Summary
Cell-mediated immunity stems from the proliferation of naïve T lymphocytes expressing T cell antigen receptors (TCR) specific for foreign peptides bound to host Major Histocompatibility Complex (MHC) molecules. Due to the tremendous diversity of the T cell repertoire, naïve T cells specific for any one peptide:MHC complex (pMHC) are extremely rare. Thus, it is not known how many naïve T cells of any given pMHC specificity exist in the body or how that number influences the immune response. Using soluble pMHCII tetramers and magnetic bead enrichment, we found that three different pMHCII-specific naïve CD4+ T cell populations vary in frequency from 20 to 200 cells per mouse. Moreover, naïve population size predicted the size and TCR diversity of the primary CD4+ T cell response after immunization with relevant peptide. Thus, variation in naive T cell frequencies can explain why some peptides are stronger immunogens than others.
Introduction
Naive T lymphocytes become activated and proliferate when their antigen receptors (TCR) bind to cognate peptides presented on Major Histocompatibility Complex (MHC) molecules (Rudolph et al., 2006; Stockinger et al., 2006). During infection, antigen-presenting cells display a multitude of unique foreign peptide:MHC complexes (pMHC) derived from the invading microbe. The extent to which naive T cells respond to these various pMHCs is influenced by how well antigen-presenting cells process antigenic peptides (Yewdell, 2006) and induce costimulatory ligands and cytokines (Jenkins et al., 2001).
It is likely that naive T cell population size also contributes to differences observed in the magnitude of the primary immune response to different antigenic peptides (Busch et al., 1998; Homann et al., 2001; McHeyzer-Williams and Davis, 1995). However, the importance of this factor is unknown because naive T cells specific for any one pMHC are very difficult to detect due to their extremely low frequency, estimated at 100–3,000 cells per mouse (Blattman et al., 2002; Butz and Bevan, 1998; McHeyzer-Williams and Davis, 1995; Stetson et al., 2002; Whitmire et al., 2006). Because of this technical limitation, most current knowledge of naive T cell activation is based on the adoptive transfer of large numbers of monoclonal TCR transgenic T cells into histocompatible hosts (Jenkins et al., 2001). Although such systems have proven their use, concerns over the effects of intraclonal competition between abnormally large numbers of identical T cells have reinforced the need to study polyclonal naïve T cells directly (Badovinac et al., 2007; Ford et al., 2007; Foulds and Shen, 2006; Hataye et al., 2006; Marzo et al., 2005). Another approach to the study of naive T cell populations relies on the use of mice in which one chain of the TCR is fixed (Malherbe et al., 2004; Zehn and Bevan, 2006). The restricted diversity of the T cell repertoire in these mice results in larger than normal populations of pMHC-specific cells, thereby facilitating their study. Although the pMHC-specific T cell populations in these mice are not monoclonal, the increased sizes of these populations may still pose caveats concerning levels of competition for ligands.
In this report, we addressed these concerns by developing a method using pMHCII tetramers and magnetic beads to study rare CD4+ T cell populations in unmanipulated mice. With this method, we directly measured the sizes of three distinct naive pMHCII-specific CD4+ T cell populations, and determined how this variable influences aspects of the primary immune response.
Results
Variable CD4+ T Cell Responses to Different pMHCs
Three distinct tetramers, each consisting of four identical biotinylated peptide:I-Ab MHC molecules complexed to a fluorochrome-labeled streptavidin core, were produced to study naive polyclonal CD4+ T cell populations. The peptides used were the 2W1S variant (Rees et al., 1999) of peptide 52–68 from the I-E alpha chain (Rudensky et al., 1991), peptide 427–441 from the FliC protein of Salmonella typhimurium (McSorley et al., 2000), and peptide 323–339 from chicken ovalbumin (Shimonkevitz et al., 1984). Each of these peptides binds to the I-Ab MHCII molecule expressed in C57BL/6 (B6) mice and is immunogenic in this strain.
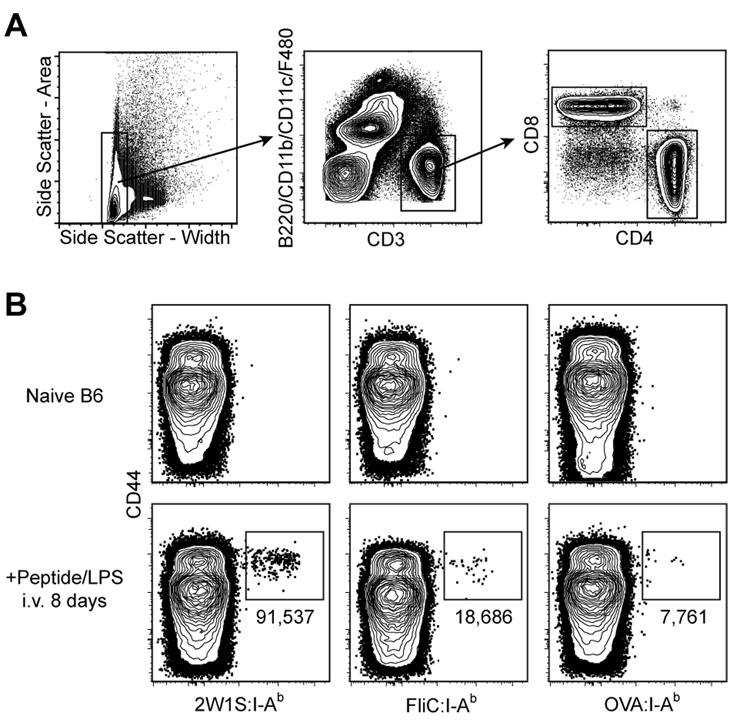
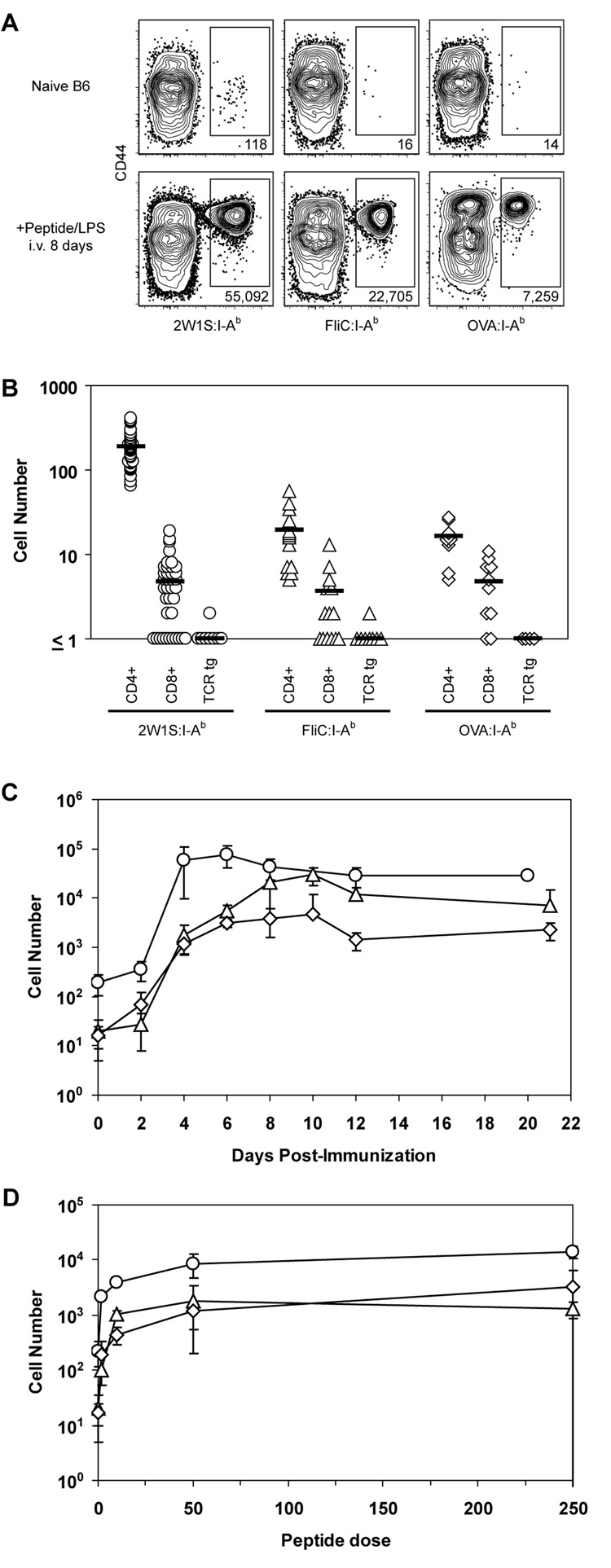
