Abstract
Initiation of X chromosome inactivation requires the presence, in cis, of the X inactivation center (XIC). The Xist gene, which lies within the XIC region in both human and mouse and has the unique property of being expressed only from the inactive X chromosome in female somatic cells, is known to be essential for X inactivation based on targeted deletions in the mouse. Although our understanding of the developmental regulation and function of the mouse Xist gene has progressed rapidly, less is known about its human homolog. To address this and to assess the cross-species conservation of X inactivation, a 480-kb yeast artificial chromosome containing the human XIST gene was introduced into mouse embryonic stem (ES) cells. The human XIST transcript was expressed and could coat the mouse autosome from which it was transcribed, indicating that the factors required for cis association are conserved in mouse ES cells. Cis inactivation as a result of human XIST expression was found in only a proportion of differentiated cells, suggesting that the events downstream of XIST RNA coating that culminate in stable inactivation may require species-specific factors. Human XIST RNA appears to coat mouse autosomes in ES cells before in vitro differentiation, in contrast to the behavior of the mouse Xist gene in undifferentiated ES cells, where an unstable transcript and no chromosome coating are found. This may not only reflect important species differences in Xist regulation but also provides evidence that factors implicated in Xist RNA chromosome coating may already be present in undifferentiated ES cells.
X chromosome inactivation in mammals leads to the cis-limited transcriptional silencing of genes on one of the two X chromosomes in females, resulting in dosage compensation between males and females (reviewed in ref. 1). This developmentally regulated process is closely associated with cellular differentiation during embryogenesis and is mirrored in vitro in female embryonic stem (ES) cells, where differentiation is accompanied by inactivation of one of two X chromosomes (2).
Classical genetic studies in human and mouse have defined a key control region, the X inactivation center (XIC), from which X inactivation initiates and spreads along the length of the X chromosome (reviewed in ref. 3). The XIC is also thought to be involved in a “counting” process whereby only a single X chromosome remains active per diploid cell, with all supplementary X chromosomes being inactivated (2). The Xist gene, which lies within the XIC region in both man and mouse, is a strong candidate for the XIC because it is expressed exclusively from the inactive X chromosome in female somatic cells, producing a long, untranslated transcript (4–7) that localizes to or “coats” the inactive X chromosome (6, 8). In the mouse, gene-targeting experiments have demonstrated that Xist is essential for X inactivation initiation in cis although it may not be involved in counting (9, 10). The Xist transcript thus has been proposed to be the cis-acting signal originating from the XIC. Transgenesis experiments have demonstrated further that murine Xist-containing yeast artificial chromosomes (YACs) and a cosmid are capable of functioning as ectopic XICs when integrated on mouse autosomes in male ES cells (11, 12) and, on condition that they are present in multicopy arrays (13), result in Xist RNA coating of autosomes and cis inactivation as well as counting.
The mechanisms underlying Xist regulation are being unraveled gradually in mice. Analysis of Xist expression in early mouse embryos and differentiating ES cells has revealed that the onset of random X inactivation is preceded by increased Xist transcript levels and localization of these transcripts to the X chromosome to be inactivated (14–16). This up-regulation in Xist expression occurs via stabilization of Xist transcripts, rather than higher transcription rates (15, 16), and more recently it has been shown that the unstable form of Xist is generated by transcription from a previously unidentified Xist upstream promoter, Po (17).
In contrast to the situation in mice, little is known concerning the regulation and function of the human XIST gene. The sequence of Xist is not highly conserved overall between man and mouse apart from some tandem repeat sequences at its 5′ end (18). It therefore is unclear to what extent the findings concerning the mouse Xist gene can be extrapolated to its human counterpart. For example, it is not known whether a human equivalent of Po exists or whether the human XIST transcript exists in an unstable form in ES cells or at any stage during development. Indeed, evidence has been found for differences in Xist regulation and X inactivation patterns between human and mouse during preimplantation development. In mice, only the paternally inherited Xist gene is expressed before implantation, and this may underlie the imprinted inactivation of the paternal X chromosome observed in extraembryonic cell lineages (15, 19). In humans, although there is also some evidence for imprinted X inactivation in extraembryonic tissues, recent findings suggest that the early expression pattern of XIST does not underlie this, because both paternal and maternal XIST genes appear to be equally transcribed in preimplantation embryos (20, 21).
To assess the cross-species conservation of XIST regulation and function, a 480-kb YAC containing the human XIST gene was introduced into mouse male ES cells. The human YAC transgene, when present in multiple copies, showed XIST expression and cis coating of mouse autosomes as visualized by RNA fluorescence in situ hybridization (FISH). Inactivation of autosomal genes linked to a XIST transgene was found, although only in a proportion of cells. Furthermore, late replication timing of transgenic autosomes was not found, suggesting that the inactivation effects were limited, in contrast to observations made with murine Xist transgenes (22). Our results suggest that although the factors responsible for cis association of the human XIST transcript with mouse chromosomes are conserved, other factors required to transform this XIST RNA-coated chromosome into an inactive state may be less well conserved. Unexpectedly, human XIST RNA coated mouse autosomes in cis in undifferentiated ES cells unlike the mouse Xist transcript, which is present only in unstable form in such cells. This may be suggestive of differences in the developmental regulation of XIST between human and mouse. It also indicates that factors necessary for Xist RNA chromosome coating are present already or can be induced in undifferentiated ES cells.
