Abstract
A cell line harboring all trans-acting elements necessary for hypermutation was transfected with a plasmid harboring the major cis-acting elements plus a green fluorescent protein gene containing a premature chain-termination codon. Transfected cells do not fluoresce unless the stop codon reverts. When a sizable cell population is purged of revertants by sorting, the frequency of mutants increases linearly with time, and there is no Luria–Delbrück fluctuation effect. Moreover, as mutant frequencies seemed to vary less than cell numbers in replicate cultures, it is suggested that hypermutation might not be coupled closely to cell division.
In 1943, Luria and Delbrück (1) pointed out that the frequency of spontaneous mutation in microorganisms was subject to great fluctuation. There are usually no mutants early in a culture started from a few cells. However, if, by chance, a mutant arises during this early period, its offspring give rise to a mutational “jackpot.” Luria and Delbrück argued that the large fluctuation in the number of bacteria surviving exposure to, for example, streptomycin, must mean that resistance was conferred by spontaneous mutation that accumulated in the culture and not by a physiological adaptation induced by streptomycin. Today, there are usually more straightforward methods, e.g., sequencing, for determining whether a variant arises by mutation. However, the fluctuation effect continues to make life difficult for those who would measure spontaneous mutation rates.
Although there have been many attempts to bypass the fluctuation effect by purely mathematical analyses, these have resulted in, at best, mitigating—not eliminating—the problem. Experimental attempts to mitigate the problem (e.g., the compartmentalization assay) have been more successful but at a heavy price: hard work (2, 3). No uncertainty principle is operative here; there is no reason why the fluctuation effect cannot be overcome experimentally. The first successful attempt along these lines was the then-heroic dissection of buds from yeast mother cells and testing them in selective medium performed by Ephrussi, L’Heretier, and Hottinguer (4) in 1949. A year later, the chemostat was invented by Novick and Szilard (5, 6). In a chemostat, effectively infinite populations of cells were grown for an effectively infinite time, but various mechanical and analytical problems limited its general usefulness, especially for eukaryotic cells.
In principle, we need only a mutant-free population that is large enough so that some tens or hundreds of mutants arise during a single generation time. Systems with relatively high mutation rates, e.g., hypermutable B cells, present no great problem. The rub is how to get a mutant-free population of that size. Because mutants inevitably will arise during the growth phase needed to attain the large population, there would seem to be only three ways to save the situation: (i) increase the mutation rate after the expansion phase; (ii) select against mutants during expansion; (iii) deplete the preexisting mutants when the population is large enough. The first alternative has been used for a long time; people working on induced mutations do not worry much about the fluctuation effect. However, for this study, we are concerned with spontaneous mutations; thus, this alternative is not available to us.
The second alternative also has been used. It requires one to be able to select for and against the same mutation. This selection was accomplished for nuclear and cytoplasmic petite mutants of Saccharomyces cerevisiae (4, 7–9). Cells from the Chinese Hamster cell line G12 with a single copy of an Escherichia coli gpt transgene (gpt+) grow in HAT medium (hypoxanthine/aminopterin/thymidine) but are killed by 6-thioguanine, whereas gpt− cells are resistant to 6-thioguanine but are killed in HAT medium (10). Rossman et al. (11) have exploited this fact by expanding a large population of G12 cells in HAT medium before measuring the rate of mutation to gpt−. Unfortunately, the use of selective agents has not yielded reliable mutation rates in lymphocyte cell lines, presumably because they grow in suspension rather than attached to a substrate (12). Resistant cells are apparently affected strongly by products leaking from dead, nonresistant cells or by their lysis. Adherent cells usually become detached from the surface when they die and, as a consequence, are easily washed off, leaving the attached, resistant cells (12).
Here, we describe a method for experimentally eliminating the fluctuation effect that is based on the third alternative, depletion of mutants after population expansion. Our method uses the green fluorescent protein (GFP), which was developed as a marker for gene expression by Prasher and colleagues (13) and which has been used by Cariello et al. (14) for assays of induced forward mutation and reversion in E. coli. The GFP is eminently suitable for use in flow cytometry (15).
