Abstract
Diploid yeast undergo meiosis under certain conditions of nutrient limitation, which trigger a transcriptional cascade involving two key regulatory genes. IME1 is a positive activator of IME2, which activates downstream genes. We report that Gcn5, a histone H3 acetylase, plays a central role in initiation of meiosis via effects on IME2 expression. An allele, gcn5–21, was isolated as a mutant defective in spore formation. gcn5–21 fails to carry out meiotic DNA replication, recombination, or meiotic divisions. This mutant also fails to induce IME2 transcription; IME1 transcription, however, is essentially normal. Further investigation shows that during wild-type meiosis the IME2 promoter undergoes an increase in the level of bound acetylated histone H3. This increase is contemporaneous with meiotic induction of IME2 transcription and is absent in gcn5–21. In contrast, the RPD3 gene, which encodes a histone H4 deacetylase and is known to be required for repression of basal IME2 transcription in growing yeast cells, is not involved in induction of IME2 transcription or IME2 histone acetlyation during meiosis. These and other results suggest that Gcn5 and Rpd3 play distinct roles, modulating transcription initiation in opposite directions under two different cellular conditions. These roles are implemented via opposing effects of the two gene products on acetylation of two different histones. Finally, we find that gcn5 and rpd3 single mutants are not defective in meiosis if acetate is absent and respiration is promoted by a metabolically unrelated carbon source. Perhaps intracellular acetate levels regulate meiosis by controlling histone acetylation patterns.
A constellation of environmental conditions triggers meiosis and spore formation in the budding yeast, Saccharomyces cerevisiae (1). Limited levels of nitrogen and a fermentable carbon source, conditions associated with starvation, and the presence of a nonfermentable carbon source (2) lead to the transcriptional activation of IME1 and IME2, two key regulators of meiosis-specific gene expression (3, 4). IME1 acts as a positive activator of IME2 transcription (4, 5). Expression of IME2 is required for meiotic DNA replication, meiotic recombination, two nuclear divisions, and spore formation (1).
Transcription of IME2 is repressed during vegetative growth (4, 5). RPD3 is required for this IME2 repression and encodes a histone deacetylase (6–8). In an rpd3Δ mutant, acetylated histone H4 levels increase specifically at the IME2 promoter, suggesting that repression is coupled to histone deacetylation at this promoter (8, 9).
In contrast, hyper-acetylation of promoter-bound histones has been implicated in transcription activation (10). GCN5 encodes a histone acetyltransferase (11) and is required for the activation of a number of genes involved in amino acid biosynthesis, respiration, and other pathways controlled by the SWI/SNF gene products (12–15). Thus far, GCN5-dependent transcription activation has been specifically correlated, in vivo, with hyperacetylation of bound histone H3 in one case, the promoter for the HIS3 gene (16).
Here we describe the identification and analysis of an allele, gcn5–21, isolated as a sporulation-defective mutant in S. cerevisiae. We find that IME2 expression requires GCN5 function. We also explore the functional interplay between a histone acetylase and a histone deacetylase acting on a single promoter under conditions of repressed and activated transcription.
MATERIALS AND METHODS
Media.
YPD is 1% yeast extract, 2% peptone, 2% glucose supplemented with 0.04% Tryptophan. YPA and YPG are the same as YPD except that 2% potassium acetate and 3% glycerol are substituted for glucose, respectively. Sporulation media (SPM) is 0.3% KOAc, 0.02% raffinose unless otherwise noted. Plates were made by adding 2% Bacto agar (Difco).
Strains.
All yeast strains were stored at −80°C and recovered by incubation at 30°C for 12–16 hr on YPG plates. Strains are derivatives of SK1 containing the markers lys2, ura3, leu2∷hisG, trp1∷hisG, arg4-nsp, arg4-bgl, his4X∷LEU2, and his4B∷LEU2 (17), or ime2-lacZ-URA3 (18). The rpd3Δ∷ KanMx allele was constructed by using the following oligos for PCR synthesis to create a KanMx4 marker (19) flanked by RPD3 sequences: 5′-ACAATTGCGCCATACAAAACATTCGTGGCTACAACTCGATATCCGTGCAGGTCGAGGGATCC-3′ and 5′-CTTTTGTTTCACATTATTTATATTCGTATATACTTCCAACTCTTTTTTTCCGCATAGGCCACTAGT-3′. The PCR product was transformed into NKY3222. Disruption of the RPD3 locus was confirmed by PCR.