MATERIALS AND METHODS
YAC Manipulation and Generation of Transgenic ES Cell Lines.
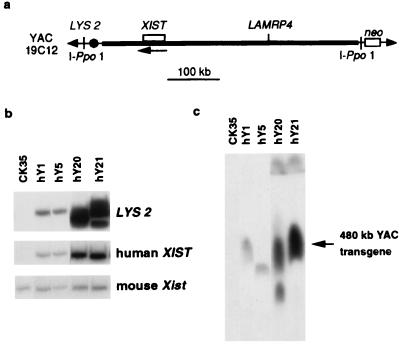
Yeast carrying YAC 19C12 was grown and manipulated according to standard protocols (23). Southern analysis of high-molecular-weight yeast DNA resolved by pulse-field gel electrophoresis (PFGE) was used to verify the size and integrity of the YAC with several XIST probes generated by PCR (corresponding to nucleotides 1–802, 10,008–10,601, 11,428–11,560 of the published sequence; ref. 7) and the 14A probe (6) as well as a LAMRP4 probe (nucleotides 197–491) (24). The integrity of XIST and its flanking sequences were verified further by Southern analysis of EcoRI- and HindIII-digested yeast DNA with the above probes. Using homologous recombination in yeast, the vector arms of YAC 19C12 were retrofitted with IPpo-1 sites and a Pgk-neoR cassette (25) (see Fig. 1a). YAC DNA was purified by preparative PFGE and lipofected into CK35 ES cells (26) as described (27). NeoR clones were selected 24 h post-YAC transfer by G418 treatment (0.25 mg/ml).
Figure 1.
Structural analysis of human XIST transgenes. (a) Human YAC 19C12 is 480 kb long (24). XIST is flanked by about 380 kb of 5′ and 70 kb of 3′ sequence. The XIST gene and LAMRP4 pseudogene are shown. The orientation of XIST transcription is indicated with an arrow. Meganuclease I-Ppo1 sites were introduced into both YAC vector arms to facilitate assessment of YAC integrity in transgenic cells (25) along with the pgk-neo-selectable marker, introduced into one arm, to enable selection for YAC uptake after lipofection into CK35 male ES cells. (b) EcoRI-digested DNA of control CK35 ES cells and transgenic clones hY1, hY5, hY20, and hY21 hybridized with probes for: LYS2, a yeast marker present in one of the YAC vector arms (Top); human XIST (6/7r in exon 1) (Middle); and mouse Xist (HF) (Bottom). Using these probes and others, transgene copy number was estimated to be 1–2 for lines hY1 and hY5, 8–10 for line hY20, and 10–14 for hY21. (c) I-Ppo1-digested DNA of transgenic clones hY1, hY5, hY20, and hY21 resolved on a pulse-field gel. Hybridization with either XIST (shown here), LAMRP4, or YAC vector probes revealed a 480-kb band corresponding to intact YAC sequences in lines hY1, hY5, hY20, and hY21. Lanes corresponding to these clones, from the same autoradiograph, have been juxtaposed. The band in hY5 appears slightly lower in size as a result of aberrant migration because of DNA-loading differences between lanes (as judged by ethidium bromide staining). In line hY20, the additional smaller fragment suggests the presence of some rearrangement in a subpopulation of the transgenic sequences.
Characterization of Transgenic ES Cell Lines.
Transgene copy number and integrity were evaluated by hybridizing Southern blots of EcoRI-digested ES cell DNA with human XIST and LAMRP4 probes (as above) and the LYS2 gene probe present on one YAC vector arm. Quantitation involved PhosphorImager analysis (Molecular Dynamics) of blots after hybridization. A mouse Xist probe (HF; ref. 4) allowed for normalization between samples. Digestion with the IPpo-1 enzyme of high-molecular-weight ES cell DNA embedded in agarose blocks was performed by using the manufacturer’s recommended conditions (Promega), and the DNA then was resolved by PFGE. Southern blots were hybridized with the probes described above to check for the presence of intact YAC transgenes.
Culture and Differentiation of ES Cells.
CK35 ES cells and transgenic derivatives were maintained in the undifferentiated state by culturing in ES medium as described (13). For differentiation into embryoid bodies (EBs), feeders first were removed by successive adsorptions on gelatinized dishes, and the ES cells then were cultivated for 3 days under adherent conditions in ES cell medium (28). Aggregates were formed after mild trypsinization and transferred to suspension culture (day 0) in EB medium as described (13). Half of the EB medium was changed every day. Four-day-old EBs were attached onto LabTek chamber slides (Nunc) for monolayer outgrowth for 3–10 days in DMEM/10% FCS. Only cells furthest from the attached EB mass, considered to be in the most advanced state of differentiation, were examined in RNA FISH analyses. For chromosome replication-timing studies (see below), BrdUrd was incorporated into undifferentiated ES cells and EBs by adding 5 μg/ml BrdUrd and 0.1 μg/ml Colcemid to the medium 4 h before chromosome preparation (29).
