Abstract
P element insertion is essentially random at the scale of the genome. However, P elements containing regulatory sequences from Drosophila engrailed and polyhomeotic genes and from the Bithorax and Antennapedia complexes show some insertional specificity by frequently inserting near the parent gene (homing) and/or near genes containing Polycomb group response elements (preferential insertion). This phenomenon is thought to be mediated by Polycomb group proteins. In this report, we describe a case of homing of P elements containing regulatory sequences of the linotte gene. This homing occurs with high frequency (up to 20% of the lines) and high precision (inserted into a region of <1 kilobase).We present evidence showing that it is not mediated by Polycomb group proteins but by a new, as yet unknown, mechanism. We also suggest that P element homing could be a more frequent phenomenon than generally assumed and that it could become a powerful tool of Drosophila reverse genetics, for which there is no other described gene targeting technique.
Transposable elements are potent and useful mutagens because they generate a detectable and reproducible lesion on insertion. In Drosophila melanogaster, the P transposable element has become particularly valuable because it moves with high frequency and can mediate germ line transformation (1) and because its transposition can be controlled by limiting the availability of transposase (2, 3). These properties, added to the use of different engineered P elements, have made this transposon one of the most powerful tools of Drosophila genetics.
Although P element insertion sites are distributed throughout the genome in a pattern indistinguishable from random if a small number of hot spots are excluded from consideration (4), some biases are known to affect P element insertion: (i) Euchromatic sites are hit more often than heterochromatin (5); (ii) some euchromatic loci are much more susceptible to P mutagenesis than others (6); (iii) within genes, there is a preference for insertion in the 5′ noncoding sequences (4); and (iv) target sites with close matches to the consensus octamer GGCCAGAC are more likely to receive P element insertions (7, 8). This broad spectrum of insertion has been used to isolate numerous mutant strains but does not allow the targeting of P elements to particular sites.
However, a few exceptions to this rule have been described. P elements including specific regulatory sequences from the Drosophila engrailed (en) gene and from the Bithorax and Antennapedia homeotic complexes (BX-C and ANT-C) have been shown to insert in the genome, in a selective manner, often near the endogenous gene (9–11). This phenomenon of insertion near the parent gene has been called “homing” (9). Moreover, these transposons, along with transposons including regulatory sequences from the polyhomeotic (ph) gene, have been shown to preferentially insert into chromosomal locations known to contain binding sites for some Polycomb group (Pc-G) proteins on polytene chromosomes (12–14). In addition to modifying the insertional specificity of P elements that contain them, these regulatory sequences also interfere with the expression of the miniwhite marker gene (15, 13, 16). Furthermore, the eye color of strains containing such P elements is modified by Pc-G genes mutations (13, 16). Finally, these P elements create new binding sites for Pc-G proteins at their chromosomal localizations (refs. 14, 17, and 18; S. Bloyer and J.-M.D., unpublished results). To account for the unusual properties of these P elements, it has been proposed that the regulatory sequences they contain are Pc-G response elements (PREs) on which Pc-G proteins can bind and form their regulatory complex (13, 14). This implies that the modification of insertional specificity of P elements including PRE could be explained by interactions between transgenic and resident PREs mediated by Pc-G proteins (13, 19).
Here, we report a case of P element homing mediated by regulatory sequences of the linotte (lio) gene (20). We show that P elements including regulatory sequences of this gene (P[lio]) insert with high frequency and precision into the lio resident gene. We also present evidence strongly suggesting that this homing occurs by a PRE-independent mechanism that does not involve the Pc-G proteins, and we propose that P element homing could be a more frequent feature than generally assumed.
MATERIALS AND METHODS
Fly Strains and Culture.
All strains were maintained on standard medium food at 25°C. All variants used are described in Lindsley and Zimm (21), except when stated in the text. The generation and identification of the lio1 and lio2 mutations have been described (20, 22). The drlP3765 (called here drlP) mutant has been described by Callahan et al. (23).
Construction of Transposons.
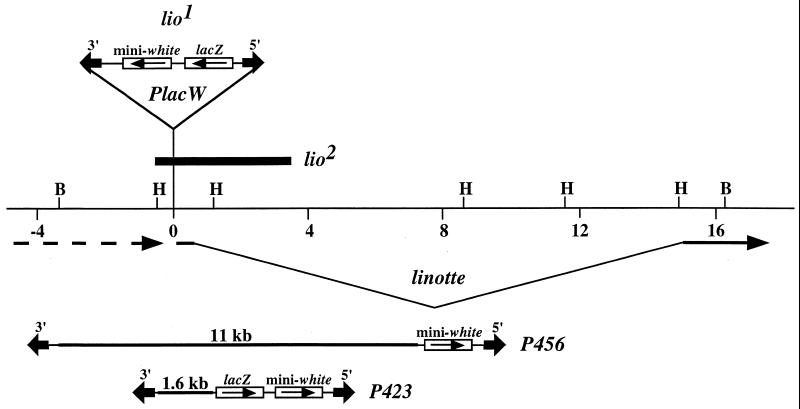
The P456 genomic insert was cloned as an 11-bilobase (kb) BamHI fragment from a CosPeR (24) genomic clone uncovering the 5′ region of the lio locus into the P[CaSpeR4] transformation vector (25). One of the flanking BamHI sites is not present in the endogenous genomic DNA, which migrates as a 19-kb BamHI fragment (Fig. 1)
Figure 1.