The following strains contain the markers ho∷LYS2, lys2, leu2∷hisG, ura3, and trp1∷hisG unless otherwise noted. NKY3198 MATa/MATα HO/′′ TRP1/′′ ste7–1/′′ gcn5–21/′′; NKY3200 MATa his4B∷LEU2 arg4-Bgl gcn5–21; NKY3201 MATα his4X∷LEU2 arg4-Nsp gcn5–21; NKY3202 MATa/MATα his4X∷LEU2/his4B∷LEU2 arg4-Bgl/arg4-Nsp gcn5–21/′′; NKY3203 MATa/MATα his4X∷LEU2/his4B∷LEU2 arg4-Bgl/arg4-Nsp gcn5–21/GCN5; NKY3210 MATa/MATα his4X∷LEU2(Bam)-URA3/his4B∷LEU2 arg4-Bgl/arg4-Nsp; NKY3212 MATa/MATα his4X∷LEU2(Bam)-URA3/ his4B∷LEU2arg4-Bgl/arg4-Nsp gcn5–21/′′; NKY3221 MATa/MATα ho∷hisG/′′ gcn5–21/′′; NKY3222 MATa/MATα ho∷hisG/′′; NKY3227 MATa/MATα ho∷hisG/′′ rpd3Δ/′′; NKY3228 MATa/MATα ho∷hisG/′′ gcn5–21/′′ rpd3Δ/′′; NKY3219 MATa/MATα TRP1/′′ his4X∷LEU2/his4 B∷LEU2 spo11∷hisG-URA3-hisG/′′ gcn5–21/′′; NKY3220 MATa/MATα TRP1 his4X∷LEU2/his4B∷LEU2 spo11∷hisGURA3-hisG/′′.
Cloning of NDT21/GCN5.
Genetic methods have been described (20). The ndt21–1 mutation was isolated in the same screen described in refs. 18 and 21. Four DNA clones able to complement the ndt21–1 sporulation-defective phenotype were recovered from a Ycp50-based yeast genomic library and share a 4.5-kb region in common (18). Confirmation that the cloned region covers the mutated segment of DNA and not a suppressor was made in two steps: First, the region in common to all four complementing clones was deleted in one wild-type (WT) clone and filled in by gap repair in a ndt21–1 mutant cell (NKY3198; ref. 22). Second, the mutant allele on the gap-repaired plasmid was used to replace the WT allele in two different unmutagenized haploid cell backgrounds by two-step allele replacement (22). Restoration of a sporulation-defective phenotype in diploids generated from these cells indicated that the NDT21 locus had been cloned. All ntd21–1 mutant strains are derived from NKY3200 and NKY3201, two haploid strains generated in this way. The Spo− phenotype conferred by ndt21–1was complemented by a 1.2-kb PstI–XhoI fragment containing GCN5 that was carried on pRS314 (23).
Cytology.
Sporulation on solid media was carried out by patching single colonies from a YPD plate to SPM and incubating at 30°C for 24–36 hr. Spore formation, scored as asci with one or more spores, was assessed by light microscopic analysis of cells resuspended in water. Cells from liquid cultures were scored similarly. Meiotic divisions were monitored by staining cells fixed in 40% ethanol with 1 μg/ml of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) and by using fluorescence and/or phase-contrast microscopy. At least 100 cells were analyzed for each cytological analysis.
Meiotic Time Courses.
The method of cell synchronization for meiotic time courses has been described (24, 25).
Nucleic Acid Techniques.
Meiotic double-strand breaks near the THR4 locus were assayed as described (26). RNA isolation and Northern blot analysis were carried out as described (27, 28). Hybond-N (Amersham Pharmacia) was used to immobilize total RNA. 32P-labeled DNA probes were prepared by using a random priming kit (Stratagene). The 240-bp HindIII–HindIII fragment containing the IME1 coding region from pPL139 (29), the BamHI–EcoRI fragment containing the IME2 coding region from pMH6 (4), and the 9-kb fragment containing the rRNA gene cluster (pOL73) were used to probe the same membrane sequentially. Probes were stripped from the membrane before each hybridization.
Analysis.
DNA replication was monitored by using a Beckton Dickinson FACScan Analyzer. Cells were prepared for flow cytometry as described (30).
Chromatin Immunoprecipitation (IP).
Chromatin IP was carried out as described (31). Anti-H3.AcLys(9/14) and anti-H4.AcLys(5/8/12/16) were purchased from Upstate Biotechnology. Cells grown in YPD were harvested at OD600 1.0. Meiotic cells were harvested 2 hr after transfer into SPM. Immunoprecipitated DNA and total preimmunopreciptiated DNA (10% of that subjected to IP) were applied to Hybond N+ (Amersham) by using a slot-blot apparatus and hybridized sequentially (32) to randomly labeled probes for the IME2 promoter region (nucleotides −751 to −124; obtained by PCR using the oligos: 5′-ACTGGTAACCCAGATACC-3′ and 5′-ATTCTCTAGACAGCCACC-3′), total genomic DNA isolated from SK1 (25) and for the IME1 promoter region (nucleotides −620 to −33, HindIII–XbaI fragment from YEpK26–7) (3). Probes were stripped from the membrane between hybridizations. A Fuji PhosphorImager was used for quantitation.