Reverse Transcription and PCR Amplification.
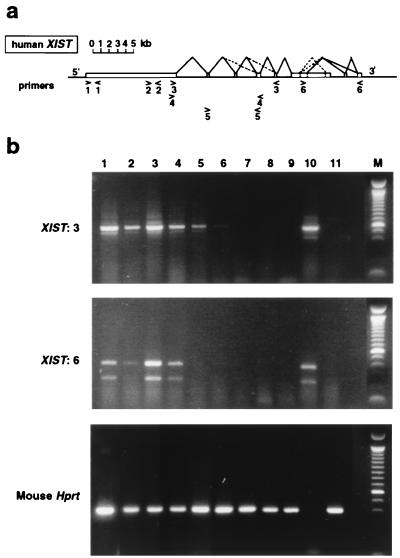
Total RNA was prepared from undifferentiated ES cells and EBs were differentiated for 8 days by using RNA-B (Bioprobe). Female human brain RNA was obtained from CLONTECH. Reverse transcription was performed on 10 μg of RNA by using 400 units of Superscript II reverse transcriptase (RT)/2 μg of random hexamer primers (GIBCO/BRL) as recommended by the manufacturer. A negative control, minus RT enzyme (RT−), was included in all experiments. After reverse transcription, 1/25th of the final reaction mix was amplified by PCR. Mouse Hprt primers NAF/NAR (14) were used to control for cDNA quality. Human XIST primers (see Fig. 2) were as described in ref. 7 for 1 (31 and 20r), 2 (6 and 7r), 3 (8 and 11r), and 6 (3 and 5r) and in ref. 20 for 4 (RT1 and RT3) and 5 (RT4 and RT5). In the case of primer pairs 1 and 2, which do not flank introns, RT− samples were amplified in parallel to control for genomic DNA contamination. All PCRs were performed under standard conditions (1 cycle of 94°C, 5 min; 30 cycles of 94°C, 30 s; 55°C, 30 s; and 72°C, 30 s; and 2 cycles of 72°C, 7 min) by using Taq polymerase (GIBCO/BRL).
Figure 2.
RT-PCR analysis of human XIST expression in transgenic mouse ES cells. (a) The human XIST gene is shown with the locations and orientations of the six primer pairs (1–6) used (see Materials and Methods). The previously determined splicing pattern (7) is shown, with less frequently observed splicing events indicated by dashed lines. (b) Amplification of cDNA in transgenic cells before and after differentiation (8-day EBs). Positive control human cDNA is female brain. Mouse controls are the host ES cell line CK35 and female kidney. Lanes: 1, hY20 undifferentiated; 2, hY20 EBs; 3, hY21 undifferentiated; 4, hY21 EBs; 5, hY1 undifferentiated; 6, hY1 EBs; 7, hY5 undifferentiated; 8, hY5 EBs; 9, CK35 undifferentiated; 10, human female brain; 11, mouse female kidney. Representative data using primer pairs 3 and 6 are outlined in a. Mouse Hprt RT-PCR was used to control the RT samples. The PCR products shown are cDNA-specific as the primers flank introns.
DNA and RNA FISH Analysis.
Interphase nuclei were prepared, and RNA and DNA FISH was performed as described (8, 13). Nuclei were mounted in Vectashield (Vector) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). A Quantix charge-coupled device camera and iplab and photoshop software were used for image acquisition and treatment. Probes (other than chromosome paints) were labeled by nick translation with Spectrum red or green dUTP (Vysis, Downers Grove, IL). The human XIST probe was a pool of approximately 3.5 kb of sequence derived from exon 1 (nucleotides 1–802, 6121–8210, and 10,008–10, 601) and 1 kb derived from exon 6 (14A; ref. 6). The mouse Xist probe was λ 510 (30). The Dhfr/Rep-3 probe was BAC Dhfr 007 C17 (Centre National de Sequencage, France). Chromosomes 2, 13, and the X were detected by using Dig-labeled paint probes (Oncor) and FITC anti-Dig antibodies (Vector). Mouse chromosome-specific YAC probes were used for transgene localization on metaphase spreads as described (31). Chromosome replication-timing studies on BrdUrd-treated cells were performed as described (29). Briefly, chromosomes were prepared and labeled by using FITC-conjugated mouse anti-BrdUrd antibodies (Becton Dickinson) under manufacturer’s recommended conditions. Metaphases containing a single BrdUrd-positive chromosome were photographed and DNA FISH was performed after chromosome denaturation to identify the late-replicating chromosome by using either chromosome (2, 13, or X) paint probes or a YAC 19C12 probe.
RESULTS
Generation and Characterization of Transgenic ES Cell Lines with YAC 19C12.
The 480-kb YAC used in this study, 19C12 (Fig. 1a), has been shown previously to contain approximately 380 kb of sequence 5′ and 70 kb of sequence 3′ to XIST (24). Before the transfer of YAC 19C12 into ES cells, the integrity of XIST and its flanking sequences were verified, and the YAC was retrofitted with a neor-selectable marker and sites for the meganuclease IPpo-1 at its extremities to facilitate assessment of YAC integrity in transgenic cells (see Materials and Methods).
