Abstract
A diverse pattern of polymorphism is defined for the paired Ig-like receptors (PIRs) that serve as activating (PIR-A) and inhibitory (PIR-B) receptors on B lymphocytes, dendritic cells, and myeloid-lineage cells in mice. The monoclonal anti-PIR antibody 10.4 is shown to recognize an allelic PIR-A/PIR-B determinant on cells from BALB/c but not C57BL/6 mice. Other strains of inbred mice also can be typed on the basis of their expression of this PIR allelic determinant. Analysis of (BALB/c × C57BL/6) F1 hybrid offspring indicates that PIR molecules bearing the paternal PIR allotype are expressed whereas PIR-A and PIR-B molecules bearing the maternal allotype are not. The monoallelic expression of the polymorphic PIR-A and PIR-B molecules, and possibly of their human Ig-like transcript/leukocyte Ig-like receptor/monocyte/macrophage Ig-like receptor and killer cell inhibitory receptor relatives, may influence innate and specific immune responses in outbred populations.
Paired Ig-like receptors (PIRs) A and B are recently identified cell surface molecules with very similar extracellular regions comprised of six Ig-like domains, D1–D6 (1, 2). The invariant PIR-B molecules differ from the variable PIR-A molecules in that PIR-B possesses cytoplasmic immunoreceptor tyrosine-based inhibitory motifs that can recruit the protein tyrosine phosphatase SHP-1 to inhibit signal transduction (3–6). Conversely, the closely related PIR-A molecules all have a short cytoplasmic tail and a polar transmembrane region that facilitates association with homodimers of the Fc receptor common γ chain to form a cell activating receptor complex (7–9). The PIR-A and PIR-B molecules are expressed on B lymphocytes, dendritic cells, monocyte/macrophages, granulocytes, and mast cells, where their expression is up-regulated during cellular maturation (9). The physiological ligands of PIR-A/PIR-B are still unknown, although it seems likely these receptors participate in the regulation of immune and inflammatory responses (1).
A single Pirb gene encodes the invariant PIR-B molecules whereas multiple Pira genes encode the different PIR-A molecules (refs. 1 and 10; data not shown). The Pira and Pirb genes belong to a subfamily of the Ig gene superfamily, members of which encode activating or inhibitory receptors (11, 12). This subfamily includes the human Ig-like transcripts (ILT) (13) [also known as leukocyte Ig-like receptors (LIR) (14), monocyte/macrophage Ig-like receptors (MIR) (15), and HM18 (16)], the killer cell inhibitory receptors (KIR) (17), and the Fc α receptor (18), the genes for which are located on human chromosome 19q13 (13, 15, 16, 19–21) in a region that is syntenic with the centromeric proximal region on mouse chromosome 7, where the Pira and Pirb genes reside (1).
Two monoclonal antibodies, 6C1 and 10.4, have been generated that recognize PIR-A and PIR-B molecules (9). In this study, we have observed that, although the 6C1 antibody appears to have universal PIR specificity, the 10.4 anti-PIR antibody recognizes a strain-specific allelic determinant on PIR-A and PIR-B molecules. When this allelic determinant was used to trace expression of the parental PIR-A and PIR-B alleles in F1 hybrid mice, we found that PIR molecules encoded by the paternal allele are expressed whereas PIR molecules for the maternal allele are not. These findings extend the principle of genomic imprinting to a multimember family of polymorphic receptors that may participate in innate and specific immune responses.
MATERIALS AND METHODS
Mice.
Young Adult A/J, BALB/cJ, C3H, C57BL/6, DBA/1, DBA/2, NZB, SJL, [C57BL/6 (female) × BALB/cJ (male)] F1, and [BALB/cJ (female) × C57BL/6 (male)] F1 mice either were obtained from The Jackson Laboratory or were derived in our animal facility.
Antibodies.
Rabbit anti-PIR D1/D2 polyclonal antibodies were prepared, and the 10.4 and 6C1 anti-PIR D1/2 monoclonal antibodies were phycoerythrin-conjugated as described (3, 9). FITC-conjugated monoclonal antibodies specific for the B220 (RA3–6B2) and Mac-1 (CD11b, M1/70) antigens were obtained from PharMingen. FITC-conjugated anti-IgMb (MB86) and biotin-conjugated anti-IgMa (RS3.1) antibodies were gifts from John F. Kearney (Univ. of Alabama at Birmingham).