Molecular map of the linotte locus. The lio1 PlacW element is inserted between two transcription units within a 1.6-kb HindIII genomic fragment that contains the transcription start site, first exon, and beginning of the first intron of the lio gene. The extent of the lio2 deficiency is represented by the black rectangle. The genomic inserts contained in the P456 and P423 transposons are represented by the black thick lines. P elements maps are not at the scale.
The P423 element was constructed by cloning the 1.6-kb HindIII fragment (Fig. 1) from a pBluescript II KS(+) (Stratagene) subclone into the pX27 transformation vector (A. Simon, F. Savary, and T. Préat, personal communication; ref. 26). The P WHZ10-I4 element used as a control was a pWHZ10 transposon containing a 249-bp intronic fragment from the suppressor of forked gene (A. Audibert, B. Benoit, and M. Simonelig, personal communication; ref. 27).
Generation of Transgenic Lines.
P element-mediated transformation was carried out essentially as described by Rubin and Spradling (1). w1118 embryos were injected with 150 mg/ml of helper plasmid [pUChsPΔ2–3 (D. Rio, personal communication)] and 300 mg/ml of construct. Transformants were identified on the basis of their eye color and were established as homozygous or as balanced heterozygous stocks. New transgenic lines were generated by mobilization of an X-linked P element to autosomes using the stable source of transposase Δ2–3 (3).
Interaction with Pc-G Genes.
The Pc-G genes alleles tested were: polyhomeotic: ph410 (21), Additional sex comb: AsxXF3 (28), Enhancer of Polycomb: E(Pc) (21), Enhancer of zeste/polycombeotic: E(z)60 (antimorphic allele) (21) and pco1902 (amorphic allele) (29), extra sex comb: escr4 (null allele; G. Struhl, personal communication), pleihomeotic: l(4)102EFc3 (21), Polycomb: Pc16 (21), Polycomb-like: Pclx21 (null or strong hypomorphic allele; R. Saint, personal communication), Posterior sex comb: Psc1 (21), Sex comb extra: Sce1 (21), Sex comb on midleg: ScmD1 (21), super sex comb: sxc1 (21).
For Pc-G mutants of the second and third chromosomes, eye color was compared between sibling flies of the same age and sex obtained after the following crosses: w1118/w1118; CyO/T(2;3)apterousXa females were crossed with mutation/Balancer males; resulting mutation/Xa males then were crossed with P[lio] females, and eye color was examined in the progeny. For the polyhomeotic ph410 allele, w ph410/w ph+ females were crossed with P[en] or P[lio] males, and the eye color was compared between ph410 (identified by their extra sex comb phenotype) and ph+ males. The fourth chromosome pleihomeotic l(4)102EFc3 mutant was tested as follows: w1118/Y, l(4)102EFc3/CiD males were crossed with P[lio] females, and eye color was examined in the progeny.
PCR and Sequence.
PCR was performed in 10-μl volumes on Drosophila genomic DNA samples extracted by the simple single-fly procedure (30). Denaturation, annealing, and extension were at 94, 58, and 72°, respectively, for 30 seconds each. The number of cycles was 30. All amplifications used Dynazyme II DNA polymerase (Finnzymes, Helsinki).