RESULTS
Isolation of gcn5–21.
A mutation, ndt21–1, was isolated in a screen for mutants defective in spore formation (21). The ndt21–1 mutation was found to segregate as a single Mendelian trait. Further analysis revealed ndt21–1 to be an allele of GCN5; it was thus renamed gcn5–21 (Materials and Methods).
A gcn5–21 Mutant Does Not Carry Out Meiotic Nuclear Divisions and Does Not Initiate Meiotic DNA Replication or Meiotic Recombination.
Events of meiosis were assayed in parallel in WT and gcn5–21 mutant cells. For each assay, a population of cells was synchronized (24). Cells cultured in rich media plus glucose (YPD) were used to inoculate rich media plus potassium acetate (YPA). Cell cultures were grown ≈13.5 hr at which point >90% of cells arrest in G1 immediately before the entry into meiosis. Cells then were transferred into SPM containing acetate after which the major events of meiosis occur. Cells prepared in this way proceed sequentially and relatively synchronously through meiotic DNA replication, recombination, divisions, and finally spore formation (24).
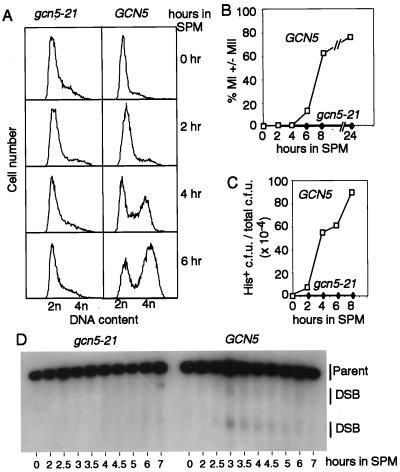
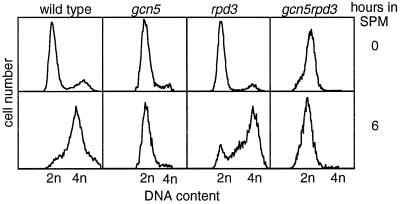
In WT meiosis, DNA replication is complete in most cells by 4 hr and in essentially all cells by 6 hr, as assayed by flow cytometry (FACS); both meiotic divisions are complete in most and all cells by ≈7 and ≈10 hr, respectively, as assayed by light microscopic examination of 4′,6-diamidino-2-phenylindole dihydrochloride-stained cells (24). In the gcn5–21 mutant, no replication was observed after 6 hr (Fig. 1A), or even after incubation in SPM for up to 24 hr (data not shown), and no nuclear divisions were observed at any time point measured through 24 hr (Fig. 1B).
Figure 1.
Events of meiosis in WT and gcn5–21 mutant cells. (A) Meiotic DNA replication in WT (NKY3221) and gcn5–21 mutant cells (NKY3222) at the indicated times points after transfer to SPM. (B) Meiotic divisions (MI ± MII) in the same time course as A. (C) HIS+ prototroph formation in GCN5/gcn5–21 cells (NKY3203) and gcn5–21/gcn5–21 mutant cells (NKY3202) returned to vegetative growth at the indicated times after transfer into SPM. Cells were plated onto synthetic complete media (SC) and onto media lacking histidine (SC-HIS). (D) Meiotic DNA double-strand breaks near the THR4 locus in gcn5–21 (NKY3198) and WT (NKY895) cells at the indicated time points after transfer to SPM assayed by Southern blot analysis.
In general, mutants defective in carrying out S phase also are defective in carrying out meiotic recombination (1), which holds for gcn5–21. Meiotic recombination was assayed in two ways. First, the frequency of His+ and Arg+ prototrophs arising from recombination between his4 and arg4 heteroalleles was measured by a “return-to-growth” assay (33). In a gcn5–21 mutant, His+ and Arg+ prototroph levels were reduced 100- and 300-fold, respectively, compared with WT, indicating no commitment to heteroallelic recombination (Fig. 1C and data not shown). Second, the occurrence of DNA double-strand breaks (DSBs), thought to be the initiating event of meiotic recombination (34), was measured. DSBs are transiently detectable in WT cells several hours after transfer into SPM, as illustrated here (Fig. 1D) for breaks at the THR4 locus (25, 35). In contrast, the gcn5–21 mutant failed to form detectable levels of DSBs at any time point after transfer into SPM (Fig. 1D).