Immunofluorescence Analysis.
Bone marrow and spleen cell suspensions were stained with fluorochrome- labeled antibodies as described (9) and were examined with a FACS Calibur flow cytometry instrument (Becton Dickinson), and the data were analyzed by using winmdi 2.7 software [The Scripps Research Institute Cytometry Software Page (http://facs.scripps.edu/software.html)].
Reverse Transcriptase–PCR and Nucleotide Sequence Analysis.
To determine the nucleotide sequences of PIR D1/D2 region transcripts, total RNA was isolated from spleen or bone marrow samples by using the TRI Reagent (Molecular Research Center, Cincinnati), converted to first-strand cDNA with oligo(dT) 12–18 primers and RNase H− reverse transcriptase (Superscript II, GIBCO/BRL) and amplified with a set of primers: the forward primer 5′-TCCCTAAGCCTATCCTCAGAG-3′ corresponding to the 5′ end of D1 and the reverse primer 5′-GAGCTCCAGGGATTCACTTG-3′ corresponding to the 3′ end of D2. Each amplification reaction involved 30 cycles of denaturing at 94°C for 30 s, annealing at 58°C for 25 s, and extension at 72°C for 40 s. A final extension was performed at 72°C for 8 min. The ≈590-bp products were electrophoresed, were purified from agarose gels by using Wizard PCR Prep (Promega) and were cloned into the pCR2.1-TOPO vector (Invitrogen). For site-directed mutagenesis of the PIR-B1 D1/D2 sequence, a similar amplification was performed by using the above forward and reverse primers in association with a mutated reverse primer, 5′-GTCACCTTAGTCCTCCTGGAGACCACAGA-3′, and a mutated forward primer, 5′-TCTGTGGTCTCCAGGAGGACTAAGGTGAC-3′, corresponding to nucleotide positions 40–69 of PIR D1. The mutated PCR products were used to produce recombinant protein as described (9). For sequence analysis of the entire PIR-B extracellular sequence (D1–D6) in A/J, C57BL/6, and SJL mice, this region was amplified from first strand splenocyte cDNA with the primers: forward primer 5′-AGATGCCATGTCCTGCACCT-3′ corresponding to the 5′ untranslated region and the reverse primer 5′-TCCTTTCCTGGTTATGGGCTCTTC-3′ corresponding to the beginning of the PIR-B cytoplasmic region. Amplification reactions were carried out by using 30 cycles of denaturing at 94°C for 1 min, annealing at 65°C for 1 min, extension at 72°C for 3 min, and a final extension at 72°C for 8 min. The ≈2-kilobase products were cloned into pCR2.1-TOPO. Sequence analysis of the insert DNA was performed by simultaneous bi-directional sequencing (Li-Cor, Lincoln, NE).
Preparation of Recombinant PIR Proteins and Enzyme-Based Immunoassay.
Recombinant PIR proteins corresponding to mutated and nonmutated versions of the first two amino-terminal extracellular domains (D1 and D2) were produced in Escherichia coli (9). Plastic microtiter plates (Costar) coated with the PIR D1/D2 recombinant proteins (10 μg/ml) were incubated sequentially with varying concentrations of purified 6C1 or 10.4 mAbs, alkaline-phosphatase labeled goat anti-rat Ig antibodies (1 μg/ml; Southern Biotechnology Associates), and the p-nitrophenyl phosphate substrate. The product of the enzyme reaction was measured by light absorbance at 405 nm by using a Titertek Multiskan Plus ELISA reader (ICN).
Immunoblot Analysis of PIR Proteins.
PIR proteins in whole cell lysates of splenocytes were isolated with the 6C1 and 10.4 anti-PIR antibodies, were resolved on SDS/PAGE, and then were transferred onto nitrocellulose membranes and immunoblotted with rabbit anti-PIR antibodies as described (9).
RESULTS
Alloreactivity of the 10.4 Anti-PIR Antibody.
