Abstract
A regulated splicing event in protein 4.1R pre-mRNA—the inclusion of exon 16–encoding peptides for spectrin-actin binding—occurs in late erythroid differentiation. We defined the functional significance of an intronic splicing enhancer, UGCAUG, and its cognate splicing factor, mFox2A, on exon 16 splicing during differentiation. UGCAUG displays cell-type–specific splicing regulation in a test neutral reporter and has a dose-dependent enhancing effect. Erythroid cells express 2 UGCAUG-binding mFox-2 isoforms, an erythroid differentiation–inducible mFox-2A and a commonly expressed mFox-2F. When overexpressed, both enhanced internal exon splicing in an UGCAUG-dependent manner, with mFox-2A exerting a much stronger effect than mFox-2F. A significant reciprocal increase in mFox-2A and decrease in mFox-2F occurred during erythroid differentiation and correlated with exon 16 inclusion. Furthermore, isoform-specific expression reduction reversed mFox-2A–enhancing activity, but not that of mFox-2F on exon 16 inclusion. Our results suggest that an erythroid differentiation–inducible mFox-2A isoform is a critical regulator of the differentiation-specific exon 16 splicing switch, and that its up-regulation in late erythroid differentiation is vital for exon 16 splicing.
Introduction
Alternative splicing allows a single gene to express multiple mRNA forms in a tissue or developmental stage–specific manner. The 80-kDa red blood cell protein 4.1R is the prototype of a diverse array of 4.1R isoforms whose expression is regulated by coupled transcription and splicing,1 use of alternative translation initiation sites,2,3 alternative pre-mRNA splicing,4,5 and posttranslational modification.6,7 The expression of exon 16, which encodes peptides critical for the spectrin-actin binding, is tightly regulated during erythroid differentiation. Exon 16 is absent from the great majority of 4.1R mRNA in preerythroid cells, but predominates in late erythroid cell 4.1R mRNA.8,9
The excision of introns from a pre-mRNA and the joining of the exons mainly depends on the recognition and usage of 5′ and 3′ splice sites by the splicing machinery.10,11 The diversity encountered in alternatively spliced pre-mRNAs depends on multiple factors that influence the recognition of splice sites through a complex network of interactions between pre-mRNA sequence elements and splicing factors.12,13 The sequence elements are generally called exonic or intronic splicing enhancers (ESEs or ISEs, respectively) and silencers (ESSs or ISSs).14 These cis-elements function by binding to proteins that stimulate or inhibit exon definition. Silencers act by recruiting specific heterogeneous ribonucleoproteins (hnRNPs).14,15 Enhancers typically function by providing binding sites for serine-arginine (SR) proteins, a family of essential splicing factors.16–18 The splicing of 4.1R's exon 16 during erythroid differentiation is regulated by a finely tuned balance of cis-elements and trans-acting factors. We have shown that exon 16 and its abutting intronic sequences contain both positive and negative elements that promote exon exclusion in predifferentiated murine erythroleukemia cells (MELCs) but allow exon inclusion as the cells differentiate.19 During maturation, an increase in SF2/ASF20 and a decrease in hnRNP A/B21 expression correlate with increased efficiency of exon 16 splicing.
A UGCAUG sequence present in triplicate in the intron downstream of exon 16 was previously identified as an ISE.19,22 This element was identified in a computational study as the most common hexanucleotide found in introns downstream of alternatively spliced exons23; UGCAUG elements influence splicing of many alternative exons.24–28 Vertebrate homologs of the Caenorhabditis elegans Fox-1 protein, and 2 Fox-1–related proteins, ataxin-2–binding protein 1 (A2BP1)/Fox-1 and Rbm9/Fxh, bind to UGCAUG.29,30 Their encoding genes give rise to a large family of isoforms through extensive alternative splicing.29,30
Neuron-28 and muscle-specific29 Fox isoforms have been shown to activate tissue-specific exons through UGCAUG-dependent enhancers. Protein 4.1R exon 16 splicing has been shown to be modulated by Fox-2 when transfected into HeLa cells.22 However, the particular Fox-2 isoforms that are expressed in erythroid cells and their physiologic importance in modulating exon 16 splicing switch during erythroid differentiation had not been determined.
We have examined the role of UGCAUG and its cognate splicing factor, mFox-2A, in exon 16 splicing during induced MELC erythroid differentiation. We identified 2 mFox-2 isoforms: an erythroid differentiation–inducible mFox-2A, and a commonly expressed mFox-2F. We also documented a correlation between a switch in mFox-2 isoforms and the increased efficiency of exon 16 splicing during late erythroid differentiation. Through both gain-of-function and loss-of-function experiments, we provide the first direct evidence that the differentiation-specific exon 16 splicing switch is mediated by a differentiation-inducible mFox-2A. Its up-regulation in late erythroid differentiation is vital for exon 16 splicing. Thus, we suggest that regulated expression of differentiation-inducible splicing factors or isoforms represents a novel splicing regulatory pathway in erythroid differentiation.