Some mutants are proficient for initiation of recombination but exhibit meiotic arrest, apparently in response to a subsequent block in the recombination process. Accordingly, this type of arrest is alleviated by mutations that block DSB formation (e.g., spo11Δ; see ref. 21). Because gcn5–21 is blocked before the initiation of meiotic recombination, it should not be subject to such alleviation. This expectation is fulfilled: WT and spo11Δ cells sporulated on solid media at 85% and 79% efficiency, respectively, whereas gcn5–21 and gcn5–21 spo11Δ cells gave spores at less than 1% efficiency (NKY3210, NKY3219, NKY3212, and NKY3220, respectively).
GCN5 Is Required for Meiotic Induction of IME2 Transcription.
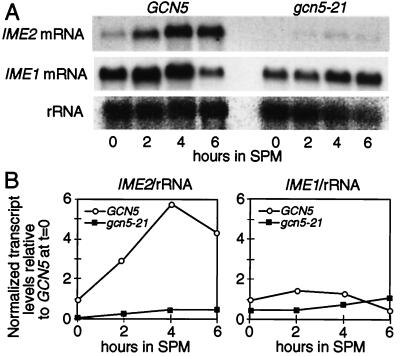
We tested whether GCN5 might be required for the activation of transcription of important meiotic regulatory gene(s) IME1 and/or IME2 (Introduction). First, IME2 transcript levels relative to rRNA were measured in WT and mutant cells at various times after transfer to SPM (Fig. 2, Left). In the gcn5–21 mutant, IME2 transcript levels were reduced 10- to 15-fold from WT levels at all time points after the transfer of cells to SPM. The same effect is observed with an IME2-lacZ transcriptional fusion integrated at the IME2 locus. In the gcn5–21 mutant, the level of β-galactosidase activity was reduced 40-fold from the WT level when measured 6 hr after transfer into SPM (data not shown).
Figure 2.
(A) Northern blot analysis of IME2 and IME1 transcripts levels in WT cells (NKY3210) and gcn5–21 mutants (NKY3212). (B) Relative transcript levels of IME2 (Left) and IME1 (Right) from WT and gcn5–21 mutant cells normalized to rRNA levels. Sporulation of WT cells occurred at 87% efficiency, and no sporulation of gcn5–21 cells occurred (<1%) after 24 hr in SPM.
In contrast, IME1 transcripts were readily detectable in both WT and gcn5–21 mutant cells after overnight growth in YPA; transcript levels did not increase on further incubation in SPM (Fig. 2, Right). Only a slight decrease in IME1 transcript levels was observed for all time points in the gcn5–21 mutant. Other studies have reported very low levels of IME1 transcript in cells grown in YPA with a >30-fold induction on transfer to SPM (e.g., ref. 4). Differences in strain background and/or premeiotic growth conditions may account for the early expression of IME1 transcript in this study.
GCN5- and RPD3-Mediated Modulation of Histone Acetylation in Vegetative and Meiotic Cells.
We wanted to determine whether GCN5 mediates its effects on IME2 expression directly via its acetylase activity and to further explore the role of RPD3 and its functional relationship to GCN5. We therefore used chromatin IP to determine the levels of acetylated histone H3 and acetylated histone H4 bound to different DNA populations in vegetative and meiotic cells, in WT and gcn5 and rpd3 single mutants.
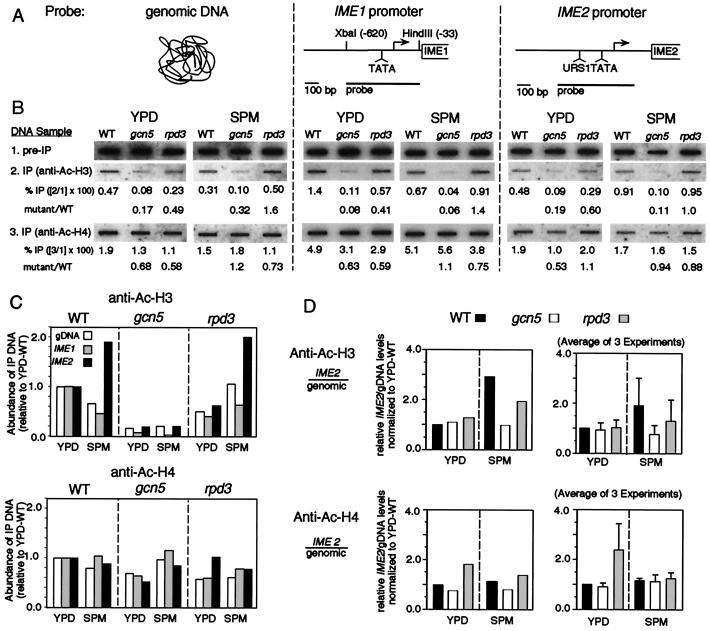
IP was carried out with antibodies specific to the acetylated forms of the two histones on sheared chromatin present in whole-cell extracts from formaldehyde-treated cultures; an aliquot of chromatin removed before IP served as a control. Formaldehyde crosslinks then were removed in both the “IP” and “preIP” samples, and DNA was isolated and applied to a nylon membrane. The membrane then was probed sequentially with three labeled DNAs corresponding, respectively, to total genomic sequences, the IME1 promoter region, and the IME2 promoter region. For each hybridization sample, the level of “bound” acetylated histone was determined by comparing the absolute hybridization signal to the signal obtained from one-tenth volume of a comparable aliquot of total DNA (IP/preIP; Fig. 3B). To simplify consideration of the signals obtained in diverse cell conditions, the efficiency of each relative IP signal was further normalized to that observed for the same probe in WT cells under vegetative growth conditions (Fig. 3C).