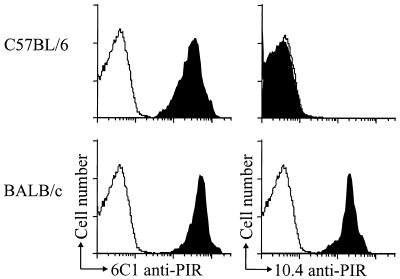
The 10.4 and 6C1 anti-PIR monoclonal antibodies were produced by heterohybridomas derived by fusion of a nonproducer mouse plasmacytoma with lymph node cells from rats hyperimmunized with a recombinant D1/D2 protein, in a region of sequence identity between BALB/c PIR-B and PIR-A1 molecules (1, 9). Variable patterns of reactivity were observed when the two antibodies were used for immunofluorescence assessment of cells from different strains of inbred mice. Although both anti-PIR antibodies reacted with myeloid and B lineage cells from BALB/c mice, only the 6C1 antibody was found to react with cells from C57BL/6 mice (Fig. 1). Because human KIR and certain ILT/LIR/MIR relatives of the mouse PIR molecules have been shown to have binding specificity for different MHC class I alleles (14, 22–26), we examined the expression of the 10.4 allotope in inbred strains of mice with a variety of MHC haplotypes to determine whether expression of the 10.4 determinant is related to the MHC haplotype. The 10.4 anti-PIR antibody was found to react with cells from BALB/c, A/J, and SJL mice and not with the other inbred strains (Table 1). The expression pattern of the 10.4 PIR allelic determinant in this panel of inbred mice thus appeared to be unrelated to the MHC haplotype. The 6C1 anti-PIR antibody by contrast was reactive with the PIR-A and PIR-B molecules in all mouse strains tested.
Figure 1.
The 10.4 anti-PIR monoclonal antibody recognizes an allelic determinant. Bone marrow granulocytes (GR-1+/Mac-1+) from BALB/c or C57BL/6 mice were analyzed by flow cytometry staining with either the 6C1 or 10.4 anti-PIR mAbs (shaded histogram) or an isotype matched antibody (open histogram). The 6C1 anti-PIR antibody reacted with cells of both strains (Left) whereas the 10.4 anti-PIR antibody only reacted with BALB/c cells (Right).
Table 1.
Strain distribution of the 10.4 PIR allelic determinant
| 10.4 anti-PIR reactivity* | Inbred mouse strain | MHC haplotype |
|---|---|---|
| Positive | A/J | a |
| Positive | BALB/c | d |
| Positive | SJL | s |
| Negative | C3H | k |
| Negative | C57BL/6 | b |
| Negative | DBA/1 | u |
| Negative | DBA/2 | d |
| Negative | NZB | d |
Tested by immunofluorescence assay.
Analysis of PIR-A and PIR-B Polymorphism.
As a first step in examining the basis for the selective 10.4 reactivity with different strains of mice, we determined the nucleotide sequences of the six extracellular Ig-like domains (D1–D6) for PIR-B in the 10.4+ A/J and SJL strains, and the 10.4− C57BL/6 strain, then compared these extracellular sequences with those determined previously for the BALB/c (10.4+) and B10.A (10.4−) mice (1, 2). This analysis confirmed the presence of an invariant PIR-B sequence in each strain. It also indicated differences in the nucleotide sequences (96–99% homology) for PIR-B between each of the inbred strains, except for the B10.A and C57BL/6 sequences, which were identical. Because the nucleotide variations noted between the 10.4+ A/J and SJL strains were silent, they shared an identical amino acid sequence. The only differences in the PIR-B extracellular regions among these 10.4+ strains of mice (BALB/c, A/J, SJL) were two amino acid residues in D3. The nucleotide homology between the extracellular PIR-B sequences for 10.4+ and 10.4− strains was ≈96%. With regard to the possible location of the 10.4 D1/D2 allelic determinant, several D1 sequence differences were evident between the 10.4+ and 10.4− strains whereas, except for position 97, the amino acid sequences for PIR-B D2 were virtually identical in all strains.