Methods
Plasmid constructs
Exon 16 minigene.
The wild-type (WT) minigene and mutant constructs (Pm, Nr, and PN) comprised of a 3 exon (13, 16, 17) splicing cassette (Figure 1A) has been described.19 The mutation construct Bss was made by a 2-step polymerase chain reaction (PCR) using WT minigenes as templates and the outer primers Ex13-clone-S, In17-clone-As as well as the inner primers Bss-S and Bss-As. The underlined sequences in the inner primers represent the mutant sequences (Table 1; exon 16 minigenes). The triple mutant PNB was constructed using the Bss inner primers and the double mutant PN as a template.
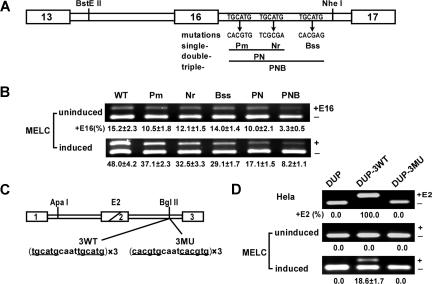
Figure 1.
UGCAUG repeats enhance exon 16 splicing and activate DUP internal exon splicing in a differentiation stage–specific manner in MELCs. (A) Schematic of the wild-type (WT) exon 16 minigene and mutations tested. The first repeat was mutated to restriction site PmlI, the second repeat to NruI, and the third repeat to BssSI. The double-mutant PN construct incorporated both the Pm and Nr mutations; the triple mutant PNB construct incorporated all 3 mutations. (B) Mutational analysis in undifferentiated and differentiated MELCs. Minigene stably transfected MELCs were induced to differentiation. Splicing products were analyzed for exon 16 inclusion by RT-PCR. +E16 indicates spliced products with exon 16; −E16, spliced products without exon 16; and +E16 (%), the percentage of spliced products includes exon 16. For each construct, 2 transfections were performed per experiment. Each experiment was repeated at least 3 times. The results shown are from 3 independent transfections. (C) Structure of the DUP minigene. DUP exon 1 is β-globin exon 1, and exon 3 is β-globin exon 2. The diagonal line in the second DUP exon indicates a fusion between the first and second β-globin exons to make a 33-nucleotide hybrid exon (E2). 3 copies of the wild-type (3WT) or mutant (3MU) sequences were inserted into the BglII site. (D) Splicing of DUP, DUP-3WT, and DUP-3MU in HeLa as well as in uninduced and induced MELCs. HeLa and MELCs were stably transfected with the vector alone (DUP), with 3 copies of the wild-type DUP-3WT, or with 3 copies of the mutated DUP-3MU. MELC stable lines were induced to differentiation with DMSO for 4 days. RNA were isolated and analyzed for E2 expression by RT-PCR. +E2 indicates spliced products with exon E2; −E2, spliced products without exon E2; and +E2(%), percentage of spliced products that include exon E2.
Table 1.
Oligonucleotides and primers used in experiments
| Target, primer names | Primer sequences (5′ to 3′) |
|---|---|
| Exon 16 minigenes | |
| Ex13-clone-S | GAAGACGAGCCACCTGAGCAA |
| In17-clone-As | CTAGGGGCAACCCAGCAGAG |
| Bss-S | CTCGTGTTGGTGAACATGAAGATATG |
| Bss-As | TTCACCAACACGAGAAAGCATTTTAAACCTAAAGTCC |
| DUP minigenes | |
| DUP-HindIII-S | GACACCAAGCTTGGTGCACCTGACTCC |