Figure 3.
Acetylation of histone H3 and histone H4 in total DNA and at the IME1 and IME2 promoters under vegetative and meiotic conditions. (A) Schematic of probes used. (B) Results of chromatin IP using antibodies to acetylated histone H3 or H4. WT cells (NKY3222) sporulated at 88% efficiency and gcn5–21 (NKY3221) and rpd3Δ (NKY3227) mutants sporulated at <1% efficiency after 24 hr in SPM. (C) Relative IP levels are shown after normalization to YPD-WT levels. (D) The ratio of precipitated IME2 fragment to precipitated genomic DNA from the experiment shown in B (Left) and from three independent experiments (± SD; Right).
At the IME2 promoter, specifically, the level of bound acetylated histone H3 increases during early meiosis.
In WT cells, the level of immunoprecipitated IME2 promoter region associated with acetylated histone H3 was about 2-fold higher in early meiotic cells than in vegetative cells (compare sample 3 with 6, Fig. 3C, Upper Left). In contrast, the levels of immunoprecipitated genomic DNA and IME1 promoter were decreased about 2-fold compared with vegetative cells (compare samples 1 and 2 with 4 and 5, Fig. 3C, Upper Left). This pattern of effects suggests that the IME2 promoter region undergoes meiosis-specific acetylation of histone H3 and that this effect is specific to the IME2 promoter region.
GCN5 mediates the IME2-specific increase in bound acetylated histone H3 and also mediates global histone H3 acetylation in both vegetative and meiotic cells.
The gcn5–21 mutant exhibited a dramatic decrease (3- to 17-fold) in the levels of acetylated H3-bound DNA for all three probed DNAs in both vegetative cells and during meiosis (Fig. 3C, Upper Center). In addition, the gcn5–21 mutant exhibits no IME2-specific increase in acetylated H3 levels during meiosis. The second effect implies that Gcn5 mediates the meiosis-specific increase in acetylated H3 levels that occurs uniquely at the IME2 promoter as seen in WT cells. The first effect implies that Gcn5 also has more global role(s) on the level of DNA-bound acetylated H3, which are strong regardless of which cellular program is occurring and which can be seen for at least one locus that is not subject to meiosis-specific modulation.
RPD3 mediates histone H4 deacetylation specifically at the IME2 promoter during vegetative growth but not during meiosis.
The levels of immunoprecipitated DNA bound to acetylated histone H4 varied little among all of the samples analyzed, with one exception: the IME2 promoter region during vegetative growth. In the rpd3Δ mutant, the acetylation level at the IME2 promoter region was specifically increased 2- to 3-fold relative to the other two probed DNAs (Fig. 3C, Lower Right, compare sample 3 with samples 1 and 2). This result confirms that RPD3-dependent histone H4 deacetylation at the IME2 promoter during vegetative growth, previously reported for haploid cells, also occurs in diploid cells. No such effect is observed during meiosis.
There also is no evidence that RPD3 directly mediates any strong global effect on histone acetylation levels analogous to that seen for GCN5. If anything, an rpd3Δ mutation causes a slight decrease in the levels of H4 acetylation for total DNA and the IME1 promoter in vegetative cells and for all three probed DNAs in meiosis. Interestingly, the rpd3Δ mutant also exhibits a slight increase in the level of bound acetylated H3 in total DNA and at the IME1 promoter.