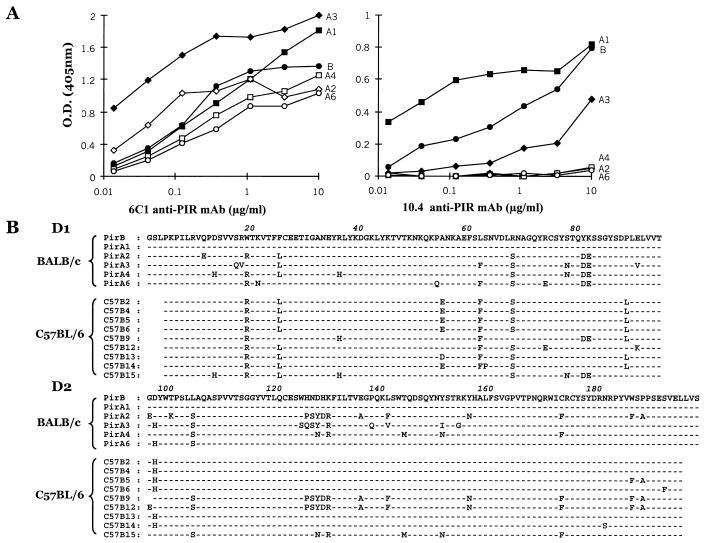
We extended this analysis to PIR-A by sequencing the D1/D2 region of seven PIR-A clones from the 10.4− strain, C57BL/6. These sequences were slightly different from their counterpart BALB/c PIR-A sequences and, for both strains, D2 exhibited greater sequence variation than D1 (Fig. 2B). The D2 variability in the different PIR-As was concentrated to three regions of the sequences that were predicted to be on loops between the β-sheets by comparative modeling [swiss-model (http://www.expasy.ch/swissmod/SWISS-MODEL.html)] based on the structure of a KIR relative (27).
Figure 2.
Analysis of 10.4 anti-PIR antibody reactivity with PIR D1/D2 recombinant proteins of BALB/c origin. (A) Binding of the 6C1 and the 10.4 anti-PIR antibodies to recombinant proteins corresponding to the first and second Ig-like extracellular domains (D1 and D2) of PIR-B (●), PIR-A1 (■), -A2 (⋄), -A3 (♦), -A4 (□), and -A6 (○) was assessed by ELISA. (B) Amino acid sequence comparison of the D1 and D2 domains of six different BALB/c cDNA clones (PIR-B and -A1 to -A6) and nine different C57BL/6 clones (C57B2 to C57B15). The C57BL/6 sequences lack the beginning of D1 and the end of D2 because of the position of the PCR primers. Amino acid identity with the PIR-B sequence (top line) is indicated by dashes (–), and changes are indicated by boldface letters. The D1 Trp residue at position 19 is highlighted in gray to indicate its importance in the likely determinant recognized by the 10.4 antibody.
This interstrain comparison of PIR-B (D1-D6) and PIR-A (D1/D2) sequences thus revealed two types of diversity. The first type was represented by allelic variations that were concentrated in the Ig C2-like D1 domain. The second type of diversity featured variable sequence motifs in the V-like D2 domain of PIR-A molecules, and these were conserved in different inbred strains. For example, the extended PSYDR-A-F-N-F-F-A motif was observed in PIR-A sequences of both BALB/c and C57BL/6 strains, as was the N-R-M-N-F motif (Fig. 2B). A third extended motif, SQSY-R-Q-V-I-G, in the BALB/c PIR-A3 sequence was not seen among the limited panel of C57BL/6 sequences. The predicted surface location of these extended sequence motifs raises the possibility of their potential role in ligand-binding specificity.
Analysis of the 10.4 Allelic Determinant.
To refine the definition of the allelic determinant recognized by the 10.4 alloreactive antibody, the 10.4 and 6C1 antibodies were tested for their capacity to bind PIR D1/D2 recombinant proteins corresponding to the PIR-A1, -A2, -A3, -A4, -A6, and PIR-B molecules of BALB/c origin. By contrast to the 6C1 antibody, which reacted with all six recombinant proteins, the 10.4 antibody reacted with PIR-B, PIR-A1, and PIR-A3 and not with other PIR-A recombinant proteins (Fig. 2A). Comparison of the six BALB/c D1/D2 amino acid sequences with those deduced from the nucleotide sequences of nine C57BL/6 cDNA clones indicated six amino acid differences: W19, F25, L62, R68, P89, and D97 in the PIR-B and PIR-A1 sequences were replaced by R19, L25, F62, S68, L89, and H97 in most of the C57BL/6 D1/D2 sequences (Fig. 2B). Among these variable residues, W19 was present only in the BALB/c PIR-B, PIR-A1, and PIR-A3 recombinant proteins that were recognized by the 10.4 antibody. The W19 residue also was found in PIR-B of the 10.4+ A/J and SJL strains and not in the 10.4− B10.A and C57BL/6 mice. This analysis implicated the D1 tryptophan residue at position 19 as a potentially important determinant of the allotope recognized by the 10.4 anti-PIR antibody. In support of this possibility, the 10.4 reactivity was reduced by ≈80% when the W19 residue was replaced with arginine by site-directed mutagenesis (data not shown).