| DUP-XbaI-As | GATGCTTCTAGATCCACGTGCAGCTTGTCAC |
| PN-3WT-S | GATCTTTTGCATGCAATTGCATGAAGGGTTTTGCATGCAATTGCATGAAGGGTTTTGCATGCAATTGCATGAAGGGTTTA |
| PN-3WT-As | GATCTAAACCCTTCATGCAATTGCATGCAAAACCCTTCATGCAATTGCATGCAAAACCCTTCATGCAATTGCATGCAAAA |
| PN-3MU-S | GATCTTTCACGTGCAATCACGTGAAGGGTTTCACGTGCAATCACGTGAAGGGTTTCACGTGCAATCACGTGAAGGGTTTA |
| PN-3MU-As | GATCTAAACCCTTCACGTGATTGCACGTGAAACCCTTCACGTGATTGCACGTGAAACCCTTCACGTGATTGCACGTGAAA |
| mFox-2 5′ RACE | |
| mEx3-RT | GGTCAGGTTATGTTCACTGGTCTGG |
| GeneRace 5′ primer | CGACTGGAGCACGAGGACACTGA |
| 5′ Race-1-As | GAGGTGGTGGGAACGGGATGGTA |
| GeneRace 5′ nested primer | GGACACTGACATGGACTGAAGGAGTA |
| 5′ Race-2-As | AGGAGTTGTTGTTGGCTCCTGGTTAC |
| Fox-2 isoform clones | |
| mEx1A-S | CGGGCGGATGGCGGAAGG |
| mEx1F-S | GGGCGGATGGAGAAAAAGAAAAT |
| mEx1D-S | CATGGACCAGCCCAGGAAC |
| mEx1E-S | CAGCGGCTCTGCCATGATGTA |
| mEx16-As | TCACGTCACTTCAGTAGGGGGCAAAT |
| hEx1A-S | ATGGCGGAGGGCGCCCAGCCGCAGCA |
| hEx1F-S | ATGGAGAAAAAGAAAATGGTAACTCAG |
| hEx16-As | GGGGTCTCACGTCACTTCAGTAG |
| Gel shift constructs | |
| Ex16-shift-S | CAATAAGCTTTTGGAGGTTTGTATGAACTTGAAGC |
| Ex16-shift-As | GAATTCTAGACAGAAGGCGAGATGTTTGCAT |
| mFox-2-shRNA constructs | |
| sh-1A130 target | GCCTGAGCAAGCGGCCACGCAC |
| sh-1A161 target | GGCCGACGGCGGGATGCAGAAC |
| sh-1A195 target | CCGGGTTATCATGGATTCCCAG |
| sh-1A204 target | CATGGATTCCCAGCAAGGGATG |
| sh-1F14 target | AATGGTAACTCAGGGTAACCAG |
| sh-1F15 target | ATGGTAACTCAGGGTAACCAGG |
| sh-1F16 target | GGTAACTCAGGGTAACCAGGAG |
| sh-1F218 target | CTTCTTCTGGTTTATGGAGAAA |
| 5′miR30-XhoI | CAGAAGGCTCGAGAAGGTATATTGCTGTTGACAGTGAGCG |
| 3′miR30-EcoRI | CTAAAGTAGCCCCTTGAATTCCGAGGCAGTAGGCA |
| Exon 16 splicing analysis | |
| mEx17-RT | CTCCAGTAGGGACTTGCCCGTTGATGTTAAGA |
| Ex13-S | AGAGCCCACAGAAGCATGGA |
| Ex17-As | GTGTGTAGATAAGCCCTTGTCCCA |
| DUP exon E2 splicing analysis | |
| DUP-ex1-S | AAGGTGAACGTGGATGAAGTTGGT |
| DUP-ex3-As | ACAGATCCCCAAAGGACTCAAAGAAC |
| mFox-2 real-time PCR | |
| mFox-2A-rt-S | AAGCGGCCACGCACAGAG |
| mFox-2A-rt-AS | CTTGCTGGGAATCCATGATAAC |
| mFox-2F-rt-S | ATGGAGAAAAAGAAAATGGTAACTCAGG |
| mFox-2F-rt-AS | ATATTCTGTGGGAATTCCATTTTGTGG |
| mFox-2-89-rt-S | GGTCCTGAGTTATATGCAGCATCC |
| mFox-2-89-rt-AS | GACAAGGGCACAGCCGC |
| mHprt-rt-S | AGCAGTACAGCCCCAAAATGGTTA |
| mHprt-rt-AS | TCAAGGGCATATCCAACAACAAAC |
DUP minigene.
DUP4-1 was kindly provided by Dr D. L. Black (University of California, Los Angeles, CA).28 The insert in DUP4-1 was amplified with DUP-HindIII-S and DUP-XbaI-As, and cloned into pcDNA3 (Invitrogen, Carlsbad, CA) to obtain neomycin resistance. DUP-3WT was constructed by inserting 3 copies of the intronic sequences, which span the first and second UGCAUG repeats downstream of 4.1R exon 16, into the DUP minigene Bgl II site (Figure 2A) using the annealed oligos PN-3WT-S and PN-3WT-As (Table 1; DUP minigenes). Similarly, DUP-3MU was constructed using PN-3MU-S and PN-3MU-As (Table 1; DUP minigenes).