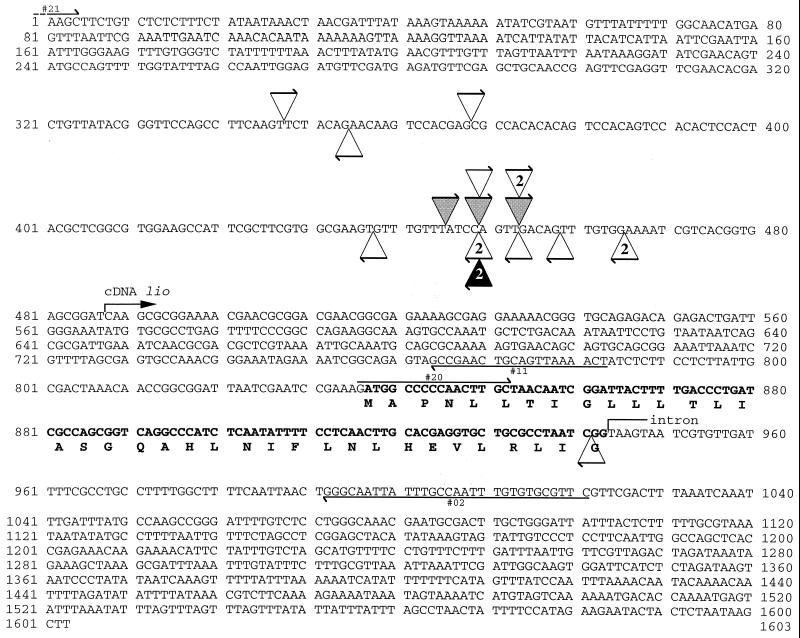
Localization of P elements within the lio gene was done with primers: #02 gaacgcacacaaattggcaaataattgccc, #11 agttttaactgcagttcggc, #21 cctattttttttttgcttctgcaagc, #20 gatggcccccaacttgc (Fig. 2), KP5′ atacttcggtaagcttcggctatcgacg, and KP3′ ctcgcacttattgcaagcatacgttaagtggatg, which extends from the 5′ and 3′ ends, respectively, of P elements in the direction of the adjacent genomic DNA. Five microliters of each PCR sample then were electrophoresed on agarose gels. As PCR products migrated as a single band, the remaining 5 μl of PCR samples were reamplified in 40-μl volumes in the same conditions as previously described using the same pair of primers and sequenced.
Figure 2.
Sequence of the 1.6-kb HindIII genomic fragment in which all of the P insertions within the lio gene have occurred. The black triangle represents P element devoid of lio regulatory sequences [lio1 (20) and drlP (23)], gray triangles represent P456 elements, and open triangles represent P423 elements. The arrow represents the 5′ to 3′ orientation of the P elements. When several independent P elements were found at the same position in the same orientation, the number of occurrences is indicated within the triangle.
DNA sequencing was done by the dye termination method using a Perkin–Elmer automatic sequencer according to the manufacturer’s instructions, with an internal primer in the P element inverted repeats: Pir gacgggaccaccttatgt for the PCR products and with different primers spread over the sequence for the 1.6-kb HindIII fragment.
RESULTS
An 11-kb Genomic DNA Insert Can Target P Element Transposition to the Resident Fragment.
In the course of the study of the lio locus (cytogenetic site 37D on chromosome 2L), we constructed the P456 transposon in which an 11-kb genomic DNA fragment was cloned into the P[CaSpeR4] transformation vector (Fig. 1). This fragment corresponds to the 3′ end of an adjacent gene and the promoter, first exon, and beginning of the first intron of the lio gene. After injection of the P456 element, 10 independent w+ strains were recovered. One of them was homozygous viable but displayed a partial pupal lethality with lengthened dead pupae identical to the phenotype observed in lio mutant stocks (data not shown). By Southern blot experiments, this particular P456 element was mapped within a 1.6-kb HindIII fragment corresponding to the 5′ end of the linotte gene (Fig. 1), near the site of the PlacW element responsible for the lio1 mutation (data not shown).
To test for the reproducibility of this phenomenon, an X-linked P456 element was mobilized to autosomes. Twenty-one independent new autosomal P456 insertions were recovered and tested for linkage with the lio1 PlacW element. For this, females trans-heterozygous for each of the new P456 insertions and the lio1 P element were generated and mated with w males. The progeny of these crosses then were screened for w individuals resulting from segregation of the two w+ marked P elements, which indicates that the P456 insertion tested was not inserted near the lio locus. If no w individual was observed, the P456 strain tested was considered as a candidate for insertion at the lio locus, and the P element was mapped. Of the 21 P456 strains, two showed linkage with the lio1 P element. By Southern blot experiments, these two P456 elements were mapped within the 1.6-kb HindIII fragment, near the lio1 PlacW element (data not shown).
To determine more accurately the orientation and localization of these insertions, PCR tests were performed by using primers in the 5′ and 3′ ends of P elements and primers in the lio region (Fig. 2). Insertion sites of the P elements then were determined by DNA sequencing of the PCR products. The three P456 elements appeared to be in the opposite 5′ to 3′ orientation relative to the original lio1 insertion and were found very close to each other with only four bp separating their different insertion sites. The lio1 PlacW element was found to be inserted at the same nucleotide as one of the P456 elements (Fig. 2).
A 1.6-kb Genomic DNA Insert Can Target P Element Transposition to the Resident Fragment.