These pMHCII tetramers were used in conjunction with flow cytometry in an attempt to detect naive pMHCII-specific CD4+ T cells. Spleen and lymph node cells were harvested from naive B6 mice or B6 mice injected intravenously (i.v.) 8 days earlier with the relevant peptides plus bacterial lipopolysaccharide (LPS) as an adjuvant. Peptides were used for immunization to minimize the effects of differential antigen processing between epitopes from different proteins, and an i.v. route of administration was chosen to provide a synchronous, systemic response. The cells were then stained with phycoerythrin (PE)-labeled pMHCII tetramers and antibodies specific for CD3, CD4, CD8, and a cocktail of non-T cell lineage-specific antibodies to allow for the identification of CD4+ T cells (CD3+, CD4+, non-T-lineage-, CD8- events) (Fig. 1A). Anti-CD44 antibody was also included in the staining cocktail to identify CD44high antigen-experienced cells (Dutton et al., 1998). Although the tetramers identified expanded CD44high populations of different sizes in the relevant peptide-injected mice, corresponding CD44low populations could not be detected in naive mice (Fig. 1B). Thus, this approach lacked the sensitivity to detect naive pMHCII-specific T cells, and therefore could not be used to determine whether the differences in the sizes of the expanded populations were related to naive T cell frequency.
Figure 1. Primary immune responses of naive CD4+ T cells vary with respect to pMHC specificity.
A, Flow cytometry gates used in all figures to identify T cells from total spleen and lymph node cells: side scatter-widthlow (left plot); CD3+, non-T cell lineage- (middle plot); and either CD4+ or CD8+ (right plot). B, Where indicated, B6 mice were injected i.v. with 50 µg of the indicated peptide plus 5 µg LPS. Eight days later, spleen and lymph node cells were harvested and stained with the indicated tetramer. Representative contour plots of CD44 versus the indicated tetramers are shown for non-T cell lineage-, CD3+, CD4+ events gated from 106 total collected events. The total number of cells for each individual mouse shown is indicated below the relevant gate. Mean values ± S.D. were 2W1S:115,000 ± 40,000; FliC: 23,000 ± 20,000; OVA: 7,000 ± 3,000 (n=4 per group).
pMHCII Tetramer-Based Enrichment
Detection of pMHCII-specific CD4+ T cells in naïve mice was unfeasible due to the limited capacity of the flow cytometer to routinely analyze more than about 106 total cells, or only 1/200th of the ~2 × 108 nucleated cells in the lymphoid organs of a mouse, at a given time. Even if such an analysis was performed using high-speed cell sorting technology, the accumulation of low frequency background events would become prohibitive to the study of rare cell populations. The solution to these problems was to concentrate all the tetramer-binding cells from a mouse into a smaller sample containing only about 106 total cells, which could then be analyzed in its entirety with a high signal-to-noise ratio. This was accomplished by performing a cell enrichment step using magnetic beads coupled to antibodies specific for the PE fluorochrome component of the tetramer.
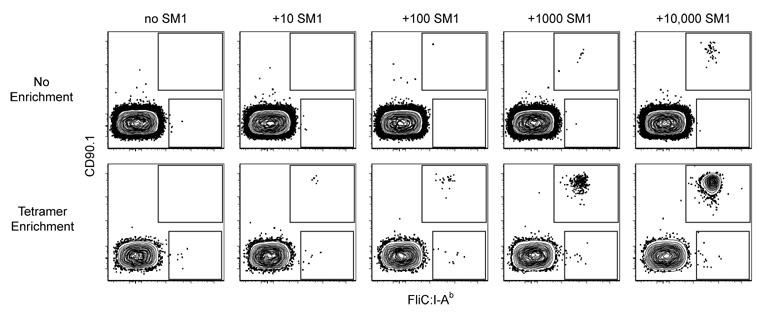
To test the efficacy of the tetramer-based enrichment, known numbers of monoclonal CD4+ T cells from SM1 TCR transgenic mice (McSorley et al., 2002), which express CD90.1 and a FliC:I-Ab-specific TCR, were mixed with 2.5 × 108 CD90.2+ spleen and lymph node cells from a B6 mouse (Fig. 2). FliC:I-Ab tetramer staining and flow cytometry was then performed with or without a preliminary FliC:I-Ab tetramer/anti-PE magnetic bead enrichment step. CD90.1+ events were not detected in samples lacking SM1 cells, demonstrating the absence of background tetramer staining. Without enrichment, SM1 cells were not detected until at least 1,000 were present in the mixture. In contrast, as few as 10 SM1 cells were detected using the enrichment method. Therefore, this tetramer-based enrichment method improved the limit of detection by about 100-fold, to the point where as few as 10 pMHCII-specific cells could be reliably detected in a background of 2.5 × 108 other cells.
Figure 2. Tetramer-based enrichment increases the sensitivity of detection of epitope-specific T cell populations.
Representative contour plots of CD90.1 versus FliC:I-Ab for CD4+ gated events from mixtures of 2.5 × 108 B6 spleen and lymph node cells and the indicated numbers of SM1 TCR transgenic T cells analyzed directly (top row) or after tetramer-based enrichment (bottom row). SM1 cells are shown in the upper gate and polyclonal FliC:I-Ab-specific CD4+ T cells in the lower gate. Data is representative of two independent experiments.