PIR Allelic Expression Pattern in F1 Hybrid Mice.
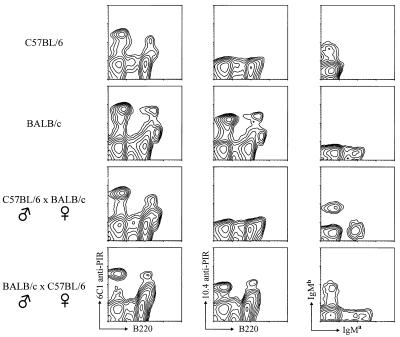
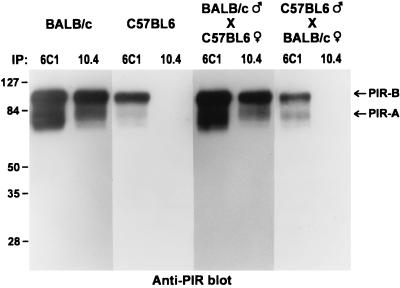
An unanticipated inheritance pattern of the 10.4+ PIR allele was observed when F1 hybrid mice derived by breeding 10.4+ BALB/c mice with the 10.4− C57BL/6 mice were examined (Fig. 3). In [BALB/c (male) × C57BL/6 (female)] F1 mice, the PIR molecules on B cells and myeloid lineage cells expressed the 10.4 BALB/c determinant as expected (Fig. 3, bottom row). In contrast, the 10.4 determinant was not found on bone marrow cells from [C57BL/6 (male) × BALB/c (female)] F1 mice, despite the fact that both parental IgM alleles were expressed and PIR-A/PIR-B expression could be verified with the 6C1 antibody (Fig. 3, third row). This pattern of monoallelic PIR expression was seen in all F1 offspring. To examine the possibility that the 10.4 reactive PIR molecules on cells in [C57BL/6 (male) × BALB/c (female)] F1 mice could simply be modulated by interaction with an unknown ligand, an immunoblot analysis of PIR molecules in lysates of splenocytes from the bi-directional (BALB/C × C57BL/6) F1 hybrids and both parent strains was performed by using the 6C1 and 10.4 anti-PIR antibodies. The PIR molecules produced by all of the mice were precipitated by the 6C1 anti-PIR antibody, whereas 10.4-reactive PIR molecules were identified only in BALB/c and the [BALB/c (male) × C57BL/6 (female]) F1 mice (Fig. 4). This analysis thus confirmed the exclusive production of PIR-A and PIR-B molecules bearing the paternal allelic marker.
Figure 3.
Immunofluorescence analysis of the inheritance pattern of the 10.4 PIR allotope expression. Bone marrow cells from C57BL/6, BALB/c, [C57BL/6 (male) × BALB/c (female)] F1, and [BALB/c (male) × C57BL/6 (female)] F1 mice were stained with a combination of either the 6C1 (Left) or the 10.4 (Middle) anti-PIR antibodies with the anti-B220 antibody. Splenocytes were stained with anti-IgMa and anti-IgMb antibodies (Right) to verify the genetic background of the mice.
Figure 4.
Immunoblot analysis of the parental origin of PIR-A and PIR-B molecules produced by F1 hybrids. Lysates of splenocytes from C57BL/6, BALB/c, [C57BL/6 (male) × BALB/c (female)] F1, and [BALB/c (male) × C57BL/6 (female)] F1 mice were immunoprecipitated with either the 6C1 or 10.4 anti-PIR antibodies. The immunoprecipitates were resolved by SDS/PAGE and were blotted onto a nitrocellulose membrane, and the PIR-A and PIR-B molecules were revealed by blotting with polyclonal anti-PIR antibodies.
DISCUSSION
The diverse PIR-A molecules pair with immunoreceptor tyrosine-based activation motif-containing Fc receptor common γ chain homodimers to achieve cell activating potential (7–9) whereas the invariant PIR-B molecules can interact with the SHP-1 protein tyrosine phosphatase to serve as an inhibitory receptor (3–6). The inhibitory function of PIR-B is dominant when both types of receptors are ligated (3). In elaborating the polymorphic features of these receptors and demonstrating their monoallelic expression, the present results provide insight into the functional potential of PIR-A and PIR-B, and possibly of their KIR and ILT/LIR/MIR relatives in humans as well.