Purified YAC DNA was transferred into CK35 XY ES cells by lipofection. Twenty clones were assessed for the presence of an intact YAC by pulse-field gel resolution of IPpo-1-digested ES cell DNA and Southern analysis by using several probes (see Materials and Methods). The integrity and copy number of the human XIST transgene in these clones also were investigated by Southern analysis of EcoRI-digested ES cell DNA by using numerous probes. Four lines (Fig. 1 b and c) were chosen for further analysis. Two of these (hY1 and hY5) contain low-copy (1–2) transgenes, and the other two contain high-copy transgenes (hY20, 8–10 copies, and hY21, 10–14 copies).
The chromosomal locations of the hY20 and hY21 transgenes were determined by DNA FISH by using mouse chromosome-specific YAC (31) and bacterial artificial chromosome probes. The hY20 transgene is located in the central portion of chromosome 2, and the hY21 transgene lies in the distal part of chromosome 13 (data not shown).
Human XIST Expression in Transgenic Mouse ES Cells Before and After Differentiation.
Human XIST expression was examined by RT-PCR analysis of the hY1, hY5, hY20, and hY21 ES cell lines, before and after in vitro differentiation (see Materials and Methods). Expression was readily detected in high-copy transgenic lines hY20 and hY21 before and after differentiation (Fig. 2). The use of primers along the length of the human gene suggested that the XIST transcript was expressed and spliced correctly, in a comparable fashion to the control human female somatic cells (e.g., primer pairs 3 and 6; Fig. 2). This was the case even in undifferentiated mouse ES cells. In lines hY1 and hY5, which contain only one to two copies of the human YAC, XIST expression was either undetectable or very low both before and after differentiation. Expression of sequences immediately upstream of the 5′ end of XIST could not be detected in any of the lines (data not shown), suggesting that transcription was initiated from the previously defined XIST start site (7) in both undifferentiated and differentiated ES cells.
RNA FISH was used to examine XIST expression in transgenic cell lines at the single-cell level. In undifferentiated ES cells, the mouse Xist gene is expressed as an unstable transcript from every X chromosome present and can be detected by using RNA FISH as a punctate signal (pinpoint) at its site of transcription (15, 16). When induced to differentiate in vitro, female mouse ES cells undergo X inactivation, the first sign of which is coating of the prospective inactive X chromosome with stabilized Xist RNA, detected by RNA FISH as a Xist RNA “domain” (15, 16). The unstable Xist transcript (Xist pinpoint) on the active X chromosome still is expressed when the Xist RNA domain first appears but disappears as differentiation progresses. In male or XO ES cells, where X inactivation does not occur normally, differentiation is accompanied by the loss of Xist expression on the single, active X, and no Xist RNA domains are ever observed (15, 16).
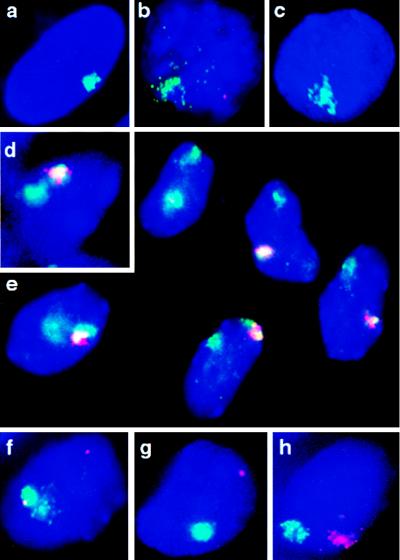
Dual-color RNA FISH was used to visualize human and murine Xist transcripts simultaneously in undifferentiated ES cells. Although no human XIST RNA signal was detectable in lines hY1 and hY5 (data not shown), in high-copy transgene lines hY20 and hY21, the human XIST transcript could be readily detected. It was, however, found to form a large domain, comparable to that seen in human female amniocytes (Fig. 3a), rather than a punctate signal in the majority of cells (94%, n = 65 for hY20, and 92%, n = 227 for hY21) (Fig. 3 b and f). No XIST signal was observed in the remaining cells. In contrast, an RNA pinpoint signal was detected with a mouse Xist probe in almost 100% of undifferentiated ES cells, in all four transgenic cell lines (example shown for line hY21 in Fig. 3b), and in control CK35 ES cells (data not shown). Formation of XIST RNA domains before differentiation appears to be specific to human XIST transgenes, because mouse transgenes (single or multicopy) in the same host ES cell line display punctate Xist RNA signals rather than domains in undifferentiated cells (13).
Figure 3.