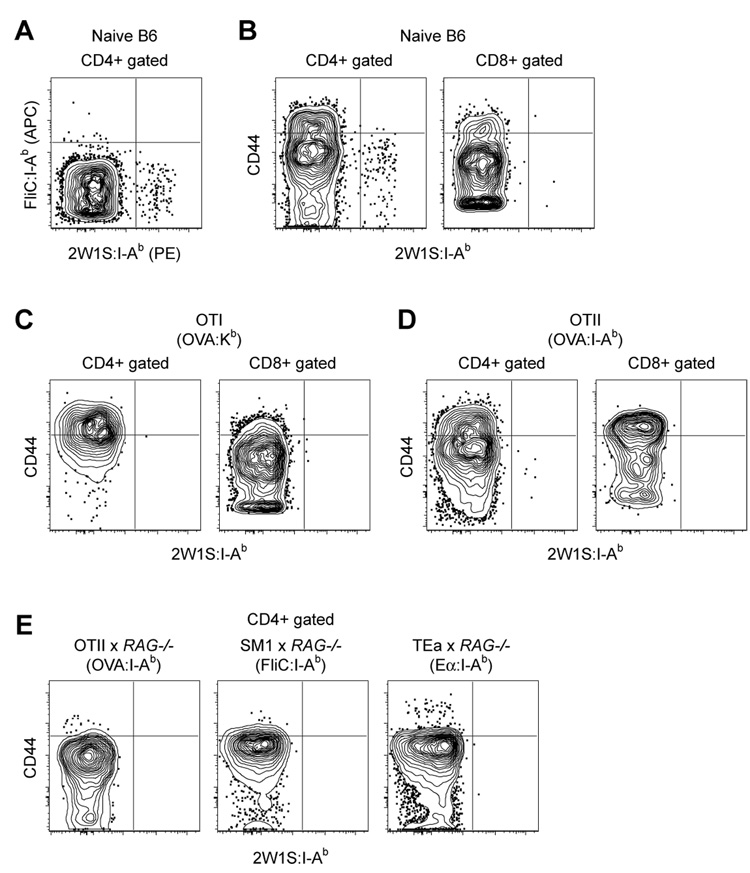
Remarkably, a small population of CD4+ FliC:I-Ab+ CD90.1− cells of polyclonal B6 origin was consistently detected in all of the tetramer-enriched samples (Fig. 2). This finding raised the possibility that this method could detect endogenous polyclonal pMHCII-specific naïve CD4+ T cells. Alternatively, the rare CD4+ T cells that bound the FliC:I-Ab tetramer may have done so non-specifically. These possibilities were tested by enriching spleen and lymph nodes cells from a naïve B6 mouse with a mixture of PE-labeled 2W1S:I-Ab and allophycocyanin (APC)-labeled FliC:I-Ab tetramers followed by anti-PE plus anti-APC magnetic beads. As shown in Fig. 3A, this experiment resulted in the detection of two mutually exclusive CD4+ populations: one that bound FliC:I-Ab but not 2W1S:I-Ab, and a larger one that bound 2W1S:I-Ab but not FliC:I-Ab. The absence of CD4+ T cells that bound both tetramers ruled out the possibility that non-specific binding was responsible for the enrichment of these distinct populations of cells.
Figure 3. Naïve pMHCII-specific CD4+ T cells can be detected using tetramer-based enrichment.
A, Representative contour plot of FliC:I-Ab tetramer (APC) versus 2W1S:I-Ab tetramer (PE) for non-T cell lineage-, CD3+, CD4+ events in a spleen and lymph node cell population from a naive B6 mouse after enrichment with FliC:I-Ab and 2W1S:I-Ab tetramers. B, Representative contour plots of CD44 versus 2W1S:I-Ab for CD4+ or CD8+ gated events following 2W1S:I-Ab tetramer enrichment of total spleen and lymph node cells from a naive B6 mouse, or C, an OT-I RAG1−/−, or D, an OT-II RAG1+/+ TCR transgenic mouse. E, Representative contour plots of CD44 versus 2W1S:I-Ab for CD4+ gated events following 2W1S:I-Ab tetramer enrichment of total spleen and lymph node cells from OT-II RAG1−/−, SM1 RAG1−/−, and TEa RAG1−/− TCR transgenic mice. Data is representative of at least 3 independent experiments.
Several other pieces of evidence indicated that tetramer-binding T cells detected by the enrichment method were truly pMHCII-specific cells. First, the 2W1S:I-Ab+ cells detected in a 2W1S:I-Ab tetramer-enriched sample from a naïve B6 mouse had the CD44low phenotype of naïve T cells (Fig. 3B). Second, few if any of these cells expressed CD8 (Fig. 3B), which marks pMHCI-specific T cells. Third, few if any 2W1S:I-Ab+ T cells were detected in the CD4+ or CD8+ T cells of a 2W1S:I-Ab tetramer-enriched sample of spleen and lymph nodes cells from an OVA 257–264:Kb-specific TCR transgenic OT-I mouse (Hogquist et al., 1994) (Fig. 3C), or an OVA 323–339:I-Ab TCR transgenic OT-II mouse (Barnden et al., 1998) (Fig. 3D). The few CD44low, CD4+, 2W1S:I-Ab+ cells detected in the OT-II sample likely expressed a 2W1S:I-Ab-specific TCR produced by rearrangement of an endogenous TCR-Vα chain, which can occur in TCR transgenic mice expressing RAG proteins (Steinmetz et al., 1989). This contention was supported by the finding that no CD4+, 2W1S:I-Ab+ cells were detected among spleen and lymph node cells from a RAG-deficient OT-II TCR transgenic mouse, or in two other TCR transgenic RAG-deficient lines containing monoclonal T cells specific for other pMHCII (Fig. 3E). Together, these results indicated that the tetramer-based enrichment method was sensitive enough to detect rare naïve CD4+ T cells specific for individual pMHCII.
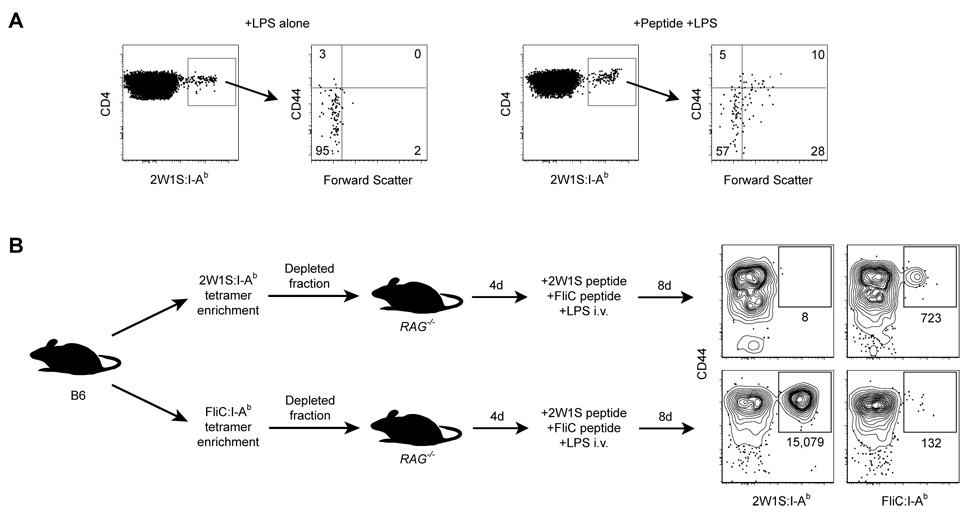
If the cells detected by tetramer-based enrichment were truly naïve pMHCII-specific CD4+ T cells, then they should show signs of activation when exposed to the relevant peptide. As shown in Fig. 4A, about 50% of the 2W1S:I-Ab+ CD4+ T cells in B6 mice injected i.v. 48 hours earlier with 2W1S peptide plus LPS expressed high levels of CD44 and/or had undergone blastogenesis compared to about 5% of the cells in mice that were injected with LPS alone. This result could not be explained by preferential expansion of a small number of cells in the peptide-injected mice because the numbers of 2W1S:I-Ab+ CD4+ T cells present in the 2 groups of mice were similar at this early time (Fig. 5C). In addition, 2W1S:I-Ab+ T cells in a dye-labeled B6 CD4+ T cell population transferred into B6 hosts did not show signs of cell division 48 hours after injection of 2W1S peptide plus LPS (data not shown). These results showed that peptide responsive naïve CD4+ T cells were detected by the tetramer enrichment method.