In summary, GCN5 promotes histone H3 acetylation at many loci, but affects the IME2 promoter uniquely, only as cells enter meiosis. RPD3 function is also specific for the IME2 promoter, but promotes a decrease in histone H4 acetylation and only during vegetative growth. These effects on IME2 are clearly seen when the ratios of immunoprecipitated IME2 promoter and total genomic DNA are plotted for each mutant for each condition in the experiment described above (Fig. 3D, Right). These effects also are seen when averaged over three independent experiments, including one using a different method to prepare whole-cell extract (ref. 36; Fig. 3D, Right).
GCN5 and RPD3 Affect, Respectively, Activated/Meiotic IME2 Transcription and Repressed/Vegetative IME2 Transcription.
The patterns of histone acetylation at IME2 described above raise the possibility that RPD3 and GCN5 might play distinct roles in IME2 transcription during vegetative growth and early meiosis, respectively. To address this possibility, IME2 transcript levels were examined in WT and single and double mutant diploid cells in both situations.
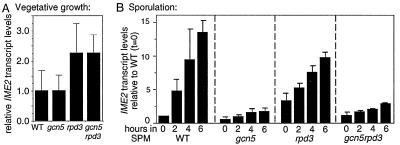
In vegetative cells, a rpd3Δ single mutant exhibits an elevated level of IME2 transcription relative to WT, as observed previously in haploid cells (8). No such increase is observed in a gcn5–21 mutant, which exhibits a WT transcript level; furthermore, a gcn5–21 rpd3Δ double mutant exhibits the same elevated transcript level as the rpd3Δ single mutant (Fig. 4A).
Figure 4.
IME2 transcript levels in vegetative and meiotic WT cells (NKY3222) and gcn5–21 (NKY3221), rpd3Δ (NKY3227), and gcn5–21 rpd3Δ (NKY3228) mutant cells. (A) Average IME2 transcript levels from Northern blot analysis (not shown) of three independent cultures (± SD) normalized to rRNA levels. (B) Average IME2 transcript levels from Northern blot analysis (not shown) of two independent meiotic time course experiments normalized to rRNA levels.
In WT meiotic cells, IME2 transcription begins just after transfer to SPM and increases more or less continuously thereafter. In a gcn5–21 mutant, IME2 transcription is drastically reduced at all time points, confirming requirements of GCN5 for meiotic induction as presented above.
In an rpd3Δ mutant, in contrast, IME2 transcript levels are higher than in WT cells at t = 0, are similar to WT at early times (t = 2 hr) and appear to be somewhat reduced as compared with WT at intermediate/late times (t = 4 and 6 hr; Fig. 4B). And in a gcn5–21 rpd3Δ double mutant, transcript levels are essentially indistinguishable from the gcn5–21 single mutant. Meiotic DNA replication patterns correlate with the presence or absence of the gcn5–21 mutation only; replication occurs in an rpd3Δ mutant but is absent in the gcn5–21 and gcn5–21 rpd3Δ double mutants (Fig. 5).
Figure 5.
Flow cytometry analysis of cells arrested before meiosis in YPA and 6 hr after the transfer of cells into SPM. Strains used are the same as in Fig. 4. After 24 hr in SPM, 61% of WT cells sporulated, and in each single and double mutant strain <1% of cells sporulated.
Complex Roles of GCN5 and RPD3 for Sporulation under Nonstandard Conditions.
In the course of these studies we found that gcn5–21 and rpd3Δ single mutants both are proficient for meiosis, yielding nearly WT levels of viable spores when they undergo meiosis on agar plates supplemented with glycerol as a nonfermentable carbon source rather than acetate (or the related carbon source, ethanol; Table 1). Furthermore, the double mutant is strongly sporulation defective (Table 1), suggesting that, under these particular nutritional conditions GCN5 and RPD3 are functionally redundant. Moreover, addition of acetate to agar plates containing glycerol restores the phenotype observed under “standard” sporulation conditions: gcn5–21 and rpd3Δ single mutants are both defective in formation of spores (Table 1; ref. 37). Thus, the presence of acetate creates more stringent requirements for GCN5 and RPD3 regardless of whether another carbon source is present or not. Notably, when no additional carbon source is present, sporulation is the same as that observed on glycerol alone (Table 1). Presumably, another carbon source in the medium is sufficient under this condition.
Table 1.
Sporulation efficiency on nonfermentable carbon sources
| Strain | Relevant genotype | Acetate | Ethanol | Glycerol | Acetate + glycerol | No carbon | Spore viability* |
|---|---|---|---|---|---|---|---|
| NKY3222 | GCN5 RPD3 | 88% | 49% | 70% | 55% | 71% | 96% |
| NKY3221 | gcn5 RPD3 | <1% | <1% | 26% | 3% | 25% | 94% |
| NKY3226 | GCN5 rpd3 | <1% | 2% | 39% | 4% | 24% | 90% |
| NKY3227 | gcn5 rpd3 | <1% | <1% | <1% | <1% | <1% | No sporulation |
Cells were grown on solid YPD media for 2 days before patching on sporulation plates. All media contain 0.2 M Mes, pH 5.5, 0.1× amino acid drop out powder, 2% agar plus 0.3% of each carbon source. Sporulation was assayed after 3 days.