Because all of the P456 insertions obtained at the lio locus occurred within the same 1.6-kb HindIII fragment, the ability of this piece of DNA to target P element transposition was tested. For this, we took advantage of the P423 transposon (Fig. 1) in which this fragment is driving the lacZ gene (A. Simon, F. Savary, and T. Préat, personal communication). An X-linked P423 insertion was mobilized to autosomes. Seventy-one independent autosomal insertions of P423 were recovered and tested for linkage with the lio1 PlacW element as previously described. Among them, 15 showed linkage, and 14 of these were shown by PCR to have inserted into the endogenous genomic 1.6-kb HindIII fragment in one orientation or the other. The majority of the P423 elements inserted at the lio locus were found clustered in the same region as the P456 elements, with 10 of them localized in a 36-bp fragment (Fig. 2).
One P423 insertion showed linkage to lio but was not localized by PCR to the endogenous genomic 1.6-kb HindIII fragment, nor could it be mapped to the 50-kb genomic region surrounding lio by Southern blot experiments (data not shown), suggesting that this insertion has occurred by a homing-independent mechanism.
As a control, an X-linked P element containing a DNA insert unrelated to lio (see Materials and Methods) was mobilized from the X chromosome to the autosomes by using the same scheme of crosses as for P[lio] elements. One-hundred and thirty-six autosomal strains were generated and tested for linkage with the lio1 PlacW element as previously described. One of them showed linkage and was tested by PCR for insertion into the lio gene by using the same strategy as for the P[lio] elements. In this strain, the insertion is not in the genomic 1.6-kb HindIII fragment in which all of the P[lio] insertions occurred. Furthermore, this insertion is not allelic to lio for the pupal lethality phenotype, indicating that it does not mutate this gene. These results show that the lio gene is not a hot spot for P elements and that the lio regulatory sequences contained in the P456 and P423 elements are responsible for the homing phenomenon.
Elimination of the Target Region of the P423 Elements Abolishes Their Homing Properties.
To assay whether the resident sequences that drive the transposition of the P423 elements to the lio locus are restricted to the 5′ part of the gene, mobilization of the X-linked P423 starter element to autosomes was done by using a second chromosome bearing the lio2 deficiency (Fig. 1). This 4-kb deletion was produced by imprecise excision of the lio1 P element and is an amorphic mutation of the lio gene deleting the 1.6-kb HindIII fragment (22) in which all of the insertions of P456 or P423 in the lio gene have occurred. Forty-two independent autosomal P423 strains with lio2 second chromosomes were established and tested for linkage with the lio1 PlacW element. None of the P423 insertions induced in the lio2 mutant background showed linkage, indicating that none of them had inserted in the linotte region. These results suggest that the resident sequences that drive the transposition of the P423 elements to the lio locus are restricted to the lio2 4-kb deficiency.
P[lio] Elements Insertions into the lio Locus Are Not Attributable to Homologous Recombination.
Several lines of evidence show that P[lio] elements have inserted into the lio locus via a P-mediated mechanism and not by homologous recombination. First, Southern blot analyses of the three P456 insertions indicate that the 11-kb BamHI fragment included in these transposons is intact and that the endogenous 19-kb BamHI fragment has been disrupted by the insertion of the transposons. In no way can homologous recombination account for such a structure. Moreover, further analyses of the P456 insertion recovered after injection indicate that the integrated construct has lost the bacterial sequences located outside of the P element in the injected plasmid, suggesting that P element functions have catalyzed its insertion into the genome. Second, PCR amplifications using primers located in both 5′ and 3′ ends of the P[lio] elements and primers in the lio gene flanking their insertion sites show that 16 of 17 inserts have intact P ends (one insert was not amplified from the 5′ end), and sequencing of these PCR products shows that they are flanked by the typical 8-bp direct duplication of genomic DNA, which is a consequence of transposition (7). Finally, when placed in a transposase-producing background (3), the P[lio] elements inserted within the lio locus show somatic mosaicism, which indicates that they can be recognized by P transposase. Moreover, one of them was used to produce w derivatives. Molecular characterization of these excision lines show that most of them correspond to internal deletions of the original element (data not shown), as described for classical P elements (7, 31, 32). Taken together, these results show that P[lio] elements inserted within the lio locus have all of the features of classical P elements and that their insertion has occurred by P transposase-mediated transposition and not by homologous recombination.
P[lio] Elements Show Neither Transvection nor Patterned Eyes.
As pairing sensitivity (i.e., eye coloration darker heterozygous than homozygous) and mosaic coloration patterns in the eye were described for P[en] and P[ph] elements (15, 13, 16), P[lio] insertions were checked for such properties. Among the 90 homozygous viable P456 or P423 elements obtained in the course of this study, none of them displays an eye coloration darker heterozygous than homozygous (data not shown), indicating that the lio regulatory regions included in the P[lio] elements do not confer pairing sensitivity. Furthermore, only one P423 strain of the 102 P[lio] produced shows an anteroposterior coloration pattern in the eye. This frequency (≈1%) is not different from that observed with classical Pw+ elements (33). These results suggest that P[lio] elements, in contrast to P[en] and P[ph] elements, do not contain any regulatory sequences interfering with miniwhite normal expression.