Figure 4. Tetramer-binding T cells are responsive to their relevant peptide.
A, 2W1S:I-Ab enrichment was performed on total spleen and lymph node cells from B6 mice injected i.v. 48 hours earlier with 5 µg LPS (left panels), or 5 µg LPS plus 250 µg 2W1S peptide (right panels). CD4+, 2W1S:I-Ab+ events were then analyzed for CD44 expression and blastogenesis (forward scatter). Numbers indicate percentages of events in each quadrant. Data is representative of at least 3 independent experiments. B, Contour plots of CD44 versus tetramer for 2W1S:I-Ab or FliC:I-Ab enriched spleen and lymph node cells from B6 RAG1−/− hosts that received either 2W1S:I-Ab or FliC:I-Ab tetramer-depleted cells from naïve B6 mice, and were then injected i.v. with a mixture of 50 µg 2W1S peptide, 50 µg FliC peptide, and 5 µg LPS 8 days before analysis. The total number of tetramer+ cells for each population is shown below the relevant gate. Data shown is representative of three independent experiments.
Figure 5. Naive CD4+ T cell populations vary in size.
A, Representative contour plots of CD44 versus tetramer for CD4+ events from total spleen and lymph node cells from naive B6 mice (top row) or B6 mice injected i.v. with 50 µg of the indicated peptide plus 5 µg LPS (bottom row) following enrichment with the indicated tetramers. B, Total numbers of tetramer+, CD4+ or CD8+ cells following tetramer enrichment of total spleen and lymph node cells from a naive B6 mouse or tetramer+, CD4+ cells from OT-II RAG1−/−, TEa RAG1−/−, or SM1 RAG1−/− TCR transgenic mice. Symbols represent individual mice; horizontal bars indicate mean values. C, Mean total number (±S.D., n=2–8) of 2W1S:I-Ab+ (circles), FliC:I-Ab+ (triangles), or OVA:I-Ab+ (diamonds) cells in B6 mice over time following i.v. injection with 50 µg of the relevant peptide plus 5 µg LPS. D, Mean total number (±S.D., n=3) of 2W1S:I-Ab+ (circles), FliC:I-Ab+ (triangles), or OVA:I-Ab+ (diamonds) cells in B6 mice 4 days following i.v. injection with 5 µg LPS plus the indicated amount of relevant peptide.
The efficiency of tetramer-based enrichment was measured by assaying its capacity to deplete a target population from a polyclonal sample. Purified CD4+ T cells (~4 × 107) from naïve B6 mice were stained with either 2W1S:I-Ab or FliC:I-Ab tetramer. The stained samples were then passed over a magnetized column, and the ~4 × 107 cells that did not bind were transferred into lymphocyte-deficient RAG1−/− mice, which were then injected intravenously with a mixture of FliC and 2W1S peptides plus LPS. Eight days later, the number of 2W1S:I-Ab+ or FliC:I-Ab+ cells in the recipient mice was measured by tetramer-based enrichment. As shown in Figure 4B, RAG1−/− mice reconstituted with 2W1S:I-Ab-depleted cells contained a population of CD44high FliC:I-Ab+ cells but not 2W1S:I-Ab+ cells, whereas RAG1−/− mice reconstituted with FliC:I-Ab-depleted cells contained a large population of CD44high 2W1S:I-Ab+ cells but very few FliC:I-Ab+ cells following immunization. These results demonstrated that most, if not all of the relevant pMHCII-specific CD4+ T cells in a naïve mouse were captured by the magnetic column.
Naive CD4+ T Cell Population Sizes
The specificity and efficiency of tetramer-based enrichment allowed for the accurate enumeration of naive CD4+ T cells specific for 2W1S:I-Ab, FliC:I-Ab, or OVA:I-Ab. Individual naïve B6 mice contained on average 190 2W1S:I-Ab-specific, 20 FliC:I-Ab-specific, and 16 OVA:I-Ab-specific CD4+ T cells in their spleen and lymph nodes (Fig. 5A, B). Each of these values was significantly greater (Student's T-test, two-tailed, P < 0.001) than the values for CD8+ cells in the same samples or irrelevant TCR transgenic T cells stained with the same tetramers (Fig. 5B). The ~5 CD8+ cells per mouse that were detected with each pMHCII tetramer may have been rare CD8+ pMHCII-specific T cells similar to those discovered in CD4−/− mice (Pearce et al., 2004; Tyznik et al., 2004). The number of 2W1S:I-Ab-specific CD4+ T cells was significantly greater than the numbers of FliC:I-Ab- or OVA:I-Ab-specific T cells (P < 0.001), which were not significantly different from each other (P = 0.48). Therefore, while the sizes of individual naïve CD4+ T cell populations were highly consistent amongst individual mice, these populations varied in size depending on their pMHCII specificity.
pMHCII-Specific CD4+ T Cell Population Sizes Following Immunization
The naive 2W1S:I-Ab-, FliC:I-Ab-, or OVA:I-Ab-specific populations were roughly proportional in size to the corresponding expanded populations observed in peptide injected mice (Fig. 1B). This relationship was explored in more detail using the enrichment method over a three week period following i.v. injection with the relevant peptides plus LPS (Fig. 5C). The 2W1S:I-Ab-specific population increased about 300-fold to a peak of ~80,000 cells by day 6. The FliC:I-Ab- and OVA:I-Ab-specific populations also increased about 300-fold to 5,000 and 3,000 cells during the same period. Since all three populations expanded at similar rates over the first 6 days after peptide injection, the larger number of 2W1S:I-Ab-specific cells present at these times was likely related to the corresponding larger naïve population. After day 6, the relationship between pre- and post-immunization frequency was less clear because the 2W1S:I-Ab-specific population was contracting while the FliC:I-Ab- and OVA:I-Ab-specific populations continued to increase.
