mg−1⋅s−1); GCR, glycerol-3-phosphate cytochrome c reductase (y = 0.5 μmol⋅mg−1⋅s−1); PDH, pyruvate dehydrogenase (y = 0.1 μmol⋅mg−1⋅s−1); KGDH, α-ketoglutarate dehydrogenase (y = 0.2 μmol⋅mg−1⋅s−1). The effect of vitamins on mitochondrial activities (i.e., interaction between vitamin and symbiosis factors) was found to be significant (∗, P < 0.05) in three of the four independent ANOVA2 tests performed.
DISCUSSION
Discovered by Pierantoni in 1927 (25) and studied by several authors (2), S. oryzae symbiosis remains poorly described at the molecular level. Using two complementary approaches (i.e., PCR–cloning–sequencing and FISH), we demonstrate clearly in this work that some Sitophilus strains harbor two separate endosymbionts: S. oryzae principal endosymbiont (SOPE), a Gram-negative bacterium belonging to the γ-proteobacteria, and a second intracellular bacterium identified as Wolbachia (α-proteobacteria) (Fig. 1). SOPE, in the Enterobacteriaceae family, shares 95% sequence homology with Escherichia coli (14). The closest intracellular symbiotic bacterium is the primary endosymbiont of S. zeamais (97.8% homology), the sibling species of S. oryzae. The Wolbachia of S. oryzae belongs to the B-group of Wolbachia (23), which causes cytoplasmic incompatibility in several insect genera such as Aedes, Drosophila, and Culex (9, 10, 26).
Histologically, Wolbachia exhibits different features from SOPE (Fig. 2). It is disseminated in tissues throughout the whole body of S. oryzae, including follicular cells, oocytes, testis, adipocytes, and intestine, and even within the bacteriocytes, where it coexists with SOPE. However, the density of Wolbachia is very tissue variable. It is highly abundant in the germ cells, but it is present at low density in other tissues. In line with this finding is the weak fluorescent signal observed in these tissues compared with the germ cells, where it is well known that Wolbachia interacts actively with the host nuclei, modifying the developing sperm in testis (possibly by means of chromatin-binding proteins) (10, 26, 27). In contrast, SOPE is tolerated only by the bacteriocytes and the female germ cells, by an as-yet-unknown mechanism. Bacteriocytes differentiate only in the presence of SOPE. SOPE-aposymbiotic insects completely lack bacteriomes, even when they are Wolbachia-symbiotic (see Fig. 2 5). The differentiation of the bacteriocytes suggests, therefore, the existence of a specific molecular signal from the SOPE to the host genome during embryogenesis. Moreover, the number of SOPE cells is controlled by the host genome (28). Therefore, a long period of SOPE–host coadaptation relative to Wolbachia seems necessary to establish such a specific interacting system. These observations tempt us to consider Wolbachia as a more recent and less integrated endosymbiont than SOPE. Paillot (29) considered parasitism a first step toward symbiosis, as exhibited by the Amoeba model (30). Wolbachia might be an attenuated parasite that only recently infected the weevils.
The coexistence of both proteobacteria in the weevil shows the metabolic and the genetic complexity existing in such eukaryotic cells. Four genomes interact, with each playing a different role in the insect’s biology. The α-proteobacterium Wolbachia seems much less involved in weevil physiology, since its elimination from the insect cells by the antibiotic treatment only slightly affects the larval development time. Reproduction is greatly impaired by the presence of Wolbachia because of cytoplasmic incompatibility between Wolbachia and the weevil nuclear genome, independent of the γ-proteobacteria principal endosymbiont (Table 2). The Wolbachia-correlated reproductive abnormality may be the result of an asynchronous condensation of parental chromatin, which leads to the formation of embryos with aneuploid or haploid nuclei (27).