Pira and Pirb Polymorphism.
Variations in the Pirb nucleotide sequences were found among representative inbred strains of mice, the sequence homology of the PIR-B extracellular region ranging from 96 to 100%. Comparison of PIR-B extracellular sequences (D1–D6) determined for C57BL/6, A/J, and SJL mice with those previously reported for 129/sv, BALB/c, and B10.A mice (1, 2, 10, 28) indicated identity only between the very closely related C57BL/6 and B10.A strains. Pira polymorphism also was indicated by comparison of the PIR-A D1/D2 sequences from BALB/c and C57BL/6 mice and by an analysis of the strain distribution for the PIR-A/PIR-B allelic determinant recognized by a monoclonal anti-PIR antibody, 10.4, raised against a BALB/c D1/D2 recombinant protein. PIR-A and PIR-B polymorphism was further indicated by the strain specificity of monoclonal antibodies raised against a D5/D6 recombinant protein (H.K., unpublished data).
In addition to the allelic polymorphism observed for the PIR-A/PIR-B receptor family, recurrent D2 motifs were recognized among the different PIR-A members. These motifs consisted of linked amino acid variations. Most of the extended D2 motifs were found among PIR-A sequences from both the BALB/c and C57BL/6 strains of mice. Computer-based modeling of PIR D1/D2 proteins based on their KIR relatives suggests that the variable amino acid sequences in the extended motifs would be on adjacent loops between β-sheets. The predicted surface location of the variable PIR-A motifs therefore could provide the potential for ligand binding diversity among the different PIR-A and PIR-B molecules in an individual.
An Allelic Determinant Shared by PIR-A and PIR-B Molecules.
The allelic determinant recognized by the 10.4 anti-PIR antibody was shared by the BALB/c, A/J, and SJL strains of mice but not by C57BL/6 and other representative inbred mice. The strain distribution pattern for the 10.4 allelic determinant did not correlate with the MHC haplotypes of these mice. Examination of 10.4 antibody reactivity with BALB/c-derived D1/D2 recombinant PIR proteins indicated that PIR-B and some PIR-A molecules (PIR-A1 and -A3) contained the allelic determinant whereas other BALB/c PIR-A members (PIR-A2, -A4, and -A6) did not. Comparison of the D1/D2 amino acid sequences for the 10.4+ and 10.4− BALB/c PIR molecules with the corresponding PIR-A/PIR-B sequences from 10.4− C57BL/6 mice suggested that a D1 tryptophan at position 19 would be an important residue in the 10.4 allelic determinant. Reactivity of the PIR-B D1/D2 recombinant protein with the 10.4 antibody was dramatically reduced by conversion of this tryptophan to arginine, thereby confirming its importance in the 10.4 allelic determinant present on of PIR-A and PIR-B molecules in BALB/c and related strains of mice. Sharing of the 10.4 allelic determinant by native PIR-A and PIR-B molecules produced by BALB/c mice was confirmed by their immunoprecipitation with the 10.4 antibody.
Monoallelic PIR-A and PIR-B Expression.
The allospecificity of the 10.4 anti-PIR antibody allowed us to examine expression of the parental alleles in F1 mice. In an analysis of the offspring derived from cross-breeding of the 10.4+ BALB/c and 10.4− C57BL/6 mice, we observed exclusive expression of the paternal PIR-A and PIR-B molecules. The maternal PIR-A and PIR-B alleles were not expressed at any stage in the differentiation of B-lineage and myeloid-lineage cells. This analysis thus provides direct evidence for the monoallelic expression of PIR-B and some of the PIR-A molecules. The subset of BALB/c PIR-A molecules for which we currently lack an allelic marker are also likely to be expressed in a monoallelic fashion. Monoallelic expression has not been observed previously for cell surface receptors that may be involved in inflammatory and immune responses. However, the Pira and Pirb genes are located on mouse chromosome 7 in a region near the centromere that includes the paternally expressed gene 3 [Peg3 (29)], the human homologue of which, Pw1, is involved in the tumor necrosis factor–NF-κB activation pathway leading to inflammatory responses (30). Imprinted genes exist in clusters, and chromosome 7 contains 20 of the 34 known imprinted genes in the mouse (31). The PIR gene family of ≈10 members significantly extends the clustering of imprinted genes in the chromosome 7 centromeric region and implies that the imprinting mechanism is likely to be complex.