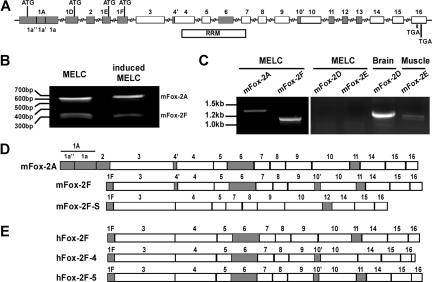
Figure 2.
Gene structure and mRNA variants of Fox-2. (A), Fox-2 gene structure. Mouse Fox-2 possesses 4 alternative promoters and contains 16 exons. Shaded boxes indicate alternatively spliced exons; open boxes, constitutive exons. Within the alternative exons, internal cryptic splice sites are used in exons 1A, 4, and 10, and generate exon variants designated as 1a′′, 1a′, 1a; 4′, 4; 10′, 10. A total of 4 alternative mutual exclusive translation initiation sites (ATG) are located in exons 1A 1D, 1E, or 1F. The RRM bar indicates the region that encodes for the RNA recognition motif (RRM). Splicing of exons 11 and 12 is mutually exclusive. (B) 5′ RACE analysis of the transcription initiation sites of Fox-2 from uninduced and induced MELCs. (C) RT-PCR analysis of mFox-2 expression in MELCs. RNAs were amplified with a primer set located at the first exon (1A, 1D, 1E, or 1F) and exon 16. MELC RNAs could be amplified with 1A or 1F primers but not with 1D or 1E primers. mRNAs from mouse brain and skeletal muscle served as positive controls for exon 1D and 1E, respectively. (D) Exon composition of Fox-2 isoforms isolated from mouse cells. mFox-2A and mFox-2F were isolated from MELCs. The same mFox-2F and an additional mFox-2F-S were isolated from C57/BL6 mouse muscle. (E) Exon composition of hFox-2 isoforms isolated from human cells. hFox-2F is expressed in HeLa cells. hFox-2F-4 and hFox-2F-5 are expressed in RD cells.
Fox-2 clones and expression constructs.
To clone or detect Fox-2 isoforms translated from alternative first exons, the primers mEx1A-S, mEx1D-S, mEx1E-S, or mEx1F-S and mEx16-As (Table 1; Fox-2 isoform clones) were used to amplify mouse Fox-2 from MELCs or C57/BL6 muscle. hEx1A-S or hEx1F-S and hEx16-As (Table 1; Fox-2 isoform clones) were used for Fox-2 from HeLa or human rhabdomyosarcoma (RD) cells. PCR products were cloned into the p3 × FLAG-CMV-14 expression vector (Sigma, St Louis, MO) in which the neo sequence was replaced with the pac sequence for puromycin resistance. mFox-2A-GST and mFox-2A-EGFP fusion constructs were prepared by subcloning the mFox-2A coding sequence into the pGEX-6p-1 (Amersham Pharmacia, Piscataway, NJ) or pEGFP vector (Clontech, Palo Alto, CA). Recombinant mFox-2A-GST protein was cleavaged with PreScission Protease (Amersham Pharmacia).
Gel mobility shift constructs.
Ex16-shift-S and Ex16-shift-As (Table 1; gel shift constructs) were used to amplify the fragments from either the WT or PNB minigene constructs and cloned into pcDNA3, generating RNAs for the gel mobility shift assays.
mFox-2-shRNA constructs.
The construction of mFox-2 isoform–specific shRNA expression vectors was as described.31 The target sequences sh-1A130, sh-1A161, sh-1A195, and sh-1A204 of mFox-2A and sh-1F14, sh-1F15, sh-1F16, and sh-1F218 of mFox-2F are listed in Table 1 (mFox-2-shRNA constructs). The synthetic short hairpin oligonucleotides composed of the target sequences, miR-30 microRNA flanking sequences, and miR-30 loop sequence were amplified with 5′miR30-XhoI and 3′miR30-EcoRI (Table 1; mFox-2-shRNA constructs), and cloned into LMP microRNA-adapted retroviral vector (Open Biosystems, Huntsville, AL). The nonsilencing shRNAmir retroviral control (Open Biosystems) was used as a negative control.