FISH analysis of human and murine Xist expression in transgenic mouse ES cells. RNA FISH on undenatured nuclei and DNA FISH after chromatin denaturation (see Materials and Methods). Nuclei are counterstained with 4′,6-diamidino-2-phenylindole (blue). (a) Human XIST RNA (green) detected in human amniocytes: a domain-like signal over the human X chromosome (8). (b) Representative example of human XIST RNA domain (green) and mouse Xist RNA pinpoint (red) signals in undifferentiated ES cells of line hY20 or hY21. (c) Human XIST RNA domain (green) in line hY21 after differentiation (8 days). (d) Human XIST RNA (red) and chromosome 2 DNA (green) in transgenic hY20 cells; overlapping signals appear yellow. (e) Multiple differentiated cells of transgenic line hY21, illustrating the degree of localization of the human XIST RNA (red) over chromosome 13 (green), which was high in the majority of cells and more restricted (e.g., top left) or dispersed (e.g., bottom left) in a minority of cells. Similar observations were made in undifferentiated hY21 cells. Overlapping signals appear yellow. (f) Human XIST RNA (green) and Dhfr/Rep-3 RNA (red) signals detected in undifferentiated hY21 nuclei. (g) Human XIST RNA (green) and Dhfr/Rep-3 RNA (red) signals detected in differentiated (10-day) hY21 cells. (h) Human XIST (red) and mouse Xist (green) RNA domains detected in differentiated (10-day) transgenic hY21 cells.
Human and murine Xist expression also was examined after 8 days of differentiation (i.e., 4 days post-EB attachment; see Materials and Methods) by dual-color RNA FISH. The appearance of the human XIST signal, when present, was unchanged after differentiation in all four transgenic lines: a large, domain-like signal continued to be observed in the majority of hY20 and hY21 cells (Fig. 3 c– e, g, and h), whereas no signal was seen in lines hY1 and hY5 (data not shown).
A small but significant decrease in the proportion of cells containing a XIST signal after differentiation was noted for both lines (χ2 test, P < 0.01). In the case of hY20, the decrease was from 94% (n = 65) to 78% (n = 144), and for hY21, it was from 92% (n = 227) to 82% (n = 583). The reasons for this are unclear, although some cell counterselection may occur as a result of XIST-induced inactivation of autosomal genes in cis upon differentiation (see below).
In the overwhelming majority of 8-day differentiated cells of lines hY20 and hY21, as in CK35 cells, the endogenous mouse Xist signal had, as expected, disappeared completely. However, in a small proportion of transgenic cells, in addition to the human XIST signal, the mouse Xist RNA pinpoint signal had been replaced by a domain over the mouse X chromosome, similar to that seen in female mouse cells (1%, n >300) (Fig. 3h). This was never observed in control CK35 ES cells (n > 1,000) and suggests that human XIST YAC transgenes may be capable of inducing some counting (inactivation of the single mouse X chromosome) as is found with mouse Xist transgenes (11–13). Because inactivation of the single X leads to nullisomy and cell death, earlier differentiation stages of lines hY20 and hY21 were examined. Xist RNA domains were not found in undifferentiated transgenic ES cells but were observed after 1–2 days of differentiation. The proportion of cells with a Xist RNA-decorated X chromosome, however, never was found to exceed 1–2% (n > 300).
Transgenic XIST RNA Coating of Mouse Autosomes and Transcriptional Inactivation of Genes in Cis.
Although the XIST signal in the majority of XIST RNA-positive cells both before and after differentiation resembled that seen to cover the inactive X chromosome in human female amniocytes, in a proportion of transgenic cells it appeared less “compact” or less tightly localized to the chromosome from which it was derived. To assess the degree of autosomal coating by the human XIST transcript more accurately, simultaneous XIST RNA/DNA FISH using paint probes for chromosome 2 (line hY20) and chromosome 13 (line hY21) was performed. Although the XIST RNA signal was found to cover a large part of the relevant autosome in the majority of cells in both lines (>60%), it rarely covered the entire chromosomal region detected by the paint probe, confirming that there was some variability in the degree of coverage of cis-linked regions by the human XIST transcript (Fig. 3 d and e). The same pattern was observed both before and after differentiation. Coating was most extensive for the chromosome 13 transgene in line hY21 (Fig. 3e).
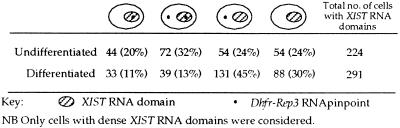
To determine whether human XIST RNA coating could lead to transcriptional inactivation, the expression of two closely linked housekeeping genes on chromosome 13 (Dhfr and Rep-3) was examined in line hY21. Given the variability in the degree of spread of XIST RNA over the autosome mentioned above, these genes were chosen for their relative proximity to the transgene locus, lying just distal to it, as assessed by DNA FISH on metaphase spreads (data not shown). Transcription from each Dhfr/Rep-3 locus was found to be detectable with approximately 50% efficiency in both undifferentiated (49%, n = 143) and differentiated (51%, n = 123) control (nontransgenic) ES cells when RNA FISH was performed by using a probe covering these genes. Dual-color RNA FISH to detect XIST and Dhfr/Rep-3 signals simultaneously was used to analyze potential cis inactivation in transgenic cells. Only cells in which a XIST RNA domain was seen (as shown in Fig. 3 f and g) were scored. In undifferentiated hY21 cells, 52% of the XIST RNA domains contained a Dhfr/Rep-3 RNA pinpoint (n = 224), as expected in the absence of transcriptional inactivation (Fig. 3f and Table 1). After differentiation, cis inactivation in a proportion of cells was suggested by a decrease in the number of XIST RNA domains containing a Dhfr/Rep-3 pinpoint to 25% (n = 291) (Fig. 3g and Table 1). When the full set of data shown in Table 1 was considered, this cis-inactivation effect was found to be highly significant (χ2 test, P < 0.01). The decrease in the proportion of cells containing two Dhfr/Rep-3 pinpoints before (32%) and after (13%) differentiation was significant (χ2 test, P < 0.01), as was the increase in the proportion of cells containing only a single Dhfr/Rep-3 pinpoint from the nontransgenic chromosome 13 before (24%) and after (45%) differentiation (χ2 test, P < 0.01). Dhfr/Rep-3 detection efficiency was controlled by examining the proportion of cells with XIST RNA domains in which the nontransgenic chromosome 13 Dhfr/Rep-3 allele was expressed and was found to be in the order of 50% (56% in undifferentiated cells and 58% in differentiated cells). Repeated analyses of undifferentiated and differentiated hY21 cells gave similar results.
