Abstract
We used 22 fragments corresponding to the reverse transcriptase domain of copia-like retrotransposons as representatives to study the organization and distribution of these elements in the rice genome. The loci detected by these 22 fragments were assigned to 47 locations in the molecular-linkage map involving all 12 chromosomes. The distributional features of copia-like retrotransposons found in the rice genome indicated that (i) the loci detected were located mainly in one arm of each chromosome; (ii) one fragment usually detected several loci that were mapped to similar locations of different chromosomes; (iii) retrotransposons sharing high identity in nucleotide sequences were usually assigned to similar locations of the chromosomes; and (iv) concurrences of multiple loci, detected by different fragments, in similar locations or stretches of different chromosomes were common in the rice genome. We also determined that the copy number of copia-like retrotransposons in rice genome may be as low as ≈100 per haploid genome. The restricted distribution, along with low copy number, suggested that copia-like retrotransposons in rice were relatively inactive during evolution compared with those in other plants. The distributional features of the copia-like retrotransposons suggested the existence of possible lineages among the rice chromosomes, which in turn suggested that chromosome duplication and diversification may be a mechanism for the origin and evolution of the rice chromosomes. The information provided by fine mapping of the retroelements in the genetic linkage map may also be useful for gene tagging and molecular cloning.
Keywords: reverse transcriptase, mapping, molecular marker
Retrotransposons are ancient components that are ubiquitous in the genomes of higher plants (1–3). Three major retroelement groups have been characterized, including copia-like retrotransposons, gypsy-like retrotransposons, and non-long-terminal-repeat (LTR) retrotransposons. These retroelement groups can be distinguished by their structure, their organization, and the amino acid sequences of the encoded enzymes, especially in the coding region of the reverse transcriptase (4, 5). It is also known that copia-like retrotransposons comprise the major group of the retrotransposons in higher plants.
Results from recent studies indicate that retrotransposons flanking plant genes may be involved in gene duplication as well as in regulation of gene expression (6). It has also been shown that transposition of retrotransposons can be induced by stress conditions, such as pathogen infection, cell culture, and wounding (7–10). The stress-induced retrotransposon amplifications frequently cause gene mutations; thus, they may be an important contributor to genetic diversity (11, 12). Such transpositions may also be used as a tool for gene tagging and isolation. In addition, DNA polymorphisms caused by the activity of retrotransposons may be used as molecular markers for genotype identification and linkage analysis (13–15).
Other studies have shown that the copy numbers of retrotransposons vary greatly in different plant species (16). Plants such as sugar beets, maize, barley, fava beans, and also a wild rice (Oryza australiensis Domin, E genome) contain extremely large numbers of retrotransposons that are distributed throughout the chromosomes (17–22). However, information is scarce concerning the genomic distribution of retroelements in plants containing relatively small numbers of the elements. Even less is known regarding the precise locations of special groups of retroelements or the distribution of particular retrotransposons in the plant genome. This lack of knowledge has limited understanding of the role of retrotransposons in genome evolution and has also made it difficult to use retrotransposons in molecular genetic studies.
Recent studies of comparative mapping indicated that chromosomes and/or chromosomal segments in distantly related organisms often share colinearity. The arrangements of genes or DNA fragments in the chromosomal segments are remarkably similar among different organisms, indicating that these chromosomes or chromosomal segments may have a common origin. For example, Moore et al. (23) found that the genomes of the grass family can be divided into 20 chromosomal segments whose syntenic relationships can be identified in all the major grass species. The researchers further hypothesized a consensus grass genome that may reflect the genome of the ancestral grass.
All the genomes of higher organisms contain multiple chromosomes. Although it can be imagined that the multiple chromosomes of a genome may be the derivatives of one or a few ancestral chromosomes, there is little knowledge of possible relationships among the chromosomes in the same genome, other than observations of duplications of certain chromosomal segments in some genomes (e.g., see ref. 24). Thus, little is known regarding the origin and evolution of the various chromosomes in the same genome.