Cell culture, transfection, retroviral infection, and induction
MELCs, HeLa cells, and RD cells were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). Cells were transfected with Lipofectamine 2000 (Invitrogen). Retroviral shRNA vectors were amplified in PirPlus DH10βF′DOT competent Escherichia coli and packaged using LinX viral packaging cell line (Open Biosystems).A total of 0.5 mL shRNA retrovirus containing medium was added to 1.5 mL fresh medium with 0.5 to 1 × 106MELCs for infection. Exon 16 minigene– and DUP minigene–transfected stable lines were selected with 800 μg/mL G-418. A total of 2.0 μg/mL puromycin was used for selection of shRNA-infected or mFox-2–expressing cell lines. MELCs were induced to erythroid differentiation as previously described.20
Erythroid precursor cell isolation and in vitro differentiation
Fetal liver cells were isolated from day-12 to -13 C57BL/6 mouse embryos as described.32 TER119− cells were purified through a StemSep column (StemCell Technologies, Vancouver, BC) and cultured for erythroid differentiation. TER119− cells were seeded in fibronectin-coated wells in Iscove modified Dulbecco medium (IMDM) containing 15% FBS, 1% de-toxified bovine serum albumin (BSA), 200 μg/mL holo-transferrin, 10 μg/mL recombinant human insulin, 2 mM l-glutamine, 10−4 M β-mercaptoethanol, and 2 U/mL erythropoietin (Epo). On the second day, this medium was replaced with erythroid-differentiation medium (EDM).32. Flow cytometry analysis was used to distinguish mouse fetal liver erythroblasts at different developmental stages stained for CD71 and TER119.32
5′ RACE and RT-PCR analyses
5′ rapid amplification of cDNA ends (RACE) was performed following the instruction of GeneRacer Kit (Invitrogen); mEx3-RT was used for reverse transcription, and sense (GeneRace 5′ Primer and 5′ Nested Primer) and antisense primers (5′Race-1-As and 5′Race-2-As; Table 1; mFox-2 5′ RACE) were used for nested PCR. Semiquantitative reverse transcription (RT)–PCR analysis of splicing products were performed as described.20 RNAs were reverse transcribed using SP6 primer for exon 16 or DUP minigenes, and mEx17-RT for mouse endogenous exon 16 (Table 1; exon 16 splicing analysis). Amplification of exon 16 products was performed with Ex13-S and Ex17-As (Table 1; exon 16 splicing analysis), respectively. The DUP minigene spliced products were amplified using DUP-ex1-S and DUP-ex3-As (Table 1; DUP exon E2 splicing analysis). For each construct, 2 transfections were performed in each experiment. Each experiment was repeated 3 times, and standard deviations were determined.
Fox-2 real-time PCR was performed on an Bio-Rad iCycler using iQ SYBR Green Supermix and analyzed with iCycler iQ Real-Time PCR Detection System Software (Bio-Rad, Hercules, CA). cDNA was generated with an oligo-dT primer. Primers mFox-2A-rt-S and mFox-2A-rt-As were used for mFox-2A; mFox-2F-rt-S and mFox-2F-rt-As were used for mFox-2F; and mFox-2-89-rt-S and mFox-2-89-rt-As were used for both mFox-2 isoforms (Table 1; mFox-2 real-time PCR). The quantitative data were calculated from the Ct values for each reaction and the average reaction efficiency for each primer set. The reaction efficiencies of each reaction were determined by linear regression analysis.33 All mFox-2 expression values were normalized to the expression values of the hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1) gene.34 All reactions were performed in triplicate.
In vitro transcription and gel mobility shift assay
RNA substrates were produced from plasmids linearized with XbaI and transcribed with T7 RNA polymerase (Novagen, Madison, WI) in the presence of a Ribo m7G Cap Analog (Promega, Madison, WI) and α-32P-UTP (Perkin Elmer, Shelton, CT). For gel mobility shift assays, 20 ng of α-32P labeled RNA and increasing amounts of purified mFox-2A recombinant protein were incubated in a 25-μL splicing reaction that contained 0.5 mM ATP, 20 mM creatine phosphate, 3.2 mM MgCl2, 7.5 μL splicing dilution buffer (20 mM Tris-HCl [pH 7.6] and 100 mM KCl), and 7.5 μL purified mFox-2A diluted in buffer D20 at 30°C for 20 minutes. The reactions were fractionated in a 5% nondenaturing polyacrylamide gel electrophoresis (PAGE) gel and visualized by autoradiography.
Indirect immunofluorescence and imaging
MELCs were transfected with either the pEGFP vector or mFox-2A-EGFP construct and subjected to immunofluorescence staining with anti-SC35 antibody (BD Biosciences, Palo Alto, CA) as described.35 TER119+ mouse fetal liver cells were stained using an anti-mFox2 antibody. The samples were viewed with a Zeiss Axiovert 200M inverted microscope (63×/1.40 oil objective, Zeiss, Oberkochen, Germany). The images were collected using SlideBook4 software (Intelligent Imaging Innovations, Denver, CO) and processed using Photoshop software (Adobe Systems, San Jose, CA).