Table 1.
XIST and Dhfr/Rep-3 expression patterns in transgenic line hY21
Because late replication timing is one of the earliest signs of inactivation (ref. 1 for review), transgenic hY20 and hY21 cells were examined to see whether the human transgene was capable of inducing late replication of the transgenic autosome, as has been observed with murine Xist YAC transgenes (22). BrdUrd incorporation and detection were performed on undifferentiated transgenic ES cells and differentiating EBs (see Materials and Methods). Although several hundred metaphases were examined for each line at various stages of differentiation, a late-replicating chromosome was detected only very rarely (<1%, n >200 per slide) and never corresponded to the transgenic chromosome, as assessed by DNA FISH (data not shown). In most cases the relatively small, late-replicating chromosome detected probably corresponded to the Y. We cannot formally rule out, however, the presence of some transgene-associated allocycly (early or late replication) that went undetected under our conditions of BrdUrd incorporation.
DISCUSSION
One approach to understanding the complex, developmentally regulated process of X inactivation is to assess the degree of functional conservation between species of the key initiator region, the XIC and the critical XIST gene lying within it. Although the human XIST promoter linked to a reporter gene has been shown to be constitutively active in mouse somatic cells (32), the regulation of the human XIST gene at the time of X inactivation, during early mouse development or in differentiating ES cells, so far has not been investigated. Here we report that a human XIST-containing YAC, when introduced into mouse ES cells, shows some of the functions found when using mouse Xist YACs in a similar assay but also shows some important differences.
The first interesting parallel between the human XIST YAC transgenic ES cell lines described here and previous reports concerning mouse Xist transgenes (11–13) was that XIST RNA coating was observed only for transgenes present in multicopy arrays. This copy-number dependence suggests that certain elements are not present in sufficient numbers in single- or low-copy Xist/XIST YACs. Such elements may be important in promoting the efficient cis association of XIST RNA, and Lyon (33) recently has proposed that interspersed repetitive elements of the LINE type, in which the X chromosome is particularly rich compared with autosomes, may play such a role. XIST transgenes integrated into relatively LINE-poor autosomal regions therefore might be incapable of efficient ectopic Xic function when present in low numbers.
Human XIST RNA was found to coat mouse autosomes (carrying multicopy transgenes) to some extent in mouse ES cells before and after differentiation, suggesting that the factors ensuring human XIST RNA localization to mouse chromatin in cis are conserved. This contrasts with reports concerning human–rodent somatic cell hybrids, where the human XIST transcript was not found to be localized correctly to the human X chromosome and where it was suggested that species-specific factors presumably were necessary for cis localization (34, 35). Because these studies concerned adult somatic cells rather than ES cells, it may be that murine factors present only during early development and in ES cells are able to ensure the correct cis localization of the human XIST transcript. In adult cells these factors may no longer be present or as efficient in performing this function. The lack of tight localization observed in a minor proportion of transgenic cells may be due to a lack of sufficient elements, enabling full XIST RNA localization to the autosomes in question, as mentioned above (31), rather than to the human origin of the transcript. Indeed, the efficiency of transgene-associated functions may well vary depending on the autosomal integration site. This would be consistent with the partial spread of X inactivation into autosomal sequences observed in certain X-autosome translocations (36).
That the human XIST transcript appeared to coat mouse autosomes even in undifferentiated ES cells, unlike the mouse Xist transcript, which is present only in an unstable form and shows no such coating before differentiation, suggests that the factors necessary for cis localization may already be present in undifferentiated ES cells, but that the mouse transcript is somehow prevented from interacting with them, perhaps via its destabilization (15). Although the stability of the human XIST transcript before and after differentiation was not analyzed directly, the size and intensity of the signal detected by RNA FISH in undifferentiated cells and the fact that this signal was unchanged after differentiation suggested that XIST was not destabilized in undifferentiated ES cells, unlike its murine counterpart. This may indicate a fundamental difference between human and mouse in the way that Xist is regulated during development. One possibility is that there is no human equivalent of the Po promoter, which, in mice, gives rise to the unstable form of the Xist transcript (17). Although our RT-PCR analysis appears to support this, we cannot exclude that such a promoter exists but cannot be used in mouse cells. However, no significant sequence homology to the mouse Po sequence has been found in a large region upstream of the human XIST gene (N. Brockdorff, personal communication). An alternative explanation for the lack of apparent destabilization of the human transcript may be the poor overall conservation of the XIST sequence (18) and an inability of the mouse factors responsible for Xist destabilization to recognize the human target sequences, whether at the RNA level or at the level of regulatory DNA sequences. The investigation of XIST expression in human female ES cell lines (37) should distinguish between these possibilities. We have preliminary evidence that in undifferentiated human teratocarcinoma cells no XIST RNA signal can be seen, suggesting that in human cells XIST expression levels indeed may be regulated differently, i.e., not by transcript destabilization.