Cultivated rice (Oryza sativa L., A genome) has become a model system of genome research for cereal plants. Previously, we examined 23 PCR fragment clones from rice, corresponding to the reverse transcriptase domain of copia-like retrotransposons; our examination identified high heterogeneity of amino acid sequences among the retrotransposons (15). This earlier study also provided evidence of distributional polymorphism for given elements in different rice varieties, indicating that retrotransposons may be a contributory factor of genetic diversity in rice.
The present study was undertaken (i) to assess the distribution patterns of the copia-like retrotransposons in the rice genome by examining their locations in the molecular-linkage map, (ii) to examine the relationship between the distributional patterns and sequence similarity among the retroelements, and (iii) to infer, based on the information provided by distributional patterns of the retrotransposons in the rice genome, possible mechanisms for the origin and evolution of the 12 chromosomes of the genome.
MATERIALS AND METHODS
Experimental Materials.
We used 22 reverse transcriptase clones of copia-like retrotransposons of rice (15) as representatives to study the distribution of this retroelement in the rice genome. The 22 clones were divided into six subgroups according to the similarity of their deduced amino acid sequences (group I: Rrt1, Rrt3, Rrt5, Rrt8, Rrt15, and Rrt16; group II: Rrt22; group III: Rrt7, Rrt12, Rrt19, and Rrt23; group IV: Rrt4, Rrt9, Rrt10, Rrt17, and Rrt18; group V: Rrt13 and Rrt14; group VI: Rrt2, Rrt11, Rrt20, and Rrt21).
The mapping population consisted of 235 F1 individuals derived from a three-way cross: [Balilla (O. sativa spp. japonica) × Dular (O. sativa spp. indica)] × Nanjing 11 (O. sativa spp. indica). Data had already been collected for 154 restriction fragment length polymorphism loci (25), giving good coverage of all 12 chromosomes by using clones from maps from both Cornell University and the Japanese Rice Genome Research Program (24, 26). A bacterial artificial chromosome (BAC) library constructed from an indica variety, Minghui 63, with a size of nine-genome equivalent (27) was used to assess the copy number of the copia-like retrotransposons.
DNA Hybridization.
Parental polymorphisms were surveyed by using each of the 22 clones as the probe in combination with six restriction enzymes (BamHI, BglII, DraI, EcoRI, EcoRV, and HindIII). The probe/enzyme combinations that detected polymorphisms between Balilla and Dular were used to assay the individuals of the mapping population. Hybridization was conducted essentially as described (28), except that the posthybridization washing was in 0.5× SSC and 0.1% SDS once for 5 min at room temperature and twice for 15 min at 65°C. The same hybridization and washing conditions were used in BAC library screening.
Data Analysis.
The data were scored by using the scheme of a back-cross population for map construction. The chromosomal locations of the 22 retrotransposon clones on the molecular-linkage map were determined by using mapmaker/exp 3.0 with a logarithm of odds threshold of 3.0 (29).
RESULTS
Distribution of Retrotransposons on the Chromosomes.