Conclusion.
In demonstrating an example of genomic imprinting for the PIR-A and PIR-B family, these studies reveal a previously unrecognized mode of monoallelic expression for a diverse set of Ig-like receptors on cells responsible for both specific and nonspecific immune responses. Unlike the Ig and T cell receptors, in which there is allelic exclusion but the choice of alleles is random, PIR-A and PIR-B molecules were found to be expressed from only one parental allele, in this case the paternal allele. The monoallelic expression of the PIR-A/PIR-B family of activating and inhibitory receptors, the inhibitory PIR-B member of which appears to be dominant, could predispose hematopoietic cells to an increased susceptibility to leukemogenesis because tumor development has been noted after monoallelic mutations of other imprinted genes that affect cell growth (31). Other potential physiological consequences of the monoallelic expression of polymorphic PIR-A and PIR-B molecules are less obvious, given the unknown nature of their ligands. However, the ligands for their human KIR relatives have been shown to be MHC class I alleles (12, 17). The nearest relatives of the murine PIR-A/PIR-B molecules are the human ILT/LIR/MIR molecules, some of which have been shown to recognize MHC class I alleles (22–26). This raises the possibility that the PIR-A/PIR-B molecules also may bind MHC class I molecules.
The genes encoding the KIR and ILT/LIR/MIR receptor families reside on human chromosome 19q13, together with the genomically imprinted Peg3/Pw1 gene, in a region syntenic with the mouse chromosome 7 region where the Pira, Pirb, and Peg3 genes reside. Given that monoallelic expression has been demonstrated for this cluster of mouse genes, as well as for the human Peg3/Pw1 gene, it seems reasonable to predict that the ILT/LIR/MIR and KIR genes will be imprinted. If so, the monoallelic expression of these receptors in outbred populations could influence maternal–fetal relationships and perhaps other innate and specific immune responses. Elucidation of the mechanism for the monoallelic expression of the PIR-A/PIR-B molecules therefore could also provide information relevant to the physiological roles of their ILT/LIR/MIR and KIR relatives.
Acknowledgments
We thank Le Hong Ho and Lanier Gartland for technical assistance, Charlie Mashburn and Ann Brookshire for help in preparing the manuscript, and Drs. Pierre-André Lazenave, Eric Vivier, Lorenzo Moretta, and Peter Burrows for helpful advice and criticism. This work was supported in part by National Institutes of Health Grants AI42127 and AI39816. M.D.C. is an Investigator of the Howard Hughes Medical Research Institute.
ABBREVIATIONS
- PIR
paired Ig-like receptors
- ILT
Ig-like transcripts
- KIR
killer cell inhibitory receptors
- LIR
leukocyte Ig-like receptors
- MIR
monocyte/macrophage Ig-like receptors
Footnotes
References
- 1.Kubagawa H, Burrows P D, Cooper M D. Proc Natl Acad Sci USA. 1997;94:5261–5266. doi: 10.1073/pnas.94.10.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayami K, Fukuta D, Nishikawa Y, Yamashita Y, Inui M, Ohyama Y, Hikida M, Ohmori H, Takai T. J Biol Chem. 1997;272:7320–7327. doi: 10.1074/jbc.272.11.7320. [DOI] [PubMed] [Google Scholar]
- 3.Bléry M, Kubagawa H, Chen C C, Vély F, Cooper M D, Vivier E. Proc Natl Acad Sci USA. 1998;95:2446–2451. doi: 10.1073/pnas.95.5.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maeda A, Kurosaki M, Ono M, Takai T, Kurosaki T. J Exp Med. 1998;187:1355–1360. doi: 10.1084/jem.187.8.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timms J F, Carlberg K, Gu H, Chen H, Kamatkar S, Nadler M J, Rohrschneider L R, Neel B G. Mol Cell Biol. 1998;18:3838–3850. doi: 10.1128/mcb.18.7.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg K L, Carlberg K, Rohrschneider L R, Siminovitch K A, Stanley E R. Oncogene. 1998;17:2535–2541. doi: 10.1038/sj.onc.1202203. [DOI] [PubMed] [Google Scholar]