SOPE greatly enhances female fertility and decreases larval development time. Furthermore, SOPE influences the behavior of weevil populations by enhancing the flight ability of the host (17). These energy-dependent performances were mediated by the SOPE effect on mitochondrial oxidative metabolism (16). Mitochondrial enzymatic complexes that form the phosphorylation chain are encoded by mitochondrial DNA (13 polypeptides) and nuclear genome (over 60 other polypeptides) (31). Nuclear proteins are imported into mitochondria, where they interact with mitochondrial polypeptides and with coenzymes, partly originating from vitamins, to construct enzymatic complexes involved in ATP synthesis. Coordination between associated genomes in the cytosol is thus crucial for aerobic energy production. This study shows that the improvement of weevil mitochondrial enzymatic activities by SOPE is essentially due to the pantothenic acid and riboflavin supply (Fig. 3). SOPE does not exhibit any effect on mitochondrial DNA replication or expression (unpublished data). Pantothenic acid and riboflavin, vitamins in low quantity in wheat, are directly involved in biosynthesis of CoA and NAD+–FAD, respectively. Both are required cofactors for mitochondrial enzyme function. We interpret SOPE to be a third genome implicated in ATP production in weevil cells.
Wolbachia and SOPE are subjected to different evolutionary constraints: SOPE (like mitochondria) is transmitted vertically only, and is strictly located in bacteriocytes and ovaries. It is present in all strains of Sitophilus and in all individuals. Wolbachia, on the other hand, is transmitted both vertically and horizontally (32), and it is disseminated throughout body cells and is at particularly high density in the ovaries and testes. It does not infect all populations.
The Sitophilus oryzae model of symbiosis is consistent with “serial endosymbiotic theory” (3). Endosymbiosis did not occur just once in eukaryotic evolution with nucleus origin (33), or even twice when an anaerobic protist acquired a respiring bacterium to give rise to the mitochondrion or hydrogenosome (34). Nor did bacterial endosymbiont acquisition occur only three times when plants acquired plastid ancestors (35). Acquisition of bacterial genomes by eukaryotic cells indeed continues “today” in the multicellular organism. Two types of acquired microbial genomes may be present in a single animal cell in addition to mitochondria. In Sitophilus, after mitochondria established in the protist ancestor, the next endosymbiont was SOPE and the third was Wolbachia. Endosymbionts contribute to the emergence of a new symbiocosm or type of cell where different genomes interact. The most important consequence is production of more viable offspring, indicating better adaptation to the environment. The colonization of cereal plants by Sitophilus was facilitated by the acquisition of SOPE, a vitamin supplier. We consider symbiosis in the rice weevil a sophisticated mechanism for acquiring new sets of genes.
Acknowledgments
We thank C. Nardon for FISH technical assistance, V. James for English corrections, and Y. Rahbé and C. Biémont for critical reading of the manuscript. This work was supported by the Institut National des Sciences Appliquées, the Institut National de la Recherche Agronomique, and the Conseil régional du Rhône.
ABBREVIATIONS
- SOPE
Sitophilus oryzae principal endosymbiont
- FISH
fluorescence in situ hybridization
Footnotes
Data deposition: The 16S rDNA sequence of Wolbachia reported in this paper has been deposited in the GenBank database (accession no. AF035160).
References
- 1.Saffo M B. In: The Unity of Evolutionary Biology. Proceedings of the 4th International Congress of Systematics and Evolutionary Biology. Dudley E C, editor. Portland, OR: Dioscorides; 1991. pp. 674–680. [Google Scholar]