Clearly, the ultimate proof of differences in XIST regulation and X inactivation between human and mouse can come only from investigation of human XIST expression during development. RT-PCR studies on preimplantation human embryos suggest that there is no differential XIST expression from paternal and maternal X chromosomes (20, 21), unlike in mice, where only paternal Xist transcripts are detectable up to the late morula stage (14, 19). RNA FISH analysis on mouse embryos has shown that this expression corresponds to stable Xist transcripts that coat the paternal X chromosome (15). This is presumed to underlie the imprinted inactivation of the paternal X in extraembryonic tissues. Unstable Xist transcripts are detected by RNA FISH from both paternal and maternal X chromosomes only from the blastocyst stage, in cells that will go on to form the embryo-proper and in which random rather than imprinted X inactivation occurs (15). Such RNA FISH studies of embryonic human XIST expression have not yet been reported and clearly would be important in revealing mechanistic differences in imprinted and random X inactivation between species.
Evidence for cis inactivation of mouse genes as a result of cis localization of the human XIST transcript was obtained, with a 50% reduction in the number of cells showing transcriptional activity at the Dhfr and Rep-3 genes linked to the hY21 XIST transgene after differentiation. That inactivation was not observed in 100% of cells may be due to resistance of autosomal genes to X inactivation, as has been found for some genes linked to a mouse Xist YAC transgene on chromosome 12 (22) and in human XIC-autosome translocations (36). Alternatively, inefficient silencing could be a result of the inability of the human transgene to recruit efficiently other aspects of the inactivation process that are downstream of XIST RNA coating, such as late replication timing. The rarity with which cells containing mouse Xist RNA domains were detected suggests that the human YAC also was inefficient in its capacity to induce counting.
The absence of Dhfr/Rep-3 inactivation in undifferentiated cells, despite XIST RNA coating, confirms that additional factors present only upon differentiation are required to establish the inactive state. This is consistent with recent findings involving preimplantation mouse embryos (15) and 5-azacytidine-treated somatic cells (where a silent Xist gene is reactivated; ref. 35), where it was demonstrated that Xist RNA chromosome coating, in itself, is not sufficient to induce X inactivation and that other, developmentally regulated factors must be necessary.
We conclude that, in both undifferentiated and differentiated mouse ES cells, a 480-kb transgene shows human XIST expression and that the XIST transcript can associate with mouse autosomes. The latter is, in itself, somewhat surprising given the ≈30% overall sequence divergence between the human and mouse Xist genes (18). It may, however, be the more highly conserved regions, such as the tandem repeats at the 5′ end of the gene, that are critical for Xist’s chromosome-coating function. The cis inactivation observed after differentiation suggests that this association is at least partially functional. Our experiments provide a potential means for identifying essential sequences within this long, functional RNA (≈15 kb in mouse and 17 kb in man) through the creation of chimeric human/mouse Xist genes that could be tested for their ability to carry out different aspects of Xist and Xic function. For example, using such an approach, the sequences underlying the ability in undifferentiated ES cells of the human XIST transcript to form a large domain and not be destabilized, unlike its mouse counterpart, could be identified. The species specificity underlying this last finding may well reflect a fundamental difference in the developmental control of X inactivation between species. Examination of XIST RNA expression patterns in human embryos and ES cells should address this issue.
Acknowledgments
We thank C. Kress and C. Babinet for CK35 ES cells, D. Le Paslier (Centre d’Étude du Polymorphisme Humain) for YAC 19C12, I. Poras (Centre National de Sequencage) for BAC clones, A. Keohane for helpful advice on replication-timing studies, and N. Brockdorff for information concerning the Xist Popromoter. This work was supported by grants from the Association Francaise contre les Myopathies and the Association de Recherche sur le Cancer.