The correspondence between naïve population size and response magnitude was further confirmed in a peptide dose response experiment. As shown in Fig. 5D, the number of 2W1S:I-Ab-specific T cells was significantly greater than the number of FliC:I-Ab- and OVA:I-Ab-specific T cells 4 days after peptide/LPS injection at all doses of peptide tested (P<0.05). Thus, the greater magnitude of the 2W1S-specific T cell response is likely due to higher precursor frequency rather than more efficient peptide presentation.
TCR Variable Segment Diversity
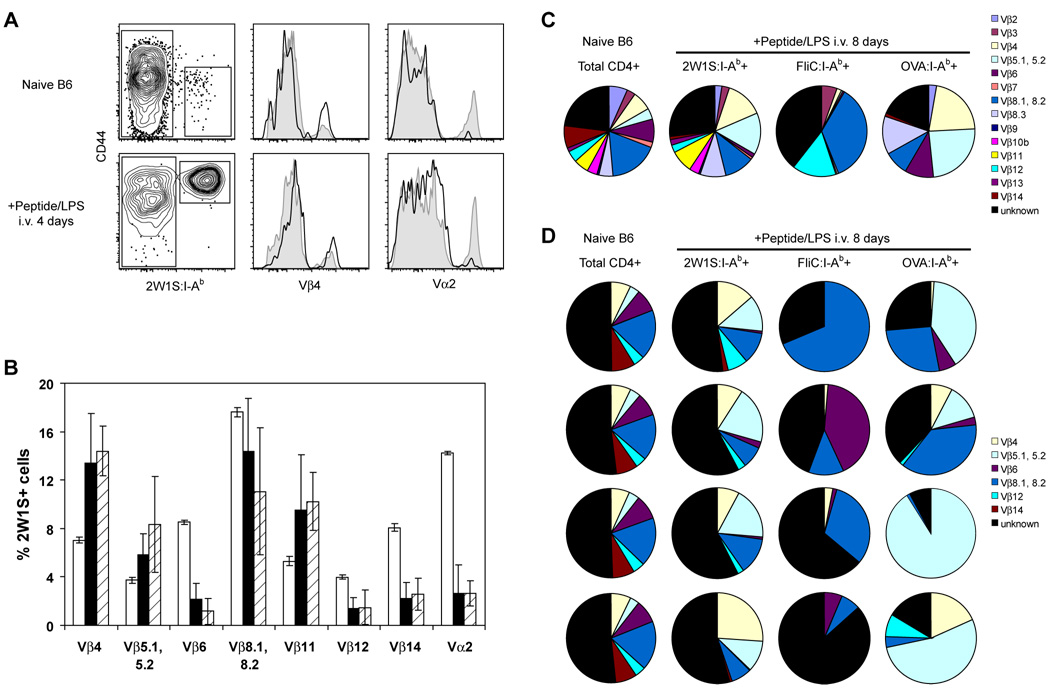
Analysis of TCR variable gene usage as a measure of clonal diversity provided additional evidence for variation in naïve population sizes. The naive 2W1S:I-Ab-specific population consistently displayed a V segment usage pattern that was distinct from that of the total CD4+ T cell population (Fig. 6A, 6B). Cells expressing Vβ4, Vβ5, or Vβ11 were overrepresented in the 2W1S:I-Ab-specific population, whereas cells expressing Vβ6, Vβ8, Vβ12, Vβ14, and Vα2 were underrepresented. This distinct V segment usage pattern was preserved in the expanded population of 2W1S:I-Ab-specific cells 4 days after intravenous injection with 2W1S peptide plus LPS (Fig. 6A, 6B). The similarity between pre- and post-immunization V segment usage indicated that most of the cells in the naïve 2W1S:I-Ab-specific repertoire proliferated during the early stages of the primary immune response.
Figure 6. Naive CD4+ T cell population diversity is related to population size.
A, Total lymph node and spleen cells from naive B6 mice (top) or B6 mice injected i.v. with 50 µg 2W1S peptide plus 5 µg LPS 4 days earlier (bottom) were stained with antibodies to TCR Vβ4 and Vα2. 2W1S:I-Ab- and 2W1S:I-Ab+ gated events are represented in histograms by shaded profiles and solid lines, respectively. B, TCR Vβ and Vα gene segment usage in total CD4+ T cells from naive mice (unfilled bars), or 2W1S:I-Ab+ CD4+ T cells from naive mice (filled bars) or mice injected i.v. with 50 µg 2W1S peptide plus 5 µg LPS 4 days earlier (hatched bars). Data are mean values ± S.D. from 4∓8 mice per group. C, Usage profile for TCR Vβ gene segments in total CD4+ T cells from naive B6 mice or tetramer+ CD4+ T cells from B6 mice injected i.v. with 50 µg of the indicated peptide plus 5 µg LPS 8 days earlier. Data are values from pooled samples taken from 4–10 mice per group, or D, samples taken from 4 individual mice per group.
These results suggested that V segment usage by an expanded T cell population is reflective of the naïve starting population. Therefore, V segment usage in the FliC:I-Ab- and OVA:I-Ab-specific naïve populations, which were too small to study directly with multiple anti-V segment antibodies, could be estimated from their corresponding expanded populations. By pooling tetramer-enriched cells from mice 8 days after peptide injection, it was possible to assess usage of all Vβ segments for which antibodies are available. The expanded population of 2W1S:I-Ab-specific T cells was very diverse (Fig. 6C), containing cells expressing each of the 14 Vβ segments tested, but with overrepresentation of Vβ4, 5, and 11 compared to the total population of CD4+ T cells. In contrast, the smaller populations of FliC:I-Ab- and OVA:I-Ab-specific cells were much less diverse. The FliC:I-Ab-specific population was dominated by cells expressing Vβ8 or Vβ12 whereas the OVA:I-Ab-specific population was comprised primarily of cells expressing Vβ4, Vβ5, or Vβ8. Since each clone expresses a single Vβ segment, the presence of fewer Vβ segments in the smaller populations is consistent with the possibility that they are comprised of fewer clones than the larger 2W1S:I-Ab-specific population.
Analysis of cells from individual mice provided additional evidence for small FliC:I-Ab- and OVA:I-Ab-specific populations (Fig. 6D). Vβ usage among these populations was much more variable than in the case of 2W1S:I-Ab-specific populations. In some individual mice, the FliC:I-Ab and OVA:I-Ab-specific populations were dominated by cells expressing only one of the preferred Vβ segments, whereas in other mice the population contained at least 4 different Vβ segments. Collectively, the limited diversity of the FliC:I-Ab- and OVA:I-Ab-specific populations and their greater susceptibility to individual variation reinforced the finding that their corresponding naive precursor frequencies are smaller than that of the 2W1S:I-Ab-specific population.