P[lio] Elements Do Not Interact with the polyhomeotic Polycomb-Group Gene, nor with Any Other Pc-G Genes.
To compare the properties of P[lio] and P[en] elements, 14 P[en] autosomal strains were generated by mobilizing the wSac35 X-linked element in which a 7.5-kb engrailed promoter fragment was cloned into the pw8 transformation vector (9) and was tested for linkage with a Pw+ element inserted at the engrailed locus. One of these lines did show linkage with en, was homozygous lethal, and did not complement the Df(2R)enSFX31 and Df(2R)enB deficiencies (21). This P[en] insertion has thus occurred in the engrailed gene, confirming that, as described (9), the wSac elements were able to produce homing.
It has been shown that mutations in some Pc-G genes could modify the eye color of some P[en] elements (13, 16). We tested the effects of ph mutants on the eye color of the 14 autosomal wSac produced. The ph410 allele (34) showed a strong effect, with 8 lines of the 14 tested having darker eyes in ph410 mutant background. For comparison, 32 P[lio] elements representing P456 and P423 elements inserted (8/32) or not (24/32) at the lio locus were tested in the same conditions. None of these P[lio] elements showed any modification of their eye color in ph410 mutant background. These results establish that, although it has a strong effect on P[en] elements, mutation in the ph Pc-G gene has no effect on P[lio] elements.
To ensure that P[lio] elements do not contain any Pc-G response element that is unresponsive to ph mutation but that may respond to other members of the Pc-G, we analyzed two P423 elements and two P456 elements (one of each inserted within the lio gene) by comparing eye color between elements in a wild-type background and in backgrounds heterozygous for one strong (null if possible) mutant allele of each of the known Pc-G genes (see Materials and Methods). None of these genes was found to modify the eye color of the P[lio] elements tested whereas a large number of them are known to affect the eye color of P[ph] and/or P[en] elements (13, 16, 35). These results show that mutations in the Pc-G genes have no effect on P[lio] elements.
DISCUSSION
linotte Regulatory Sequences Mediate Very Precise P Element Homing.
In this study, we describe the behavior of P elements containing either 11 kb (P456) or 1.6 kb (P423) of lio regulatory sequences. When mobilized from the X chromosome, the P456 and P423 elements show as many as 10% (2/21) and 20% (14/71), respectively, of insertions into the lio resident gene. In contrast, a P element containing a DNA insert unrelated to lio mobilized and screened under the same conditions as P[lio] elements does not show any insertion into the lio gene (0/136). It should be noted that two transposons that do not include any lio regulatory sequences (PlacW and P[etau-lacZ]) responsible for lio1 (20) and drlP (23) mutations, respectively, are inserted in the lio gene. They were derived from screens of >4,000 independent lines (20, 23), indicating that the lio gene is hit at a frequency of ≈0.05% by P elements devoid of lio regulatory sequences. Thus, the exceptionally high rate of P[lio] homing depends on the presence of lio sequences in the transgenes and cannot be explained by the lio gene being a particular hot spot for neutral P element insertions.
Moreover, the precise mapping of the P[lio] elements inserted in the lio gene reveals that most of these insertions (14/17) have occurred within a 36-bp fragment, with some of them having the same target sites (Fig. 2). The PlacW and P[etau-lacZ] elements inserted in the lio gene also were mapped within this 36-bp fragment at the same target site as four P[lio] elements (Fig. 2). The target sites used by several independent P elements match imperfectly the consensus for P target sites (7, 8) but lie near the lio transcription start site, as expected (4). Thus, the precise insertion of P[lio] elements within the lio gene seems to depend on P biases for transcription start sites rather than on lio sequences included in P[lio] elements.
We also have observed that a 4-kb deficiency removing the 5′ part of the lio gene prevents the homing of P[lio] elements. This implies that the sequences that drive P[lio] elements to the resident lio gene are contained within this 4-kb deficiency and are not spread over the lio locus. It is likely that these sequences are restricted to the 1.6-kb HindIII fragment, which is sufficient to promote P[lio] homing.
Taken together, these results suggest a two-step mechanism for P[lio] elements homing: (i) P[lio] elements are targeted to the lio resident gene, in the very close vicinity of the promoter, by interactions with regulatory elements contained in the 1.6-kb HindIII fragment of the lio resident gene; and (ii) P[lio] elements select a target within the lio promoter according to a P[en]dependent mechanism.