Twelve four-spore asci were dissected from glycerol-containing media onto YPD plates to measure spore viability.
DISCUSSION
GCN5 Is Required for Entry into Meiosis and for Induction of IME2 Expression.
The gcn5–21 mutation confers a block on meiosis at a very early step, before any DNA replication detectable by flow cytometry. This block explains the absence of meiotic recombination initiation, meiotic divisions, and spores, which are known not to occur if S phase is blocked.
The gcn5–21 mutation also confers a strong block to induction of IME2 transcription. This block is likely because of a direct role of GCN5 in establishing an active IME2 promoter. GCN5 is a histone H3 acetylase; the IME2 promoter, uniquely, exhibits an early meiosis-specific increase in H3 acetylation; and no such increase occurs in the mutant.
The gcn5–21 defect in IME2 transcription should be sufficient to explain the failure of mutant cells to enter meiosis. It cannot be excluded, however, that other defects (e.g., altered bulk chromatin status) also contribute and/or also would by themselves be sufficient. In support of this idea, the gcn5–21 mutation is not an exact phenocopy of ime2Δ; whereas gcn5–21 cells do not carry out meiotic DNA replication, ime2Δ cells eventually do (ref. 38; S.M.B. and N.K., unpublished work). In addition, effects on histone actylation by Gcn5 extend to chromosomal loci that are not also affected for transcription (e.g., the IME1 locus).
GCN5 and RPD3, Independently, Mediate Programmed, Locus-Specific, Histone Subunit-Specific Modulation of Histone Acetylation at the IME2 Promoter with Resultant Effects on Transcription Initiation.
The results presented above suggest that induction of IME2 transcription during meiosis requires the presence of GCN5-acetylated histone H3 and is independent of RPD3-mediated deacetylation of histone H4. Conversely, repression of basal IME2 transcription during vegetative growth requires RPD3-dependent deacetylation of histone H4, as shown previously, and is independent of GCN5.
These results provide an additional example of a positive correlation between histone acetylation and transcriptional activity. More particularly, regulation of the IME2 promoter involves several highly specialized features. Two different specific constellations of bound histones are achieved at the same promoter in two different cell types (vegetative and meiotic). In both cases, the acetylation level of one particular histone subunit was critical, but with two different subunits being relevant in the two different cell types. Also, locus-specific modulation of bound histone status was achieved over and above the background of general histone acetylation, implying that locus-specific factors must participate in setting up the observed arrays. Presumably, different sets of such factors would be important in different cell types, e.g., IME1 in the case of meiotic activation of IME2.
Do GCN5 and RPD3 Play Multiple Roles during Meiosis?
Under standard conditions, GCN5 is required for entry into the meiotic program (above); RPD3, in contrast, is required at later step(s) (39), and it is unknown whether GCN5 does or does not participate in those events. On SPM lacking acetate (or a metabolically related compound), however, a different set of gcn5 and rpd3 mutant phenotypes is observed. These findings imply that the nature of the nonfermentable carbon source present during meiosis affects the roles of these gene products.
The ability of acetate to uncover an early role for GCN5 in meiosis, even in the presence of glycerol, is also interesting because the substrate molecule for GCN5-mediated histone acetylation, acetyl Co-A, is a direct metabolite of acetate (40). This result raises the possibility of a direct link between the presence of acetate in the medium and the activation of IME2 expression. This idea is further supported by the finding that ethanol has the same effect as acetate, because metabolism of ethanol also leads directly to acetyl Co-A (whereas metabolism of glycerol does not; ref. 40). In this regard, it may be relevant that the SNF1-kinase-dependent pathway(s), which enable cells to use acetate, appear to act at the same two steps of meiosis affected by the gcn5 and rpd3 mutations (41).
During meiosis in the absence of acetate, GCN5 and RPD3 are functionally redundant: gcn5 and rpd3 single mutants are proficient whereas only the double mutant is defective. This situation is quite paradoxical given that the two gene products are thought to have opposing effects on histone acetylation status. We find that GCN5 plays a role in bulk chromatin acetylation both during vegetative growth and during early meiosis in the presence of acetate. Thus it is likely that acetylation of bulk chromatin also is affected in the gcn5 mutant in no-acetate meiosis. We are intrigued by the possibility that, in the presence of acetate, the target of Gcn5 is the status of the IME2 promoter, whereas no-acetate meiosis reveals a role for acetylation of bulk chromatin structure. In particular, the presence of hypoacetylated bulk chromatin in meiosis in the gcn5 mutant might uncover or create an additional role for RPD3.