Discussion
Since Klinman’s pioneering studies on the frequency of naïve B cells (Klinman, 1972), numerous efforts have been made to address this issue with respect to epitope-specific naïve T cells. Collectively, past work produced estimates ranging from 100 to 3,000 cells depending on the epitope in question (Blattman et al., 2002; Butz and Bevan, 1998; McHeyzer-Williams and Davis, 1995; Stetson et al., 2002; Whitmire et al., 2006). Most of the methods used to arrive at these values relied on indirect means of detection. For example, one approach involved titration of TCR transgenic T cells into normal mice, which were then exposed to the relevant antigen (Blattman et al., 2002; Butz and Bevan, 1998; Whitmire et al., 2006). The number of transferred cells that produced an antigen expanded population equal to that produced from the host was assumed to reflect the number of host precursors. In addition, some of these studies involved sampling only a small fraction of the total lymphoid cells in a mouse, thereby necessitating large extrapolation factors to calculate total cell numbers. Finally, all of the early studies produced estimates for only one epitope-specific population, precluding a definitive comparison of different population sizes.
Many of these limitations were overcome in the current study. Identification of epitope-specific CD4+ T cells was based directly on pMHCII binding. A novel enrichment step allowed comprehensive sampling of the entire secondary lymphoid compartment of a mouse, thereby avoiding the need for large extrapolation factors in the calculation of T cell frequency. The use of exclusion gating and relevant negative controls allowed detection of as few as 5 cells per mouse. For these reasons, the values obtained here for naive CD4+ T cell population sizes are likely the most accurate obtained to date, with the caveat that they may be slight underestimates due to cell loss during manipulations and the fact that blood and mucosal-lymphoid tissues were not sampled.
The values reported here for the number of epitope-specific CD4+ T cells allows for estimation of the number of epitopes that could be recognized by the naïve repertoire. Assuming that each pMHCII-specific CD4+ T cell population consists of about 100 cells and that there are about 3 × 107 naive CD4+ T cells in a mouse, it follows that at least 3 × 105 unique pMHCII specificities exist within the naive repertoire. However, this is likely a minimum value because individual T cells have been shown to recognize more than one pMHC (Evavold et al., 1995; Felix et al., 2007).
Although the naïve populations studied in this report clearly contained multiple clones as evidenced by diverse Vβ usage, the precise number of clones in each is not clear. Extensive TCR sequence analysis by Casrouge et. al. showed that individual mice contain about 2 × 106 distinct naïve αβ TCR clones of 10 cells each in the spleen (Casrouge et al., 2000). Our findings indicated that the naive populations of FliC:I-Ab- and OVA:I-Ab-specific T cells numbered only about 20 per mouse, and that in some individual mice these populations contained cells expressing one of at least 4 different Vβ segments (Fig. 6D). Therefore, because each T cell expresses only one Vβ segment, these populations must consist of at least 4 clones each. Indeed, it is possible that each cell in each population was a unique clone in which case our results would be more in line with the recent finding that 50–550 distinct CD8+ T cell clones recognize each epitope in individual mice (Kedzierska et al., 2006).
Our results indicate that polyclonal CD4+ T cell populations expand in proportion to the frequency of their naïve progenitors, at least during the early phase of the primary immune response. Thus, at least for certain peptides, variations in naïve CD4+ T cells population size may account for the observation that the number of T cells generated at the peak of the primary immune response to infection varies for different pMHCII (Busch et al., 1998; Homann et al., 2001; Masopust et al., 2006).
Notably, not all of the 2W1S:I-Ab tetramer-binding cells in naïve mice showed signs of activation 48 hours after peptide immunization. Our findings that pMHCII tetramer-binding cells were not detected in CD8+ populations or in mice containing irrelevant monoclonal T cells provides strong evidence that tetramer binding was indeed TCR-specific. Thus, the tetramer-binding cells were unlikely to be background events. On the contrary, the non-responsive tetramer-binding cells may have simply not encountered a stimulatory antigen-presenting cell in the 48 hour time frame. This is supported by our finding that the TCR diversity of the naive 2W1S:I-Ab-specific T cell population was preserved after four days of antigen-induced proliferation, implying that a high percentage of this naive repertoire eventually participated in the response. This conclusion is in agreement with a previous study on another pMHCII-specific CD4+ T cell population (Malherbe et al., 2004). Alternatively, the non-responders may have possessed TCRs with too low an affinity to become activated at the peptide dose administered. Finally, these cells may have been recent thymic emigrants that have been reported to be inherently hyporesponsive to antigenic stimulation (Boursalian et al., 2004).
Naive CD8+ T cell populations that vary in size have been reported to peak at the same time following immunization (Busch et al., 1998; Homann et al., 2001). In contrast, we found that a relatively large naive CD4+ T cell population peaked earlier than two smaller populations, as predicted by earlier experiments with adoptively transferred TCR transgenic T cells (Hataye et al., 2006). Because the large and small populations increased with the same initial kinetics, it is possible that the large population became numerous enough to compete for limiting pMHCII, resulting in an earlier cessation of the response. This is plausible because unlike CD8+ T cells (Kaech and Ahmed, 2001; Mercado et al., 2000; van Stipdonk et al., 2001), CD4+ T cells only continue to proliferate in the presence of pMHC (Obst et al., 2005). Differences in pMHCII persistence at later times after immunization may explain why the FliC:I-Ab- and OVA:I-Ab-specific populations eventually diverged.
Our findings indicate that large naïve populations contain more distinct clones than small populations, rather than more copies of each clone. Large naïve populations may exist for those foreign pMHCII that have properties, for example charge and hydrophobicity, which are conducive to recognition by many structurally distinct TCRs. In contrast, the properties of other foreign pMHCII, for example similarity to self pMHCII, may be conducive to recognition by only a small number of TCRs due to strong thymic negative selection. Our results agree with other studies suggesting that such populations can be small enough that it becomes unlikely that the random process of TCR gene rearrangement will produce identical sets of clones in individual mice (Bousso et al., 1998). Immune responses that depend on very small naïve populations may be inherently variable and at risk for extinction under conditions where total lymphocyte numbers are reduced, such as aging, chemotherapy, and HIV infection (Prlic and Jameson, 2002).
Experimental Procedures
Mice
Six-8 week old C57BL/6 mice were purchased from the National Cancer Institute. C57BL/6 RAG1−/−, SM1 RAG1−/− CD90.1+, OT-II RAG1−/− and TEa RAG1−/− mice were bred in our facilities. All mice were housed under specific pathogen-free conditions in accordance with University of Minnesota and NIH guidelines.
Antibodies
Pacific Blue-conjugated anti-B220, CD11b, F4/80 (Caltag), CD11c (eBiosciences); Pacific Orange-conjugated CD8 (eBiosciences); FITC-conjugated CD3, Vβ2, Vβ3, Vβ4, Vβ5.1/5.2, Vβ6, Vβ7, Vβ8.1/8.2, Vβ8.3, Vβ9, Vβ10b, Vβ11, Vβ12, Vβ13, Vβ14 (BD Pharmingen); PerCP-conjugated CD4 (BD Pharmingen); PE-Cy7-conjugated CD3 (eBiosciences); and AlexaFluor 700-conjugated CD44 (eBiosciences) antibodies were purchased from the indicated sources.