- 2.Nardon P, Grenier A-M. Ann Soc Entomol Fr. 1993;29:113–140. [Google Scholar]
- 3.Margulis L. Symbiosis in Cell Evolution. 2nd Ed. New York: Freeman; 1993. [Google Scholar]
- 4.Taylor F J R. Ann N Y Acad Sci. 1987;503:1–16. doi: 10.1111/j.1749-6632.1987.tb40595.x. [DOI] [PubMed] [Google Scholar]
- 5.Buchner P. Endosymbiosis of Animals with Plant Microorganisms. New York: Wiley Interscience; 1965. [Google Scholar]
- 6.Nardon P, Grenier A-M. In: Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. Margulis L, Fester R, editors. Cambridge, MA: MIT Press; 1991. pp. 153–169. [PubMed] [Google Scholar]
- 7.Baumann P, Lai C-Y, Baumann L, Rouhbaksh D, Moran N A, Clark M A. Appl Environ Microbiol. 1995;61:1–7. doi: 10.1128/aem.61.1.1-7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumann P, Moran N A, Baumann L. Bioscience. 1997;47:12–20. [Google Scholar]
- 9.Breeuwers J A J, Werren J H. Nature (London) 1990;346:558–560. doi: 10.1038/346558a0. [DOI] [PubMed] [Google Scholar]
- 10.O’Neill S L, Karr T L. Nature (London) 1990;348:778–780. doi: 10.1038/348178a0. [DOI] [PubMed] [Google Scholar]
- 11.Stouthamer R, Breeuwer J A J, Luck R F, Werren J H. Nature (London) 1993;361:66–68. doi: 10.1038/361066a0. [DOI] [PubMed] [Google Scholar]
- 12.Rigaud T, Souty-Grosset C, Raimond R, Mocquard J P, Juchault P. Endocyt Cell Res. 1991;7:259–273. [Google Scholar]
- 13.Min K T, Benzer S. Proc Natl Acad Sci USA. 1997;94:10792–10796. doi: 10.1073/pnas.94.20.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heddi A, Charles H, Khatchadourian C, Bonnot G, Nardon P. J Mol Evol. 1998;47:52–61. doi: 10.1007/pl00006362. [DOI] [PubMed] [Google Scholar]
- 15.Wicker C. Comp Biochem Physiol. 1983;76A:177–182. [Google Scholar]
- 16.Heddi A, Lefèbvre F, Nardon P. Insect Biochem Mol Biol. 1993;23:403–411. [Google Scholar]
- 17.Grenier A-M, Nardon C, Nardon P. Entomol exp Appl. 1994;70:201–208. [Google Scholar]
- 18.Gasnier-Fauchet F, Gharib A, Nardon P. Comp Biochem Physiol. 1986;85B:245–250. [Google Scholar]
- 19.Gasnier-Fauchet F, Nardon P. Insect Biochem. 1987;17:17–20. [Google Scholar]
- 20.Charles H, Condemine G, Nardon C, Nardon P. Insect Biochem Mol Biol. 1997;27:345–350. [Google Scholar]
- 21.Charles H, Heddi A, Guillaud J, Nardon C, Nardon P. Biochem Biophys Res Commun. 1987;239:769–774. doi: 10.1006/bbrc.1997.7552. [DOI] [PubMed] [Google Scholar]
- 22.Nardon P. C R Acad Sci Paris. 1973;277D:981–984. [Google Scholar]
- 23.Werren J H, Zhang W, Guo L R. Proc R Soc London Ser B. 1995;261:55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- 24.Amann R I, Ludwig W, Schleifer K H. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierantoni U. Rend R Accad Sci Fis e Mat Napoli Ser 3a. 1927;35:244–250. [Google Scholar]
- 26.Werren J H. Annu Rev Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- 27.Callaini G, Dallai R, Riparbelli M G. J Cell Sci. 1997;110:271–280. doi: 10.1242/jcs.110.2.271. [DOI] [PubMed] [Google Scholar]
- 28.Nardon P, Grenier A-M, Heddi A. Symbiosis. 1998;25:237–250. [Google Scholar]
- 29.Paillot A. L’infection chez les Insectes—Immunité et Symbiose. Trévoux, France: Patissier; 1933. [Google Scholar]
- 30.Jeon K W. Int Rev Cytol. 1983;14:29–47. [Google Scholar]
- 31.Tsagoloff A, Myers A M. Ann Rev Biochem. 1986;55:249–288. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- 32.Grenier S, Pintureau B, Heddi A, Lassablière F, Jager C, Louis C, Khatchadourian C. Proc R Soc London. 1998;265:1441–1445. [Google Scholar]
- 33.Lake J A, Rivera M C. Proc Natl Acad Sci USA. 1994;91:2880–2881. doi: 10.1073/pnas.91.8.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sogin M L. Curr Biol. 1997;7:R315–R317. doi: 10.1016/s0960-9822(06)00147-3. [DOI] [PubMed] [Google Scholar]
- 35.Martin W, Somerville C C, Loiseaux-de Goër S. J Mol Evol. 1992;35:385–404. doi: 10.1007/BF00162973. [DOI] [PubMed] [Google Scholar]