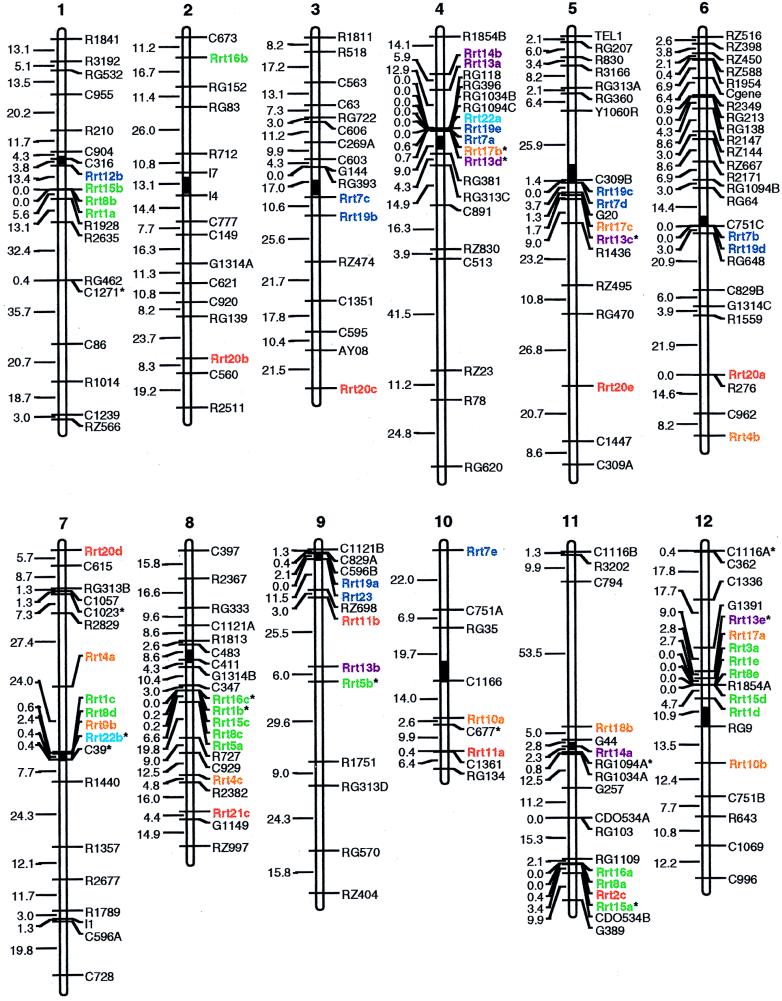
All the 22 reverse transcriptase clones of rice copia-like retrotransposons detected polymorphisms with at least one of the six restriction enzymes among the three parents of the mapping population, resulting in a total of 70 polymorphic bands. Of these, 60 were assigned to 47 locations in the 12 chromosomes of the molecular-linkage map by mapmaker analysis. The 10 remaining bands could not be assigned to any linkage group. The distributions of the retrotransposon elements represented by the polymorphic bands are illustrated in Fig. 1, from which a number of features emerged, as follows.
Figure 1.
Distribution of copia-like retrotransposon loci (Rrt) on the molecular-linkage map of rice. The retroelement loci appearing in the same color belong to the same subgroup. A lower case letter following a retrotransposon locus indicates that multiple copies were detected by the same clone. All of the loci were assigned by using a logarithm of odds score of 3.0, except those marked with asterisks (∗), which were assigned by using a logarithm of odds score of 2.0. The map distances are presented in centimorgans to the left of the chromosomes. The solid black bars represent the relative locations of the centromeres according to the results of Singh et al. (30) with reference to the information from three published molecular-linkage maps (24, 26, 34). The orientation of the chromosomes was presented according to Singh et al. (30) with the short arm placed on top.
Most of the Retroelements Existed in Multiple Loci That Were Scattered on Different Chromosomes.
The majority of the clones detected multiple loci; also, loci detected by the same clone were often scattered on different chromosomes. For example, clones Rrt5, Rrt10, Rrt11, Rrt14, and Rrt22 each detected two loci that were located on two different chromosomes. Similarly, each of three clones, Rrt4, Rrt16, and Rrt17, detected loci located on three different chromosomes. Moreover, fragments homologous to each of the three clones, Rrt1, Rrt13, and Rrt15, were detected on four different chromosomes. Finally, the loci detected by clones Rrt7, Rrt8, Rrt19, and Rrt20 showed the widest distribution; each of these four clones detected loci on five different chromosomes.
However, there were also cases in which a single given clone detected different loci on the same chromosome. Examples are Rrt13, which detected two loci on chromosome 4, and Rrt1, which detected two loci on chromosome 12.
The Retroelements Were Distributed in One Arm of the Chromosome.
A very interesting feature (shown in Fig. 1) is that the retroelements usually were distributed in only one arm of each chromosome, according to the approximate locations of centromeric regions determined by Singh et al. (30). The retroelements were located on the long arms of chromosomes 1, 3, 5, 6, 8, 9, and 11 and on the short arms of chromosomes 4, 7, and 12. The only exceptions were chromosomes 2 and 10, in which two or three retrotransposon loci, respectively, were found in both arms. The locations of the retrotransposons within the arms ranged from the centromeric regions on chromosomes 7, 9, and 11, to pericentromeric regions (within 10 centimorgans from the centromeric regions) on chromosomes 1, 3, 4, 5, 6, and 12 and to the ends of the chromosomes (chromosomes 4, 7, and 10).