Plasmid Construction
pRMHa-3 vectors containing the alpha and beta chains of I-Ad under the control of the metallothionein promoter were provided by N. Glaichenhaus (Malherbe et al., 2000). These constructs included C-terminal fusions to acidic and basic leucine zipper domains to force heterodimerization (Scott et al., 1996), as well as a 6x His epitope tag on the beta chain construct to facilitate purification, and an IgG2a-Fc domain on the alpha chain. The I-Ad sequences in both constructs were replaced with corresponding I-Ab alpha and beta sequences generated by PCR amplification. The Fc domain on the alpha chain was replaced with a BirA biotinylation signal sequence (Beckett et al., 1999) generated by oligonucleotide synthesis. Sequences encoding antigenic peptides (FliC 427–441: VQNRFNSAITNLGNT, 2W1S: EAWGALANWAVDSA, OVA 323–339: ISQAVHAAHAEINEAGR) were fused to the N-terminus of the beta chain via a flexible polyglycine linker (Kozono et al., 1994).
Tetramer Production
Peptide:I-Ab molecules were expressed in Drosophila S2 cells using the Drosophila Expression System kit (Invitrogen). Cells were co-transfected via calcium phosphate with plasmids encoding the I-Ab alpha chain, the I-Ab beta chain, and a blasticidin resistance gene at a molar ratio of 9:9:1. Transfected cells were selected in blasticidin-containing media for 2 weeks at 28° C, passaged into serum-free media, and scaled up to 1 liter cultures in 3 liter spinner flasks. When cell densities exceeded 107/ml, expression was induced by the addition of 0.8 mM copper sulfate. Peptide:I-Ab heterodimers were purified from supernatants 4–6 days later using an anti-MHC II (Y3P) affinity column. Bound peptide:I-Ab molecules were eluted with 0.1 M glycine pH 2.8, exchanged into 10 mM Tris pH 8, and biotinylated with BirA enzyme according to manufacturer's instructions (Avidity). Biotinylated peptide:I-Ab molecules were then separated from free biotin using a Sephacryl S-300 size exclusion column (GE Healthcare Bio-Sciences). Tetramers were created by mixing biotinylated peptide:I-Ab molecules with PE-conjugated streptavidin (Prozyme) at a molar ratio of 5–8:1 for 1 h at room temperature, and then spinning the mixture through a 100 kD molecular weight cut-off concentration filter (Millipore). The final concentration of tetramer was then calculated by measuring the absorbance of PE at 565 nm.
Peptide:MHC II Tetramer-based Enrichment Protocol
The spleen and inguinal, axillary, brachial, cervical, mesenteric, and periaortic lymph nodes were harvested for each mouse analyzed. A single cell suspension was prepared in 0.2 ml of Fc block (supernatant from 2.4G2 hybridoma cells grown in serum-free media + 2% mouse serum, 2% rat serum, 0.1% sodium azide). PE-conjugated tetramer was added at a concentration of 4–50 nM, depending on the saturating dose for each production batch, and the cells were incubated at room temperature for 1 h, followed by a wash in 15 ml of ice-cold sorter buffer (PBS + 2% fetal bovine serum, 0.1% sodium azide). In some cases, PE- and APC-conjugated tetramers were added together at this step.
The tetramer-stained cells were then resuspended in a volume of 0.4 ml of sorter buffer, mixed with 0.1 ml of anti-PE antibody conjugated magnetic microbeads (Miltenyi Biotech) (and in some cases with anti-APC antibody conjugated beads), and incubated on ice for 20 min, followed by two washes with 10 ml of sorter buffer. The cells were then resuspended in 3 ml of sorter buffer and passed over a magnetized LS column (Miltenyi Biotech). The column was washed with sorter buffer and then removed from the magnetic field. The bound cells were obtained by pushing 5 ml of sorter buffer through the column with a plunger.
The resulting enriched fractions were resuspended in 0.1 ml of sorter buffer, and a small volume was removed for cell counting while the rest of the sample was stained with a cocktail of fluorochrome labeled antibodies specific for B220, CD11b, CD11c, F4/80, CD3, CD8, CD4, or CD44. The entire stained sample was then collected on an LSR II flow cytometer (BD Immunocytometry Systems) and analyzed using FlowJo software (Treestar). The percentage of tetramer positive events was multiplied by the total number of cells in the enriched fraction to calculate the total number of tetramer positive cells in the mouse.
For TCR-Vβ segment analysis, tetramer-enriched fractions from pooled or individual mouse samples were split into multiple tubes and stained as described above with the addition of Vβ-specific antibodies.
Cell transfer
CD4+ T cells were purified from several naïve B6 mice using a CD4+ T cell isolation kit (Miltenyi Biotech). Half of the purified CD4+ T cells were stained with 2W1S:I-Ab tetramer and the other half with FliC:I-Ab tetramer followed by anti-PE magnetic beads as described above. Cells (~3 × 107) from each group that did not bind to the magnet were injected intravenously into RAG1−/− mice. Four days later, these mice were injected intravenously with a mixture containing 50 µg 2W1S peptide, 50 µg FliC peptide, and 5 µg LPS. Eight days later, individual recipient mice were analyzed for 2W1S:I-Ab- and FliC:I-Ab-specific CD4+ T cells by tetramer enrichment.
Acknowledgments
We thank Kristin Hogquist and Daniel Mueller for helpful discussions and Jennifer Walter for technical assistance. This work was supported by grants from the National Institutes of Health (to J.J.M. and M.K.J.). The authors declare that they have no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badovinac VP, Haring JS, Harty JT. Initial T cell receptor transgeniccell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity. 2007;179:53–63. doi: 10.1016/j.immuni.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattman JN, Anita R, Sourdive DJ, Wang X, Kaech SM, Murali-Krishna K, Altman JD, Ahmed R. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J Exp Med. 2002;195:657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursalian TE, Golob J, Soper DM, Cooper CJ, Fink PJ. Continued maturation of thymic emigrants in the periphery. Nat Immunol. 2004;5:418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- Bousso P, Casrouge A, Altman JD, Haury M, Kanellopoulos J, Abastado JP, Kourilsky P. Individual variations in the murine T cell response to a specific peptide reflect variability in naive repertoires. Immunity. 1998;9:169–178. doi: 10.1016/s1074-7613(00)80599-3. [DOI] [PubMed] [Google Scholar]
- Busch DH, Pilip IM, Vijh S, Pamer EG. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity. 1998;8:353–362. doi: 10.1016/s1074-7613(00)80540-3. [DOI] [PubMed] [Google Scholar]
- Butz EA, Bevan MJ. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity. 1998;8:167–175. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casrouge A, Beaudoing E, Dalle S, Pannetier C, Kanellopulos J, Kourilsky P. Size estimate of the alpha beta TCR repertoire of naive mouse splenocytes. J Immunol. 2000;164:5782–5787. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- Dutton RW, Bradley LM, Swain SL. T cell memory. Annu Rev Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- Evavold BD, Solan-Lancaster J, Wilson KJ, Rothbard JB, Allen PM. Specific T cell recognition of minimally homologous peptides: evidence for multiple endogenous ligands. Immunity. 1995;2:655–663. doi: 10.1016/1074-7613(95)90010-1. [DOI] [PubMed] [Google Scholar]