Western blot
Anti-mFox2 polyclonal antibody was raised against a synthetic peptide (CENSADADRAREKLH) located on mFox-2 exon 6. Cysteine was added to the N-terminus of the peptides, conjugated to the carrier protein keyhole limpet hemocyanin (KLH), and used as an antigen to raise antibody in rabbit. mFox-2 proteins were detected with anti-Flag antibody (Sigma) for Flag-tagged mFox-2 or with anti–mFox-2 antibody for endogenous mFox-2 using the ECL detection kit (Amersham Pharmacia). Anti–β-actin antibody (Sigma) served as a loading control.
Results
Intronic splicing enhancer UGCAUG facilitates erythroid differentiation stage–specific 4.1R exon 16 splicing switch in a dose-dependent manner
In a previous study,19 we showed that the intronic splicing regulatory element, UGCAUG, enhances 4.1R exon 16 splicing. To further assess their significance in exon 16 splicing, we evaluated the relative contribution of each intrinsic repeat individually (Figure 1A; Pm, Nr, Bss) or in combination (Figure 1A; PN and PNB) by incorporating their respective mutations into the exon 16 minigene construct and transfecting into MELCs. Stable transformants were analyzed for exon 16 expression by RT-PCR. The WT 4.1R exon 16 minigene mimics endogenous exon 16 splicing patterns in uninduced and induced MELCs (Figure 1B; WT). A single mutation in the UGCAUG repeats showed a moderate reduction of exon 16 in undifferentiated (Figure 1B; uninduced, lanes Pm, Nr, and Bss) and exerted a stronger effect in differentiated MELCs (Figure 1B; induced, lanes Pm, Nr, and Bss). A more pronounced effect was observed for the double mutant, PN (Figure 1B; uninduced and induced, lane PN). The greatest decrease was observed when mutations were introduced in all repeats (Figure 1B; uninduced and induced, lane PNB). These results suggest that UGCAUG is an ISE functioning in a dose-dependent manner.
UGCAUG repeats activate an internal exon in a test “neutral” reporter system in a cell type–specific and differentiation stage–specific manner
To investigate the direct effect of UGCAUG on the expression of exon 16 in erythroid cells, we examined it in the context of a neutral reporter system, DUP4-1.28 DUP4-1 presents a very simple binary splicing pattern. An exon 1 and 3 product in the absence of added splicing enhancers is obtained (Figure 1C). Inclusion of exon 2 (E2) requires addition of enhancer sequences. DUP4-1 thus provides an assay for the inserted enhancers that promote internal exon E2 inclusion.
Transfection of DUP resulted in the complete exclusion of the internal chimeric exon in both HeLa cells and MELCs (Figure 1D; lane DUP). The insertion of 3 copies of the WT sequences spanning the first and second repeats promoted full inclusion of E2, whereas mutated sequences exerted no effect in HeLa cells (Figure 1D; HeLa, lanes DUP-3WT and DUP-3MU). UGCAUG enhanced inclusion of E2 in a differentiation stage–specific manner in MELCs (Figure 1D; uninduced and induced, lane DUP-3WT) but mutant sequences did not (Figure 1D; uninduced and induced, lane DUP-3MU). UGCAUG thus exerts cell type–specific and differentiation stage–specific splicing regulation.
Cell type–specific expression of Fox-2 isoforms
Fox-1 and Fox-2 proteins bind to the UGCAUG element.29,30 We found no Fox-1 mRNA in MELCs, even though it could be detected in mouse muscle and HeLa cells (data not shown). We did detect Fox-2 isoforms (Figure 2C). Fox-2, also known as RBM9,30 fxh,36 or RTA,37 contains a single RNA recognition motif (RRM)–type RNA-binding domain.22,30,38 Fox-2 encodes tissue-specific isoforms that are derived from use of alternative promoters and alternative splicing.30
Translation of mFox-2 can be initiated from alternative start sites located within the mutually exclusive alternative exons 1A, 1D, 1E, or 1F (Figure 2A). We performed a 5′ RACE to investigate which exons are expressed in MELCs. Only 2 species of mFox-2 were found containing either exons 1A or 1F from both uninduced and induced MELCs (Figure 2B). Using sequence-specific primers, exons 1A and 1F were detected in MELCs, while exons 1D and 1E were not (Figure 2C). Consistent with the GenBank database, exons 1D and 1E were detected in mouse brain and skeletal muscle (Figure 2C). Within exon 1A of mFox-2A, an internal 3′ and 5′ splice site was apparently used to exclude 54-bp sequences (Figure 1A; 1a′) and join the flanking sequences together (Figure 1A; 1a′′ and 1a). The mFox-2F isoform is identical in exons 3 to 16 with mFox-2A (Figure 2D; mFox-2A) except for 4 additional amino acids in exon 10′ (Figure 2D; mFox-2F). mFox-2A and -2F (Figure 2D) are representative of the 2 major classes in MELCs, but there may be other isoforms present with different internal splices.