Acknowledgments
We thank S. Honigberg, J. Dekker, O. Hughes, V. Boerner, and J. Henle for discussion and comments on the manuscript. This work and M.A. were supported by a grant to N.K. from the National Institutes of Health (RO1-GM44794). S.M.B. was supported by the Helen Hay Whitney Foundation and a Science Scholar Fellowship from the Bunting Institute of Radcliffe College.
ABBREVIATIONS
- YPD
yeast extract/peptone/glucose
- YPA
yeast extract/peptone/potassium acetate
- SPM
sporulation media
- IP
immunoprecipitation
- WT
wild type
- DSB
double-strand break
References
- 1.Kupiec M, Byers B, Esposito R E, Mitchell A P. In: The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae. Pringle J, Broach J, Jones E, editors. Vol. 3. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 889–991. [Google Scholar]
- 2.Freese E B, Chu M, Freese E. J Bacteriol. 1982;149:840–851. doi: 10.1128/jb.149.3.840-851.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kassir Y, Granot D, Simchen G. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- 4.Smith H E, Mitchell A P. Mol Cell Biol. 1989;9:2142–2152. doi: 10.1128/mcb.9.5.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida M, Kawaguchi H, Sakata Y, Kominami K, Hirano M, Shima H, Akada R, Yamashita I. Mol Gen Genet. 1990;221:176–186. doi: 10.1007/BF00261718. [DOI] [PubMed] [Google Scholar]
- 6.Bowdish S, Mitchell A P. Mol Cell Biol. 1993;13:2172–2181. doi: 10.1128/mcb.13.4.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 8.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Nature (London) 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 9.Kadosh D, Struhl K. Mol Cell Biol. 1998b;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 11.Brownell J E, Zhou J, Ranall T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 12.Guarente L. Trends Biochem Sci. 1995;20:517–521. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]
- 13.Georgakopoulos T, Thireos G. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollard K J, Peterson C L. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Martin J, Johnson A D. Mol Cell Biol. 1998;18:1049–1054. doi: 10.1128/mcb.18.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su S S, Mitchell A P. Genetics. 1993;133:67–77. doi: 10.1093/genetics/133.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wach A, Brachat A, Pohlmann R, Philippsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 20.Rose M, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 21.McKee A H Z, Kleckner N. Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothstein R. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 23.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padmore R, Cao L, Kleckner N. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 25.Cao L, Alani E, Kleckner N. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 26.Keeney S, Kleckner N. Proc Natl Acad Sci USA. 1995;92:11274–11278. doi: 10.1073/pnas.92.24.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collart M A, Oliviero S. In: Current Protocols in Molecular Biology. Ausubel F M, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. New York: Wiley; 1993. pp. 13.12.1–13.12.2. [Google Scholar]
- 28.Sagerstrom C G, Sive H. In: A Laboratory Guide to RNA: Isolation, Analysis, and Synthesis. Krieg P A, editor. New York: Wiley; 1996. pp. 83–103. [Google Scholar]
- 29.Surosky R T, Esposito R E. Mol Cell Biol. 1992;12:3948–3958. doi: 10.1128/mcb.12.9.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sazer S, Sherwood S W. J Cell Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- 31.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 32.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esposito R E, Esposito M S. Proc Natl Acad Sci USA. 1974;71:3172–3176. doi: 10.1073/pnas.71.8.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith K N, Nicolas A. Curr Opin Genet Dev. 1998;8:200–211. doi: 10.1016/s0959-437x(98)80142-1. [DOI] [PubMed] [Google Scholar]
- 35.Goldway M, Sherman A, Zenvirth D, Arbel T, Simchen G. Genetics. 1993;133:159–169. doi: 10.1093/genetics/133.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 37.Vidal M, Gaber R F. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foiani M, Nadjar-Boger E, Capone R, Sage S, Hashimshoni T, Kassir Y. Mol Gen Genet. 1996;253:278–288. doi: 10.1007/s004380050323. [DOI] [PubMed] [Google Scholar]
- 39.Hepworth S R, Friesen H, Segall J. Mol Cell Biol. 1998;18:5750–5761. doi: 10.1128/mcb.18.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fraenkel D. In: Molecular Biology of the Yeast Saccharomyces cerevisiae. Strathern J, Jones E, Broach J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1983. pp. 1–37. [Google Scholar]
- 41.Honigberg S M, Lee R. Mol Cell Biol. 1998;18:4548–4555. doi: 10.1128/mcb.18.8.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]