The Locations for Most of the Retroelements Were Consistent Across Chromosomes.
Another interesting feature is that the locations of the loci detected by each clone on different chromosomes were usually consistent. For example, Rrt20 detected five loci distributed on chromosomes 2, 3, 5, 6, and 7. All of these five loci were mapped to the terminal or near terminal regions of these chromosomes, and four of the five loci detected by Rrt19 were located in the centromeric or pericentromeric regions. Additionally, the loci detected by the rest of the clones that identified multiple loci were also mapped to similar locations of different chromosomes.
Distribution Patterns of Loci Detected by Clones of Different Groups.
There was clearly a tendency for loci detected by clones whose DNA sequences were more similar to each other to map to similar locations of the chromosomes. For example, the large number of loci detected by the six clones of subgroup I were located on seven different chromosomes (1, 2, 7, 8, 9, 11, and 12); many of these loci appeared in clusters or cosegregated with each other. The only clone in subgroup II, Rrt22, detected certain loci that were distributed in the centromeric regions of chromosomes 4 and 7. The loci detected by clones of subgroup III were all located in the centromeric or pericentromeric regions (chromosomes 1, 3, 4, 5, 6, and 9), except that one locus detected by Rrt7 mapped to the tip of chromosome 10. The loci detected by the five clones of subgroup IV were dispersed on eight chromosomes (4, 5, 6, 7, 8, 10, 11, and 12); of these loci, those resolved by Rrt9, Rrt17, and Rrt18 were all located in centromeric or pericentromeric regions. The loci detected by the two clones of subgroup V, Rrt13 and Rrt14, were widely dispersed on five chromosomes (4, 5, 9, 11, and 12). Loci resolved by clones of subgroup VI, Rrt2, Rrt11, Rrt20, and Rrt21, showed the widest distribution range on nine chromosomes (2, 3, 5, 6, 7, 8, 9, 10, and 11); all these loci, except one detected by Rrt11 on chromosome 9, appeared at or near the terminal regions of the chromosomes.
Concurrence of the Loci Resolved by Different Retroelements.
A striking feature was the simultaneous occurrence of the loci detected by different clones across different chromosomes, including clones belonging to different groups. For example, loci detected by clones Rrt1 and Rrt8 occurred simultaneously on chromosomes 1, 7, 8, and 12, either cosegregating or tightly linked. Similar concurrence also was observed between loci detected by Rrt8 and Rrt15 on chromosomes 1, 8, 11, and 12, as well as loci detected by Rrt13 and Rrt17 on chromosomes 4, 5, and 12 and loci detected by Rrt7 and Rrt19 on chromosomes 3, 4, 5, and 6.
The concurrence of multiple loci over long chromosomal stretches across different chromosomes was even more striking. The four loci detected by Rrt7, Rrt17, Rrt13, and Rrt19 provided an example of such concurrence. These four loci were clustered in the centromeric region of chromosome 4 and were also present in a tightly linked block in the centromeric region of chromosome 5. A more remarkable example is a chromosomal block, formed of three loci detected by Rrt7, Rrt19, and Rrt20, that occurred on chromosomes 3, 5, and 6. In this block, the loci detected by Rrt7 and Rrt19 were located in the centromeric or pericentromeric regions, and loci detected by Rrt20 appeared in terminal or subterminal regions, a structure that was well conserved across the chromosomes.
Copy Number of the copia-Like Retrotransposons.