- Felix NJ, Donermeyer DL, Horvath S, Walters JJ, Gross ML, Suri A, Allen PM. Alloreactive T cells respond specifically to multiple distinct peptide-MHC complexes. Nat Immunol. 2007;8:388–397. doi: 10.1038/ni1446. [DOI] [PubMed] [Google Scholar]
- Ford ML, Koehn BH, Wagener ME, Jiang W, Gangappa S, Pearson TC, Larsen CP. Antigen-specific precursor frequency impacts T cell proliferation, differentiation, and requirement for costimulation. J Exp Med. 2007;204:299–309. doi: 10.1084/jem.20062319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds KE, Shen H. Clonal competition inhibits the proliferation and differentiation of adoptively transferred TCR transgenic CD4 T cells in response to infection. J Immunol. 2006;176:3037–3043. doi: 10.4049/jimmunol.176.5.3037. [DOI] [PubMed] [Google Scholar]
- Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Homann D, Teyton L, Oldstone MB. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat Med. 2001;7:913–919. doi: 10.1038/90950. [DOI] [PubMed] [Google Scholar]
- Jenkins MK, Khoruts A, Ingulli E, Mueller DL, McSorley SJ, Reinhardt RL, Itano A, Pape KA. In vivo activation of antigen-specific CD4 T cells. Annu Rev Immunol. 2001;19:23–45. doi: 10.1146/annurev.immunol.19.1.23. [DOI] [PubMed] [Google Scholar]
- Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat Immunol. 2001;2:415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierska K, Day EB, Pi J, Heard SB, Doherty PC, Turner SJ, Perlman S. Quantification of repertoire diversity of influenza-specific epitopes with predominant public or private TCR usage. J Immunol. 2006;177:6705–6712. doi: 10.4049/jimmunol.177.10.6705. [DOI] [PubMed] [Google Scholar]
- Klinman NR. The mechanism of antigenic stimulation of primary and secondary clonal precursor cells. J Exp Med. 1972;136:241–260. doi: 10.1084/jem.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozono H, White J, Clements J, Marrack P, Kappler J. Production of soluble MHC class II proteins with covalently bound single peptides. Nature. 1994;369:151–154. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- Malherbe L, Filippi C, Julia V, Foucras G, Moro M, Appel H, Wucherpfennig K, Guery JC, Glaichenhaus N. Selective activation and expansion of high-affinity CD4+ T cells in resistant mice upon infection with Leishmania major. Immunity. 2000;13:771–782. doi: 10.1016/s1074-7613(00)00075-3. [DOI] [PubMed] [Google Scholar]
- Malherbe L, Hausl C, Teyton L, McHeyzer-Williams MG. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity. 2004;21:669–679. doi: 10.1016/j.immuni.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Marzo AL, Klonowski KD, LeBon A, Borrow P, Tough DF, Lefrancois L. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat Immunol. 2005;6:793–799. doi: 10.1038/ni1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, Murali-Krishna K, Ahmed R. Quantitating the magnitude of the LCMV-specific CD8 T cell response: it's even bigger than we thought. J Virol. 2006 doi: 10.1128/JVI.01459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHeyzer-Williams MG, Davis MM. Antigen-specific development of primary and memory T cells in vivo. Science. 1995;268:106–111. doi: 10.1126/science.7535476. [DOI] [PubMed] [Google Scholar]
- McSorley SJ, Asch S, Costalonga M, Reinhardt RL, Jenkins MK. Tracking Salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:71–83. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- McSorley SJ, Cookson BT, Jenkins MK. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J Immunol. 2000;164:986–993. doi: 10.4049/jimmunol.164.2.986. [DOI] [PubMed] [Google Scholar]
- Mercado R, Vijh S, Allen SE, Kerksiek K, Pilip IM, Pamer EG. Early programming of T cell populations responding to bacterial infection. Immunol. 2000;165:6833–6839. doi: 10.4049/jimmunol.165.12.6833. [DOI] [PubMed] [Google Scholar]
- Obst R, van Santen HM, Mathis D, Benoist C. Antigen persistence is required throughout the expansion phase of a CD4(+) T cell response. J Exp Med. 2005;201:1555–1565. doi: 10.1084/jem.20042521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Shedlock DJ, Shen H. Functional characterization of MHC class II-restricted CD8+CD4− and CD8−CD4− T cell responses to infection in CD4‒/‒ mice. J Immunol. 2004;173:2494–2499. doi: 10.4049/jimmunol.173.4.2494. [DOI] [PubMed] [Google Scholar]
- Prlic M, Jameson SC. Homeostatic expansion versus antigen-driven proliferation: common ends by different means? Microbes Infect. 2002;4:531–537. doi: 10.1016/s1286-4579(02)01569-1. [DOI] [PubMed] [Google Scholar]
- Rees W, Bender J, Teague TK, Kedl RM, Crawford F, Marrack P, Kappler J. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc Natl Acad Sci U S A. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudensky A, Rath S, Preston-Hurlburt P, Murphy DB, Janeway CA., Jr On the complexity of self. Nature. 1991;353:660–662. doi: 10.1038/353660a0. [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Scott CA, Garcia KC, Carbone FR, Wilson IA, Teyton L. Role of chain pairing for the production of functional soluble IA major histocompatibility complex class II molecules. J Exp Med. 1996;183:2087–2095. doi: 10.1084/jem.183.5.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimonkevitz R, Colon S, Kappler JW, Marrack P, Grey HM. Antigen recognition by H-2-restricted T cells. II. A tryptic ovalbumin peptide that substitutes for processed antigen. J Immunol. 1984;133:2067–2074. [PubMed] [Google Scholar]
- Steinmetz M, Bluthmann H, Ryser S, Uematsu Y. Transgenic mice to study T-cell receptor gene regulation and repertoire formation. Genome. 1989;31:652–655. doi: 10.1139/g89-119. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Mohrs M, Mallet-Designe V, Teyton L, Locksley RM. Rapid expansion and IL-4 expression by Leishmania-specific naive helper T cells in vivo. Immunity. 2002;17:191–200. doi: 10.1016/s1074-7613(02)00363-1. [DOI] [PubMed] [Google Scholar]
- Stockinger B, Bourgeois C, Kassiotis G. CD4+ memory T cells: functional differentiation and homeostasis. Immunol Rev. 2006;211:39–48. doi: 10.1111/j.0105-2896.2006.00381.x. [DOI] [PubMed] [Google Scholar]
- Tyznik AJ, Sun JC, Bevan MJ. The CD8 population in CD4-deficient mice is heavily contaminated with MHC class II-restricted T cells. J Exp Med. 2004;199:559–565. doi: 10.1084/jem.20031961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Stipdonk MJ, Lemmens EE, Schoenberger SP. Naive CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat Immunol. 2001;2:423–429. doi: 10.1038/87730. [DOI] [PubMed] [Google Scholar]
- Whitmire JK, Benning N, Whitton JL. Precursor frequency, nonlinear proliferation, and functional maturation of virus-specific CD4+ T cells. J Immunol. 2006;176:3028–3036. doi: 10.4049/jimmunol.176.5.3028. [DOI] [PubMed] [Google Scholar]
- Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Zehn D, Bevan MJ. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity. 2006;25:261–270. doi: 10.1016/j.immuni.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]