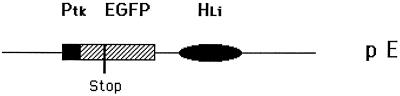
Cell line Δμ, a derivative of the A-MuLV-transformed pre-B cell line 18-81, has all of the trans-acting components necessary for hypermutation (3). Plasmid pE has the major cis-acting components, i.e., a thymidine kinase promoter and the large intron enhancer of the Ig heavy chain (16), but it is missing the 3′ enhancer, which is also necessary for full-blown hypermutation (17, 18). Plasmid pE, which is diagrammed in Fig. 1, has a GFP gene containing a TAG chain-termination codon. In clones of Δμ that are stably transfected with pE, the stop codon is hypermutable to varying degrees. For the experiments reported here, we have used a clone that was more hypermutable than most, presumably because it is integrated at a site supplying the function of the missing 3′ enhancer.
Figure 1.
Diagram of plasmid pE. The SalI–XbaI fragment of plasmid BS/V81x-tk, containing a rearranged exon encoding the variable region, was replaced by the Eco47III–MluI fragment from plasmid pEGFP-C1 (CLONTECH). The color-shifted GFP gene (EGFP) is driven by Ptk, the thymidine kinase promoter (Stratagene) and the large intron enhancer (Hli) of the Ig heavy-chain gene. There is a poly(A)-addition site between the EGFP gene and the large intron. The construct was linearized with NotI before transfection (electroporation).
Culture vessels (n = 5), each containing 100 ml of RPMI medium 1640, were each inoculated with about 0.75 million cells of the same stably transfected clone. These vessels were incubated until there were 20–40 million cells per culture. Then, on day 0, they were sorted in a Moflo high speed sorter (Cytomation, Fort Collins, CO), and 3–5 million nonfluorescent (i.e., nonmutant) cells were seeded into 100 ml of fresh medium. The volume of the culture was slowly expanded up to 800 ml over the course of 4 days. On each day, a sample was removed, and the numbers of fluorescent (i.e., mutant) and nonfluorescent (i.e., nonmutant) cells among a million viable cells were analyzed by flow cytometry. On day 4, 1–1.2 million viable cells were removed from three of the cultures and inoculated into 200 ml of fresh medium; on day 6 these cells were analyzed by flow cytometry. The results are shown in Tables 1 and 2 and Fig. 2.
Table 1.
Fluorescent mutants arising during expansion of cell populations of clone 4-19 cells sorted on day 0 for nonfluorescence
| Culture vessel | Mutant frequency, n × 106
|
||||||
|---|---|---|---|---|---|---|---|
| Day 0 | Day 1 | Day 2 | Day 3 | Day 4 | Day 6 | Σ* | |
| A | 3 | 111 | 155 | 210 | 240 | 310 | |
| B | 3 | 115 | 160 | 215 | 249 | 297 | |
| C | 3 | 92 | 128 | 201 | 243 | 308 | |
| D | 6 | 79 | 156 | 210 | 246 | — | |
| E | 6 | 76 | 119 | 173 | 233 | — | |
| Mean | 4.2 | 94.6 | 143.6 | 201.8 | 242.2 | 305.0 | 991.4 |
| SEM | 0.7 | 8.0 | 8.4 | 7.5 | 2.7 | 4.1 | |
| Var | 2.7 | 320.3 | 350.3 | 284.7 | 37.7 | 49.0 | 1044.7 |
This column tests the expectation that the mean is numerically equal to the variance (Var) if the data have been sampled from a Poisson distribution. The closeness of the sum of the means to the sum of the variances suggests that there is no systematic deviation from the Poisson expectation.
Table 2.