- 7.Maeda A, Kurosaki M, Kurosaki T. J Exp Med. 1998;188:991–995. doi: 10.1084/jem.188.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita Y, Ono M, Takai T. J Immunol. 1998;161:4042–4047. [PubMed] [Google Scholar]
- 9.Kubagawa H, Chen C C, Ho L H, Shimada T S, Gartland L, Mashburn C, Uehara T, Ravetch J V, Cooper M D. J Exp Med. 1999;189:309–318. doi: 10.1084/jem.189.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita Y, Fukuta D, Tsuji A, Nagabukuro A, Matsuda Y, Nishikawa Y, Ohyama Y, Ohmori H, Ono M, Takai T. J Biochem (Tokyo) 1998;123:358–368. doi: 10.1093/oxfordjournals.jbchem.a021945. [DOI] [PubMed] [Google Scholar]
- 11.Vély F, Vivier E. J Immunol. 1997;159:2075–2077. [PubMed] [Google Scholar]
- 12.Long E O. Annu Rev Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 13.Samaridis J, Colonna M. Eur J Immunol. 1997;27:660–665. doi: 10.1002/eji.1830270313. [DOI] [PubMed] [Google Scholar]
- 14.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu M L. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 15.Wagtmann N, Rojo S, Eichler E, Mohrenweiser H, Long E O. Curr Biol. 1997;7:615–618. doi: 10.1016/s0960-9822(06)00263-6. [DOI] [PubMed] [Google Scholar]
- 16.Arm J P, Nwankwo C, Austen K F. J Immunol. 1997;159:2342–2349. [PubMed] [Google Scholar]
- 17.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari M C, Moretta L. Annu Rev Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 18.Maliszewski C R, March C J, Schoenborn M A, Gimpel S, Shen L. J Exp Med. 1990;172:1665–1672. doi: 10.1084/jem.172.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupont B, Selvakumar A, Steffens U. Tissue Antigens. 1997;49:557–563. doi: 10.1111/j.1399-0039.1997.tb02802.x. [DOI] [PubMed] [Google Scholar]
- 20.Torkar M, Norgate Z, Colonna M, Trowsdale J, Wilson M J. Eur J Immunol. 1998;28:3959–3967. doi: 10.1002/(SICI)1521-4141(199812)28:12<3959::AID-IMMU3959>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Wende H, Colonna M, Ziegler A, Volz A. Mamm Genome. 1999;10:154–160. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 22.Colonna M, Navarro F, Bellon T, Llano M, Garcia P, Samaridis J, Angman L, Cella M, Lopez-Botet M. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borges L, Hsu M L, Fanger N, Kubin M, Cosman D. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- 24.Colonna M, Samaridis J, Cella M, Angman L, Allen R L, O’Callaghan C A, Dunbar R, Ogg G S, Cerundolo V, Rolink A. J Immunol. 1998;160:3096–3100. [PubMed] [Google Scholar]
- 25.Vitale M, Castriconi R, Parolini S, Pende D, Hsu M L, Moretta L, Cosman D, Moretta A. Int Immunol. 1999;11:29–35. doi: 10.1093/intimm/11.1.29. [DOI] [PubMed] [Google Scholar]
- 26.Navarro F, Llano M, Bellon T, Colonna M, Geraghty D E, Lopez-Botet M. Eur J Immunol. 1999;29:277–283. doi: 10.1002/(SICI)1521-4141(199901)29:01<277::AID-IMMU277>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 27.Fan Q R, Mosyak L, Winter C C, Wagtmann N, Long E O, Wiley D C. Nature (London) 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- 28.Alley T L, Cooper M D, Chen M, Kubagawa H. Tissue Antigens. 1998;51:224–231. doi: 10.1111/j.1399-0039.1998.tb03096.x. [DOI] [PubMed] [Google Scholar]
- 29.Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li L L, Tada M, Suzuki R, Yokoyama M, Shiroishi T, Wakana S, et al. Nat Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- 30.Relaix F, Wei X J, Wu X, Sassoon D A. Nat Genet. 1998;18:287–291. doi: 10.1038/ng0398-287. [DOI] [PubMed] [Google Scholar]
- 31.Falls J G, Pulford D J, Wylie A A, Jirtle R L. Am J Pathol. 1999;154:635–647. doi: 10.1016/S0002-9440(10)65309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]