The striking precision of P[lio] homing is different from what has been observed with P[en] elements whose insertions at the en locus are spread over a 200-kb region (9, 12, 16). P element homing within the BX-C also has been described (11), and P elements containing a 7-kb fragment from the bxd/iab2 region were found distributed over the entire BX-C. These observations suggest that sequences that drive insertion of these transposons are spread over the resident locus.
In addition to homing, P[en] elements also show preferential insertion near genes expressed in stripes during embryogenesis (12). For example, several independent P[en] insertions have been obtained in the wingless and escargot genes (36), suggesting that the resident sequences that drive P[en] insertions are also present in other genes. Compilation of the results published by Hama (9) and Kassis (15) reveals that 37% (23/62) of their P[en] elements are inserted at chromosomal sites that also are known to be binding sites for polycomb (PC) or polyhomeotic (PH) proteins (18). In contrast, a random sample of P element localizations extracted from Flybase show coincident localizations with PC or PH binding sites in only 13% (18/137) of the cases. Preferential insertion to PC or PH binding sites also has been described for P elements containing regulatory elements from the BX-C (14) and from the ph gene (13). These results strongly suggest that the resident sequences that attract P elements containing regulatory sequences from en, ph, and BX-C are the PREs. It should be noted at this point that the 2D, 48A, 84AB, and 89E bands, in which ph, en, ANT-C, and BX-C, respectively, are located, are binding sites for the PC and PH proteins (18). This suggests that the homing of P elements containing regulatory sequences from these genes is a particular case of preferential insertion into PRE containing loci.
In contrast, the chromosomal localization of the lio gene (37D), which is the target of P[lio] elements, has not been described as a binding site for Pc-G proteins (18) on polytene chromosomes of salivary glands. Assuming that this result can be extended to the germ line, this suggests that P[lio] elements homing cannot be explained by preferential insertion into PRE containing loci.
P[lio] Elements Do Not Contain any PRE.
P elements containing binding sites for Pc-G proteins (P[PRE]) display unusual properties: (i) They often show pairing sensitivity; (ii) many strains containing P[PRE] elements display a mosaic coloration of the eye; (iii) the eye color of P[PRE]-containing strains is modified by mutations of the Pc-G genes; and (iv) P[PRE] elements create new Pc-G protein binding sites at their chromosomal localizations when they do not coincide with endogenous sites.
All of the P elements for which homing or preferential insertion have been reported are P elements containing PREs (9, 10, 12–14). Indeed, P[en] elements often show pairing sensitivity, mosaic coloration of the eye, and enhanced coloration in Pc-G mutant background (15, 13, 16). The same properties have been described for P[ph] elements (13). Moreover, P[ph] elements create new binding sites for PC and PH proteins on polytene chromosomes (S. Bloyer and J-.M.D., unpublished results). P elements carrying regulatory regions of ANT-C and BX-C also possess PRE activity and create new binding sites for PC proteins (17, 18, 14).
In contrast, the results presented in this report clearly establish that the lio regulatory sequences included in P[lio] elements do not confer any of these properties. Furthermore, because the endogenous lio locus is not a target for Pc-G proteins on polytene chromosomes, it is very unlikely that P[lio] elements would create a new binding site for these proteins. Finally, a conserved sequence motif in PREs (CNGCCATNDNND) recently has been observed (37, 38). This motif is not present in the 1.6-kb HindIII fragment contained in the P423 elements (Fig. 2). Taken together, these results strongly suggest that P[lio] elements do not contain any PRE.
P[lio] Elements Homing Occurs by a New, PRE-Independent Mechanism.
The observation that P[PRE] elements preferentially insert into PRE containing loci has led to the hypothesis that Pc-G proteins complexes bound to the transgenic and endogenous PREs could interact together and bring the transposon to the vicinity of the locus into which preferential insertion occurs (13, 19). The variable composition of the Pc-G proteins complex at different PREs (39) could account for variable behavior of different P[PRE] elements. Furthermore, it has been shown that PREs are spread over 100-kb around the en gene (40) and all over the 340 kb of the BX-C (39). This could explain why the homing of P[PRE] elements is region-specific rather than site-specific (9, 12).
This model does not apply to P[lio] elements homing because no PRE has been detected at the lio locus or in P[lio] elements. We suggest that P[lio] elements homing is not mediated by Pc-G proteins complex but by a new, as yet unknown protein or complex of proteins whose features should be the following: (i) expressed in the germ line as transposition events occur in this tissue; (ii) capable of specific binding to the 1.6-kb HindIII fragment, which is sufficient to promote P423 elements homing; and (iii) able to mediate interactions directly or indirectly between two pieces of DNA on which it is bound and to bring them together. Sequence analysis of the 1.6-kb HindIII fragment does not reveal any candidate for mediating P[lio] elements homing. Whatever this putative protein is, these results show that other kinds of protein–protein interactions than those mediated by the Pc-G proteins are able to target the insertion of transposon constructs.