Total cell numbers during expansion of cell populations of clone 4-19 sorted on day 0 for nonfluorescence
| Culture vessel | Total cell numbers, n × 106
|
|||||
|---|---|---|---|---|---|---|
| Day 0 | Day 1 | Day 2 | Day 3 | Day 4 | Day 6 | |
| A | 3.0 | 12.0 | 40.0 | 160.0 | 840.0 | 13.0 |
| B | 3.0 | 6.5 | 41.0 | 190.0 | 960.0 | 14.0 |
| C | 3.0 | 8.0 | 23.0 | 88.0 | 430.0 | 14.0 |
| D | 5.0 | 17.0 | 69.0 | 290.0 | 1,162 | |
| E | 5.0 | 16.0 | 66.0 | 230.0 | 1,134 | |
| Mean | 3.8 | 11.9 | 47.8 | 191.6 | 905.2 | 13.7 |
| SEM. | 0.5 | 2.1 | 8.7 | 33.8 | 132.5 | 0.3 |
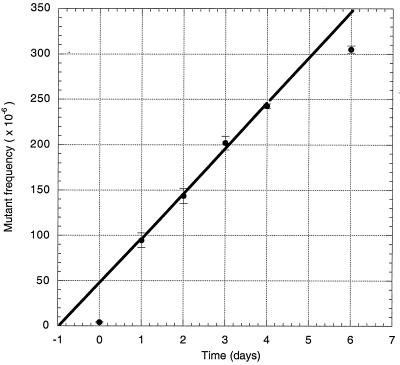
Figure 2.
Increase in the frequency of fluorescent reversions with time. Each point and its associated error bars are means ± SEM; data were taken from Table 1. The curve, which was fitted by the program kaleidagraph (Synergy Software, Reading, PA) to the points for days 1–4 only (see text), corresponds to a mutation rate of about 50 per million cells per day or 25 per million cells per generation time.
An average of 4.2 fluorescent cells per million remained in the cultures immediately after sorting (Table 1, day 0). On day 1, the number of mutants jumped to an average of 94.6 per million. This jump is largely the result of phenotypic lag, i.e., we sort out only those cells that express GFP but not those mutant cells that have not yet started to express GFP. From day 1 to day 4, the frequency of mutant cells increased linearly (Fig. 2). Perhaps because of the shock of being placed in fresh medium, the increase from day 4 to day 6 was somewhat less than expected from extrapolating the curve.
The remarkable feature of these results can be seen by looking at the means and variances shown in Table 1. For a Poisson distribution, the mean is equal to the variance, and this variance is the minimum possible for an experiment such as this one. Unfortunately, the sampling error for a variance with only five (or three) samples is relatively high, and in the data shown in Table 1, some means are much greater than the corresponding variances, but other means are quite lower. As can be seen from the rightmost column of Table 1, the sum of all the variances is very close to the sum of all the means, which would be expected if the number of mutants per culture is Poisson distributed; i.e., no Luria–Delbrück fluctuation is evident.
Another remarkable, but unexpected feature of these results is evident when comparing Table 2 with Table 1. On days 1–4, the total number of cells ranges from a factor of two to more than three. It is not clear why the cell numbers in duplicate cultures should vary so much; we estimate that the cell numbers (counted in a Neubauer chamber) in Fig. 2 ought to be accurate to within ±20%. Thus, the variation in cell numbers seems to be much greater than the variation in the frequency of mutant cells. If these variations are accurate, they suggest that mutation is not highly coupled to cell division in these cells. Clearly, this point requires further investigation. We note, however, that the notion of hypermutation not being associated with genome replication has been suggested previously (19, 20). In any event, for practical purposes, it probably would be better to express hypermutation rates on a per-hour rather than on a per-generation time basis.
The method described here was developed to study hypermutation in cell lines of the B lymphocyte lineage in the absence of Luria–Delbrück fluctuation but could be adapted easily to study mutation in other systems. Although there are few systems involving cell lines with spontaneous back-mutation rates high enough to replicate the present experiments, forward mutations (i.e., from fluorescent to nonfluorescent cells) could be studied with a vector lacking the stop codon. Moreover, the ease of automating the readout should make the method attractive for studying induced mutations.
It would be interesting to investigate whether the pE construct can also be used in vivo. In the germinal centers of a pE-transgenic mouse, the GFP gene should be a target for the endogenous hypermutation machinery. Thus, foci of fluorescent cells should be detectable in the germinal centers. Taking such an approach even further, one could envision using GFP expression as the readout for somatic mutations induced by genotoxic agents. A mouse transgenic for the construct used here should have foci of one or more fluorescent cells in various tissues, and the number of foci would increase after exposure to genotoxic agents for tissues for which the agent is a potential carcinogen. Because of the development of emission-altered GFPs, one could even generate a mouse transgenic for several similar constructs, each requiring a different molecular event for reversion to the expression of proteins, each fluorescing with its own characteristic color. Such a multiply transgenic mouse would represent an “intermediate-term” test for genotoxicity.