Copy number of the copia-like retrotransposons in the rice genome was estimated by examining the presence of the elements in a rice (BAC) library with coverage equivalent to nine genomes. When Rrt21, sharing 74–96% nucleotide identity to other clones of the same group (Rrt2, Rrt11, and Rrt20), was used as the probe, 140 positive BAC clones were detected in the library. We subsequently selected one clone from each of the six groups (Rrt3, Rrt22, Rrt19, Rrt4, Rrt13, and Rrt21, sharing 74–97% nucleotide identity to other clones within the respective groups). Equal amounts of DNA from these clones were mixed to provide a probe for screening the library, resulting in ≈800 positive BAC clones. Of these, 38 were chosen at random, digested with HindIII, and subjected to Southern blot analysis by using the same mixture as the probe. No hybridization signal was detected in 3 of the 38 clones; one positive fragment was detected in 25 clones; and two positive fragments were observed in the remaining 10 clones with the size of the positive fragments ranging from 1.5 kb to 10 kb. These results suggest that the majority of the positive BAC clones contained only one copy of the retroelement. Assuming the genome is randomly represented in the BAC library, the copy number of the copia-like retrotransposons is ≈100 per haploid genome in rice.
DISCUSSION
The results of this study identify several distinct features of copia-like retrotransposons in rice. The main feature is the highly restricted distribution of the retroelements in the rice genome, in which the copia-like retrotransposons, although found on all the 12 chromosomes, appeared mainly in one arm of each chromosome. Such localized distribution is clearly distinct from all other plant species studied thus far, including sugar beet, maize, barley, fava bean, and a wild rice (O. australiensis Domin), in which retroelements were detected throughout the chromosomes, except certain segments such as centromeric, telomeric, or nucleolus organizer regions (17–22).
The copy number of the copia-like retrotransposon in rice, ≈100 per haploid genome, represents another feature of the rice genome that is also distinct from other plant species. This number of copia-like retrotransposons is much smaller than those in the genomes of other plant species. For example, it was estimated that LTR-retrotransposons make up at least 50% of the maize genome (20). The genome of the sugar beet contains 2–5% copia-like retroelements and long interspersed nuclear elements (18). There are at least 3 × 104 copies of a copia-like retrotransposon, BARE-1, in barley (21). The copy number of copia-like retroelements in fava bean is ≈106 (19). In a wild rice (O. australiensis Domin), with genome size ≈2.1 times that of the cultivated rice, there are about 8 × 104 copies of a copia-like element, RIRE1 (22).
By screening a genomic DNA library with an oligonucleotide probe complimentary to the primer binding site of LTR-retrotransposons, Hirochika et al. (31) estimated that there were ≈1,000 copies of retroelements in cultivated rice. The discrepancy between their estimate and ours probably resulted from the fact that both copia-like and gypsy-like retrotransposons contain the primer binding site and the latter is also known to exist in the rice genome (3). Moreover, the oligonucleotide probe used by this group was highly homologous to plant initiator methionine tRNA genes (32). Thus, the actual copy number of the copia-like retroelements should certainly be much less than 1,000.
It has been suggested that the sizes of eukaryotic genomes are determined, to a large extent, by the amounts of repetitive DNA sequences (22, 33). It is also known that retroelements constitute a major class of repetitive DNA sequences in plants (20, 22). In rice (O. sativa L.), there are, in addition to the small number of copia-like retrotransposons as we estimated, also a small number of non-LTR retroelements, long interspersed nuclear elements (S.W., unpublished data). The small number of retroelements is clearly one of the determinants for the small genome size of rice.
Copia-like retrotransposons of plants are usually inactive. However, they can be activated under certain environmental conditions (14), and each activation of a retroelement increases the number of the element by one copy. Thus, as ancient elements, copy numbers of copia-like retrotransposons in various plant species may serve as indicators for the relative activity of the elements in different genomes during evolution. The restricted distribution along with the relatively small copy numbers of the copia-like elements in rice suggest that these elements became inactivated in the early stage of the genome evolution and have remained to be highly inactive in the process of evolution.