Could P Element Homing Be a Frequent Phenomenon?
The existence of two distinct systems leading to P element homing raises the question of how frequent this phenomenon is. P element homing was first described in the course of the study of the en regulatory regions by linking the en promoter, or portions of it, to a reporter gene in a P element vector (9). Because these regions are complex (41), the only strains that reproduced the correct en pattern were insertions into the en resident gene. The same results have been obtained with regulatory sequences of the ANT-C (10). On the other hand, in most cases of promoter studies, the correct expression pattern is readily reproduced, and only a few transgenic lines are made and studied. This implies that, even if homing could occur, it would be missed very easily.
In this study, the homing of P[lio] elements was detected because 1 of the 10 P456 strains obtained after injection displayed a partial pupal lethality with lengthened dead pupae identical to that observed in lio mutant stocks. This phenotype is specific to lio mutants and is displayed even by hypomorphic lio alleles. If P element homing occurs with a gene whose mutations do not display such a specific phenotype (i.e., total or partial lethality but without any obvious morphological defects), the phenotype observed in a strain in which P element homing has occurred would be attributed to random insertion in a gene leading to the same nonspecific phenotype, and the phenomenon would be easily missed. Thus, we believe that P element homing could have gone unnoticed and may constitute a more frequent feature than generally assumed.
This hypothesis is supported by the report of Cherbas et al. (42), who coined the term “parahomologous” targeting to describe the frequent insertion of transfected DNA near the endogenous gene in Drosophila cultured cells. These insertional events are not caused by homologous recombination and have been attributed to an efficient, homology-dependent pathway of illegitimate recombination. However, it is tempting to speculate that there is a common molecular basis for this parahomologous targeting and for the homing of P[lio] elements. In this latter case, the properties of P transposition would add precision and efficiency to this phenomenon.
For this reason, we suggest that P mutagenesis designed to obtain insertions in a particular gene should be done with P elements including regulatory sequences of this gene. Indeed, if these sequences can mediate high-frequency homing, as is the case for the lio gene, the use of such P elements will increase the probability of insertion in this gene, and the isolation of a mutant strain will be much easier than with neutral P elements. Consequently, P element homing could provide a new, powerful tool to produce mutants of cloned genes and facilitate Drosophila reverse genetics, for which there is otherwise no technique for targeted gene disruption.
Acknowledgments
We are grateful to Judith Kassis for stimulating discussions and encouragement. We thank Judith Kassis and Kathy Matthews at the Indiana Drosophila Stock Center for providing stocks. We thank Anne Simon, Fabrice Savary, and Thomas Préat for the P423 stock and also Agnés Audibert, Béatrice Benoit, and Martine Simonelig for the PWHZ10-I4 stock. We are particularly grateful to Roger Karess, François Schweisguth, and Leonie Ringrose for useful comments on the manuscript. E.T. is supported by a fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche, from the Ligue Départementale de l’Essonne, and from the Association pour la Recherche contre le Cancer. This work was supported by grants from the Association pour la Recherche contre le Cancer (Grant 6786), the Centre National de la Recherche Scientifique (ATIPE 7 and ACC-SV 4 “Biologie du Développement”), and from the University Paris XI-Orsay (URA 2227).
ABBREVIATIONS
- BX-C
Bithorax homeotic complex
- ANT-C
Antennapedia homeotic complex
- ph gene
polyhomeotic gene
- Pc-G
Polycomb group
- PRE
Pc-G response element
- lio gene
linotte gene
- kb
kilobase
- PC
Polycomb protein
- PH
Polyhomeotic protein
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF147883).