The University of California Santa Cruz (UCSC) Genome Browser40 shows a number of Fox-2 clones with a full-length exon 1A in both human and mouse. However, none of the clones had a 54-nucleotide internal deletion within exon 1A as mFox-2A isolated from MELCs. To examine whether mFox-2A and -2F isoforms are uniquely expressed in MELCs, we cloned Fox-2 mRNA species from murine and human cells by RT-PCR using primer sets located at either exon 1A or exon 1F and the last exon. The mFox-2A isoform, containing an internal deleted exon 1A, could be detected only in MELCs but not in any other tissue or cell lines tested.
In C57BL/6 mouse muscle tissue, mFox-2F and mFox-2F-S were both translated from exon 1F, but differences in internal exon compositions were noted (Figure 2D; mFox-2F and mFox-2F-S). All human major isoforms are translated from exon 1F; hFox-2F (GenBank accession no. CR456559) was identified in HeLa cells (Figure 2E; hFox-2F), and hFox-2F-4 and hFox-2F-5 (GenBank accession no. AF229058) were identified in RD cells (Figure 2E; hFox-2F-4 and hFox-2F-5). All forms identified have correct reading frames. The novel sequences were deposited in GenBank: mFox-2A (accession no. AY904025), mFox-2F (accession no. DQ017388), mFox-2F-S (accession no. DQ017389), and hFox-2F-4 (accession no. AY960684). Our survey suggests that tissue- or cell-type–specific expression of the Fox-2 isoform is a commonly occurring phenomenon, and that the unique expression of mFox-2A may contribute to cell-type– and differentiation stage–specific enhancing activity of the UGCAUG element in MELCs (Figure 1D).
mFox-2A enhances 4.1R exon 16 and DUP exon E2 splicing in a UGCAUG-dependent manner
Since MELCs express both mFox-2A and mFox-2F, we analyzed the effect of these isoforms on exon 16 splicing in uninduced cells. Introduction of mFox-2A or mFox-2F enhanced endogenous exon 16 inclusion with differential activity (Figure 3A; endo, lanes mFox-2A and mFox-2F). The mFox-2A isoform (Figure 3A; endo, lane mFox-2A) exhibited the greatest enhancement: 4 times more than the vector alone (Figure 3A; endo, lane vector) and 2 times more than mFox-2F (Figure 3A; endo, lane mFox-2F).
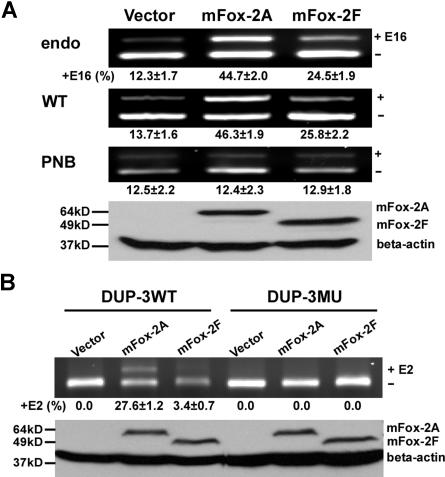
Figure 3.
mFox-2 enhances 4.1R exon 16 and DUP exon E2 splicing in an UGCAUG-dependent manner. (A) mFox-2 isoforms exhibit differential enhancing activities on exon 16 splicing. MELCs or exon 16 WT or PNB minigene stably expressing MELCs were transfected with vector alone, mFox-2A, or mFox-2F. Exon 16 splicing products were analyzed by semiquantitative RT-PCR. endo indicates endogenous; WT, wild-type; and PNB, mutant minigene PNB. Western blot analysis confirmed the expression of transfected mFox-2A or mFox-2F isoforms from WT-minigene–expressing MELCs by anti-Flag antibody; β-actin was used as a loading control. +E16 indicates spliced products with exon 16; −E16, spliced products without exon 16; and +16 (%), percentage of spliced products with standard deviation that include exon 16. The results shown are from 3 independent transfections. (B) mFox-2 isoforms exhibit differential enhancing activities on exon E2 splicing in the DUP system. DUP-3WT or DUP-3MU minigenes stably expressing MELCs were transfected with vector alone, mFox-2A, or mFox-2F and analyzed for exon E2 expressions. Western blot analysis confirmed the expression of mFox-2A or mFox-2F isoforms. +E2 indicates spliced products with exon E2; −E2, spliced products without exon E2; and +E2 (%), percentage of spliced products that include exon E2.