The distribution patterns of the copia-like retrotransposons in the rice genome have a number of important implications in the evolution of the rice genome. Our results suggest the existence of two major groups of chromosomes, according to the distribution of loci detected by various retrotransposon clones used in this study. The first group includes chromosomes 1, 7, 8, 11, and 12 and is characterized by the clusters of loci that were detected by Rrt1, Rrt5, Rrt8, Rrt15, and Rrt16 and that are located in the pericentromeric regions of all these chromosomes, except chromosome 11. The other group, represented by chromosomes 3, 5, and 6, is characterized by the concurrence of the three loci that were detected by Rrt7, Rrt19, and Rrt20 over long stretches of the chromosomes. Such distributional similarity of the retroelements on different chromosomes suggests certain syntenic relations or lineages among the chromosomes (or segments of the chromosomes). Thus, chromosomes 1, 7, 8, 11, and 12 may be in one lineage, and chromosomes 3, 5 and 6 may be in a different lineage.
Many hypotheses based on the distributional similarity of the retroelements on different chromosomes can be formulated regarding the evolution of the rice genome, especially the origin and evolution of the chromosomes. One obvious hypothesis is that the 12 rice chromosomes may have originated from one or a few ancient chromosomes. The chromosomes in the same lineage may have a common origin, created most likely by duplication of the entire chromosome followed by diversification. Most parts of the chromatin were diversified into various genes, whereas the retrotransposons that were inactivated remained to serve as the relics of the evolutionary events. If this speculation is correct, such duplication–diversification processes might have been completed long before the branching of the grass family into various genera. The distributional similarity of the retroelements also indicates that the spreading of these elements in the genome occurred largely by vertical transmission through the process of chromosome duplication, whereas the occasional occurrence of the same element on different locations of the same chromosome or different chromosomes may be an indication of horizontal transmission of the element, possibly by transposition.
It should be noted that all of our data were obtained on the basis of the bands that were polymorphic between the parents of the mapping population. Consequently, many of the retrotransposon loci may not have been detected because of the lack of polymorphisms between the parents. It should also be pointed out that some of the Rrt bands that were detected by different clones but mapped to the same location may actually represent the same element, because the sequences are highly homologous among some of the Rrt probes.
The retroelements may provide a useful tool for gene tagging and isolation. It is known that plant genes are frequently flanked by retroelements (6, 20); thus, retrotransposons may be useful molecular markers for gene isolation. Transposition of retroelements under various conditions, which have been found in both the original genome from which the element was isolated and in heterologous species into which the element was introduced by transformation (8, 11), may be used as a means for gene tagging (6, 13). In particular, the relatively small number of copia-like retrotransposons in the rice genome may provide an excellent opportunity for using retroelements for tagging particular genes.
Acknowledgments
This project was supported by National Natural Science Foundation of China Grant 39670384 and by a grant from the Rockefeller Foundation.
ABBREVIATIONS
- LTR
long terminal repeat
- BAC
bacterial artificial chromosome
References
- 1.Flavell A J, Dunbar E, Anderson R, Pearce S R, Hartley R, Kumar A. Nucleic Acids Res. 1992;20:3639–3644. doi: 10.1093/nar/20.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voytas D F, Cummings M P, Konieczny A, Ausubel F M, Rodermel S R. Proc Natl Acad Sci USA. 1992;89:7124–7128. doi: 10.1073/pnas.89.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suoniemi A, Tanskanen J, Schulman A H. Plant J. 1998;13:699–705. doi: 10.1046/j.1365-313x.1998.00071.x. [DOI] [PubMed] [Google Scholar]