Acknowledgments
This paper is dedicated to the memory of Salva E. Luria and Max Delbrück. We thank Marilia Cascalho, Matthias Wabl, Matthias Haury, and Ed Palmer for critically reading the manuscript. The Basel Institute for Immunology was founded and is supported by F. Hoffmann La-Roche. Work in San Francisco was supported by National Institutes of Health Grant 1R01 GM37699. Work in Edmonton was supported by an operating grant from the Natural Sciences and Engineering Research Council of Canada.
References
- 1.Luria S E, Delbrück M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Borstel R C, Cain K T, Steinberg C M. Genetics. 1971;69:17–27. doi: 10.1093/genetics/69.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wabl M, Burroughs P D, von Gabain A, Steinberg C M. Proc Natl Acad Sci USA. 1985;82:479–482. doi: 10.1073/pnas.82.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ephrussi B, L’Heritier P, Hottinguer H. Ann Inst Pasteur (Paris) 1949;77:64–83. [Google Scholar]
- 5.Novick A, Szilard L. Science. 1950;112:715–716. doi: 10.1126/science.112.2920.715. [DOI] [PubMed] [Google Scholar]
- 6.Novick A, Szilard L. Proc Natl Acad Sci USA. 1950;36:708–719. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ephrussi B, Hottinguer H. Nature (London) 1950;166:956. doi: 10.1038/166956a0. [DOI] [PubMed] [Google Scholar]
- 8.Ogur M, St. John R, Ogur S, Mark A M. Genetics. 1959;44:483–496. doi: 10.1093/genetics/44.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Borstel R C. Methods Cell Biol. 1978;20:1–24. doi: 10.1016/s0091-679x(08)62005-1. [DOI] [PubMed] [Google Scholar]
- 10.Klein C B, Rossman T G. Environ Mol Mutagen. 1990;16:1–12. doi: 10.1002/em.2850160102. [DOI] [PubMed] [Google Scholar]
- 11.Rossman T G, Goncharova E I, Nadas A. Mutat Res. 1995;328:21–30. doi: 10.1016/0027-5107(94)00190-g. [DOI] [PubMed] [Google Scholar]
- 12.Wabl M, Jack H M, Meyer J, Beck-Engeser G, von Borstel R C, Steinberg C M. Immunol Rev. 1987;96:91–107. doi: 10.1111/j.1600-065x.1987.tb00511.x. [DOI] [PubMed] [Google Scholar]
- 13.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 14.Cariello N F, Narayanan S, Kwanyuen P, Muth H, Casey W M. Mutat Res. 1998;414:95–105. doi: 10.1016/s1383-5718(98)00043-6. [DOI] [PubMed] [Google Scholar]
- 15.Ropp J D, Donahue C J, Wolfgang-Kimball D, Hooley J J, Chin J Y, Hoffman R A, Cuthbertson R A, Bauer K D. Cytometry. 1995;21:309–317. doi: 10.1002/cyto.990210402. [DOI] [PubMed] [Google Scholar]
- 16.Bachl J, Olsson C, Chitkara N, Wabl M. Proc Natl Acad Sci USA. 1998;95:2396–2399. doi: 10.1073/pnas.95.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachl J, Wabl M. Proc Natl Acad Sci USA. 1996;93:851–855. doi: 10.1073/pnas.93.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachl J, Wabl M. Immunogenetics. 1996;45:59–64. doi: 10.1007/s002510050167. [DOI] [PubMed] [Google Scholar]
- 19.MacPhee D G. Genetica (The Hague) 1996;97:183–195. doi: 10.1007/BF00054625. [DOI] [PubMed] [Google Scholar]
- 20.Bertocci B, Quint L, Delbos F, Garcia C, Reynaud C A, Weill J C. Immunity. 1998;9:257–265. doi: 10.1016/s1074-7613(00)80608-1. [DOI] [PubMed] [Google Scholar]