References
- 1.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 2.Cooley L, Berg C, Spradling A. Trends Genet. 1988;4:254–258. doi: 10.1016/0168-9525(88)90032-7. [DOI] [PubMed] [Google Scholar]
- 3.Robertson H M, Preston C R, Phillis R W, Johnson S D, Benz W K, Engels W R. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spradling A, Stern D, Kiss I, Roote J, Laverty T, Rubin G. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg C A, Spradling A C. Genetics. 1991;127:515–524. doi: 10.1093/genetics/127.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmons M J, Raymond J D, Johnson N A, Fahey T M. Genetics. 1984;106:85–94. doi: 10.1093/genetics/106.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Hare K, Rubin G M. Cell. 1983;34:25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- 8.Preston C, Sved J, Engels W. Genetics. 1996;144:1623–1638. doi: 10.1093/genetics/144.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hama C, Ali Z, Kornberg T. Genes Dev. 1990;4:1079–1093. doi: 10.1101/gad.4.7.1079. [DOI] [PubMed] [Google Scholar]
- 10.Engström Y, Schneuwly S, Gehring W. Roux’s Arch Dev Biol. 1992;201:65–80. doi: 10.1007/BF00420417. [DOI] [PubMed] [Google Scholar]
- 11.Hudson A, O’Connor M, McCall K, Bender W. Annu Drosophila Res Conf. 1995;36:240. [Google Scholar]
- 12.Kassis J, Noll E, VanSickle E, Odenwald W, Perrimon N. Proc Natl Acad Sci USA. 1992;89:1919–1923. doi: 10.1073/pnas.89.5.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fauvarque M-O, Dura J-M. Genes Dev. 1993;7:1508–1520. doi: 10.1101/gad.7.8.1508. [DOI] [PubMed] [Google Scholar]
- 14.Chiang A, O’Connor M, Paro R, Simon J, Bender W. Development (Cambridge, UK) 1995;121:1681–1689. doi: 10.1242/dev.121.6.1681. [DOI] [PubMed] [Google Scholar]
- 15.Kassis J, VanSickle E, Sensabaugh S. Genetics. 1991;128:751–761. doi: 10.1093/genetics/128.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kassis J. Genetics. 1994;136:1025–1038. doi: 10.1093/genetics/136.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zink D, Engström Y, Gehring W, Paro R. EMBO J. 1991;10:153–162. doi: 10.1002/j.1460-2075.1991.tb07931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeCamillis M, Cheng N, Pierre D, Brock H. Genes Dev. 1992;6:223–232. doi: 10.1101/gad.6.2.223. [DOI] [PubMed] [Google Scholar]
- 19.Pirrotta V. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- 20.Dura J-M, Préat T, Tully T. J Neurogenet. 1993;9:1–14. doi: 10.3109/01677069309167272. [DOI] [PubMed] [Google Scholar]
- 21.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 22.Dura J M, Taillebourg E, Preat T. FEBS Lett. 1995;370:250–254. doi: 10.1016/0014-5793(95)00847-3. [DOI] [PubMed] [Google Scholar]
- 23.Callahan C A, Muralidhar M G, Lundgren S E, Scully A L, Thomas J B. Nature (London) 1995;376:171–174. doi: 10.1038/376171a0. [DOI] [PubMed] [Google Scholar]
- 24.Tamkun J, Schwarzbauer J, Hynes R. Proc Natl Acad Sci USA. 1984;81:5140–5144. doi: 10.1073/pnas.81.16.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirrotta V. In: Vectors for P-Mediated Transformation in Drosophila. Rodriguez R, Denhardt D, editors. Boston: Butterworths; 1988. pp. 437–456. [DOI] [PubMed] [Google Scholar]
- 26.Segalat L, Berger G, Lepesant J. Mech Dev. 1994;47:241–251. doi: 10.1016/0925-4773(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 27.Hiromi Y, Gehring W. Cell. 1987;50:963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- 28.Jones R S, Gelbart W M. Genetics. 1990;126:185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips M D, Shearn A. Genetics. 1990;125:91–101. doi: 10.1093/genetics/125.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gloor G, Engels W. Drosophila Inf Serv. 1992;71:148–149. [Google Scholar]
- 31.Tsubota S, Schedl P. Genetics. 1986;114:165–182. doi: 10.1093/genetics/114.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engels W R, Johnson-Schlitz D, Eggleston W B, Sved J. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- 33.Netter S, Fauvarque M-O, Corral R D D, Dura J-M, Coen D. Genetics. 1998;149:257–275. doi: 10.1093/genetics/149.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dura J, Randsholdt N, Deatrick J, Erk I, Santamaria P, Freeman J, Weddell D, Brock H. Cell. 1987;51:829–839. doi: 10.1016/0092-8674(87)90106-1. [DOI] [PubMed] [Google Scholar]
- 35.Fauvarque M O, Zuber V, Dura J M. Mech Dev. 1995;52:343–355. doi: 10.1016/0925-4773(95)00412-t. [DOI] [PubMed] [Google Scholar]
- 36.Whiteley M, Noguchi P, Sensabaugh S, Odenwald W, Kassis J. Mech Dev. 1992;36:117–127. doi: 10.1016/0925-4773(92)90063-p. [DOI] [PubMed] [Google Scholar]
- 37.Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 38.Mihaly J, Mishra R K, Karch F. Mol Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- 39.Strutt H, Paro R. Mol Cell Biol. 1997;17:6773–6783. doi: 10.1128/mcb.17.12.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strutt H, Cavalli G, Paro R. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drees B, Ali Z, Soeller W, Coleman K, Poole S, Kornberg T. EMBO J. 1985;6:2803–2809. doi: 10.1002/j.1460-2075.1987.tb02576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherbas L, Cherbas P. Genetics. 1997;145:349–358. doi: 10.1093/genetics/145.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]