ABBREVIATIONS
- XIC
X inactivation center
- FISH
fluorescence in situ hybridization
- RT
reverse transcription
- YAC
yeast artificial chromosome
- ES cell
embryonic stem cell
- EB
embryoid body
References
- 1.Heard E, Clerc P, Avner P. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 2.Rastan S, Robertson E J. J Embryol Exp Morphol. 1985;90:379–388. [PubMed] [Google Scholar]
- 3.Lyon M. Nature (London) 1996;379:116–117. doi: 10.1038/379116a0. [DOI] [PubMed] [Google Scholar]
- 4.Borsani B, Tonlorenzi R, Simmler M-C, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzni D, Lawrence C, et al. Nature (London) 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 5.Brockdorff N, Ashworth A, Kay G F, Cooper P, Smith S, McCabe V M, Norris D P, Penny G D, Patel D, Rastan S. Nature (London) 1991;35:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 6.Brown C J, Ballabio A, Rupert J L, Lafrenière R G, Grompe M, Tonlorenzi R, Willard H F. Nature (London) 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 7.Brown C J, Hendrich B D, Rupert J L, Lafrenière R G, Xing Y, Lawrence R G, Willard H F. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 8.Clemson C M, McNeil J A, Willard H F, Lawrence J B. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdorff N. Nature (London) 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 10.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 11.Lee J T, Strauss W M, Dausman J A, Jaenisch R. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- 12.Herzing L B K, Romer J T, Horn J M, Ashworth A. Nature (London) 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 13.Heard E, Mongelard F, Arnaud D, Avner P. Mol Cell Biol. 1999;19:3156–3166. doi: 10.1128/mcb.19.4.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kay G F, Penny G D, Patel D, Ashworth A, Brockdorff N, Rastan S. Cell. 1993;72:171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- 15.Sheardown S A, Duthie S M, Johnston C M, Newall A E T, Formstone E J, Arkell R M, Nesterova T B, Alghisi G C, Rastan S, Brockdorff N. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- 16.Panning B, Dausman J A, Jaenisch R. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- 17.Johnston C M, Nesterova T B, Formstone E J, Newall A E, Duthie S M, Sheardown S A, Brockdorff N. Cell. 1998;94:809–817. doi: 10.1016/s0092-8674(00)81739-0. [DOI] [PubMed] [Google Scholar]
- 18.Hendrich B D, Brown C J, Willard H F. Hum Mol Genet. 1993;2:663–672. doi: 10.1093/hmg/2.6.663. [DOI] [PubMed] [Google Scholar]
- 19.Kay G F, Barton S C, Surani M A, Rastan S. Cell. 1994;77:639–650. doi: 10.1016/0092-8674(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 20.Daniels R, Zuccotti M, Kinis T, Serhal P, Monk M. Am J Hum Genet. 1997;61:33–39. doi: 10.1086/513892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray P F, Winston R M L, Handyside A H. Hum Mol Genet. 1997;6:1323–1327. doi: 10.1093/hmg/6.8.1323. [DOI] [PubMed] [Google Scholar]
- 22.Lee J T, Jaenisch R. Nature (London) 1997;386:275–278. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 23.Rose M D, Winston F, Hieter P. in Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 24.Lafreniere R G, Brown C J, Rider S, Chelly J, Taillon-Miller P, Chinault A C, Monaco A P, Willard H F. Hum Mol Genet. 1993;2:1105–1115. doi: 10.1093/hmg/2.8.1105. [DOI] [PubMed] [Google Scholar]
- 25.Fairhead C, Heard E, Arnaud D, Avner P, Dujon B. Nucleic Acids Res. 1995;23:4011–4012. doi: 10.1093/nar/23.19.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camus A, Kress C, Babinet C, Barras J. Mol Reprod Dev. 1996;45:255–263. doi: 10.1002/(SICI)1098-2795(199611)45:3<255::AID-MRD1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.Lamb B T, Sangram S S, Lawler A M, Slunt H H, Kitt C A, Kearns W G, Pearson P L, Price D P, Gearhart J D. Nat Genet. 1993;5:22–29. doi: 10.1038/ng0993-22. [DOI] [PubMed] [Google Scholar]
- 28.Robertson E J. in Teratocarcinomas and Embryonic Stem Cells: A Practical Approach. Oxford: IRL; 1987. pp. 71–112. [Google Scholar]
- 29.Keohane A M, O’Neill L P, Belyaev N D, Lavender J S, Turner B. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- 30.Rougeulle C, Colleaux L, Dujon B, Avner P. Mamm Genome. 1994;5:416–423. doi: 10.1007/BF00357001. [DOI] [PubMed] [Google Scholar]
- 31.Mongelard F, Poras I, Usson Y, Batteux B, Robert-Nicoud M, Avner P, Vourc’h C. Genomics. 1996;38:432–434. doi: 10.1006/geno.1996.0649. [DOI] [PubMed] [Google Scholar]
- 32.Hendrich B D, Plenge R M, Willard H F. Nucleic Acids Res. 1997;25:2661–2671. doi: 10.1093/nar/25.13.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyon M F. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- 34.Hansen R S, Canfield T K, Staneck A M, Keitges E A, Gartler S M. Proc Natl Acad Sci USA. 1998;95:5133–5138. doi: 10.1073/pnas.95.9.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clemson C M, Chow J C, Brown C J, Lawrence J B. J Cell Biol. 1998;142:13–23. doi: 10.1083/jcb.142.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White W M, Willard H F, Van Dyke D L, Wolff D J. Am J Hum Genet. 1998;63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson J A, Itskovitz-Eldor J, Shapiro S S, Waknitz M A, Swiergiel J J, Marshall V S, Jones J M. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]