- 4.Doolittle R F, Feng D-F, Johnson M S, McClure M A. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 5.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White S E, Habera L F, Wessler S R. Proc Natl Acad Sci USA. 1994;91:11792–11796. doi: 10.1073/pnas.91.25.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirochika H. EMBO J. 1993;12:2521–2528. doi: 10.1002/j.1460-2075.1993.tb05907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreau-Mhiri C, Morel J-B, Audeon C, Ferault M, Grandbastien M-A, Lucas H. Plant J. 1996;9:409–419. [Google Scholar]
- 9.Vernhettes S, Grandbastien M-A, Casacuberta J M. Plant Mol Biol. 1997;35:673–679. doi: 10.1023/a:1005826605598. [DOI] [PubMed] [Google Scholar]
- 10.Takeda S, Sugimoto K, Otsuki H, Hirochika H. Plant Mol Biol. 1998;36:365–376. doi: 10.1023/a:1005911413528. [DOI] [PubMed] [Google Scholar]
- 11.Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Proc Nalt Acad Sci USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wessler S R. Curr Biol. 1996;6:959–961. doi: 10.1016/s0960-9822(02)00638-3. [DOI] [PubMed] [Google Scholar]
- 13.Grandbastien M-A, Spielmann A, Caboche M. Nature (London) 1989;337:376–380. doi: 10.1038/337376a0. [DOI] [PubMed] [Google Scholar]
- 14.Hirochika H. Plant Mol Biol. 1997;35:231–240. [PubMed] [Google Scholar]
- 15.Wang S, Zhang Q, Maughan P J, Saghai Maroof M A. Plant Mol Biol. 1997;33:1051–1058. doi: 10.1023/a:1005715118851. [DOI] [PubMed] [Google Scholar]
- 16.Bennetzen J L. Trends Microbiol. 1996;4:347–353. doi: 10.1016/0966-842x(96)10042-1. [DOI] [PubMed] [Google Scholar]
- 17.Moore G, Cheung W, Schwarzacher T, Flavell R. Genomics. 1991;10:469–476. doi: 10.1016/0888-7543(91)90334-b. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt T, Kubis S, Heslop-Harrison J S. Chromosome Res. 1995;3:335–345. doi: 10.1007/BF00710014. [DOI] [PubMed] [Google Scholar]
- 19.Pearce S R, Harrison G, Li D, Heslop-Harrison J S, Kumar A, Flavell A J. Mol Gen Genet. 1996;250:305–315. doi: 10.1007/BF02174388. [DOI] [PubMed] [Google Scholar]
- 20.SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, et al. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 21.Suoniemi A, Anamthawat-Jonsson K, Arna T, Schulman A H. Plant Mol Biol. 1996;30:1321–1329. doi: 10.1007/BF00019563. [DOI] [PubMed] [Google Scholar]
- 22.Uozu S, Ikehash H, Ohmido N, Ohtsubo H, Ohtsubo E, Fukui K. Plant Mol Biol. 1997;35:791–799. doi: 10.1023/a:1005823124989. [DOI] [PubMed] [Google Scholar]
- 23.Moore G, Devos K M, Wang Z, Gale M D. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- 24.Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, Wu J, Antonio B A, Shomura A, Shimizu T, Lin S Y, et al. Nat Genet. 1994;8:365–372. doi: 10.1038/ng1294-365. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Liu K D, Xu C G, Li X H, Zhang Q. Theor Appl Genet. 1998;97:407–412. [Google Scholar]
- 26.Causse M A, Fulton T M, Cho Y G, Ahn S N, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald P C, Harrington S E, et al. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng K, Zhang H, Zhang Q. Acta Bot Sin. 1998;40:1108–1114. [Google Scholar]
- 28.Liu K D, Wang J, Li H B, Xu C G, Liu A M, Li X H, Zhang Q. Theor Appl Genet. 1997;95:809–814. [Google Scholar]
- 29.Lincoln S, Daly M, Lander E. Constructing Genetic Maps withmapmaker/exp 3.0. Cambridge, MA: Whitehead Inst.; 1992. [Google Scholar]
- 30.Singh K, Ishii T, Parco A, Huang N, Brar D S, Khush G S. Proc Natl Acad Sci USA. 1996;93:6163–6168. doi: 10.1073/pnas.93.12.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirochika H, Fukuchi A, Kikuchi F. Mol Gen Genet. 1992;233:209–216. doi: 10.1007/BF00587581. [DOI] [PubMed] [Google Scholar]
- 32.Sprinzl, M., Hartman, T., Meissner, F., Moll, J. & Vorderwulbecke, T. (1987) Nucleic Acids Res.15, Suppl., r53–r188. [DOI] [PMC free article] [PubMed]
- 33.Flavell R B. Annu Rev Plant Physiol. 1980;31:569–596. [Google Scholar]
- 34.Xiong L Z, Wang S, Liu K D, Dai X K, Saghai Maroof M A, Zhang Q. Acta Bot Sin. 1998;40:605–614. [Google Scholar]