We then examined whether the mFox-2 enhancing effect was UGCAUG specific using both exon 16 and DUP minigene systems. We introduced mFox-2A and mFox-2F into WT or mutant PNB exon 16 minigenes and analyzed for exon 16 expression. In keeping with the endogenous exon 16 splicing patterns just noted, mFox-2A enhanced WT minigene exon 16 splicing approximately 4-fold, while mFox-2F exhibited a 2-fold increased effect when compared with that of the vector (Figure 3A; WT, lanes vector, mFox-2A, and mFox-2F). However, neither form had any effect on exon 16 splicing on mutant PNB (Figure 3A; PNB, lanes vector, mFox-2A, and mFox-2F). Expression of transfected mFox-2 isoforms was confirmed by a Western blot of WT cell lysate with an anti-Flag antibody (Figure 3A; Western, lanes vector, mFox-2A, and mFox-2F).
DUP testing yielded similar results when mFox-2A was introduced. mFox-2A enhanced E2 inclusion in DUP-3WT (Figure 3B; DUP-3WT, lanes vector and mFox-2A) but not in DUP-3MU pre-mRNAs in stably transfected MELCs (Figure 3B; DUP-3MU, lanes vector and mFox-2A). On the other hand, mFox-2F has very little effect on E2 inclusion in DUP-3WT (Figure 4B; DUP-3WT, lanes vector and mFox-2F) and no effect on DUP-3MU minigenes (Figure 4B; DUP-3MU, lanes vector and mFox-2F). These results suggest that overexpression of mFox-2A or mFox-2F enhanced alternative spliced exon inclusion in a UGCAUG-dependent manner; however, mFox2A exerted a much stronger enhancing activity than mFox-2F.
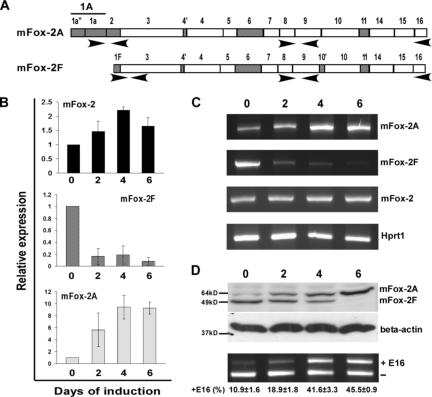
Figure 4.
Analysis of mFox-2 isoform expression and its effect on exon 16 splicing switch during MELC differentiation. (A) Schematic diagram of primer sets used in expression analysis. Primer set locations are indicated with arrows. (B) Real-time PCR analysis of total mFox-2 (mFox-2), mFox-2F, and mFox-2A expression during MELC differentiation. 0, 2, 4, and 6 indicate the days after DMSO-induced differentiation. All data are presented as mean plus or minus SD. Day 0 was taken as 1. (C) Full-length Fox-2 mRNA expression detected during MELC differentiation. mFox-2 indicates total mFox-2; Hprt1 serves as an internal control. (D) mFox-2 isoform expression switch correlates with exon 16 splicing switch during MELC erythroid differentiation. 0, 2, 4, and 6 indicate the days of induced differentiation. Top panel shows endogenous mFox-2 isoforms expression was detected by anti–mFox-2 antibody. Middle panel shows β-actin was used for loading control. Bottom panel shows exon 16 splicing was analyzed by semiquantitative RT-PCR from 3 independent transfections.
mFox-2 isoforms expression is differentially regulated during erythroid differentiation
We asked whether mFox-2 expression correlates with exon 16 inclusion during induced MELC differentiation. Using the MELC culture system that supports terminal erythroblast proliferation and differentiation, we have shown that exon 16 is preferentially included in mature 4.1R mRNA in induced MELCs.20 We cultured MELCs for 0, 2, 4, and 6 days after DMSO-induced differentiation and analyzed the timing of mFox-2 isoforms expression by real-time PCR (Figure 4A). We noted an approximately 1.5-fold increase in total mFox-2 expression by day 2 and a 2-fold increase by days 4 to 6 (Figure 4B; mFox-2). Surprisingly, the expression of mFox-2F was drastically reduced throughout the MELC differentiation process (Figure 4B; mFox-2F). The increase in total mFox-2 expression was thus most likely attributed to an increase in mFox-2A (Figure 4B; mFox-2A). RT-PCR amplification of full-length mFox-2A and mFox-2F during the induction process also revealed reciprocal expression of A and F isoforms (Figure 4C; mFox-2A and mFox-2F).
Reciprocal expression of mFox-2A and mFox-2F was confirmed by a Western blot with an anti–mFox-2 antibody (Figure 4D; mFox-2A and mFox-2F); mFox-2F was abundantly expressed in uninduced cells, whereas mFox-2A was expressed in great quantities in induced cells. The increase in mFox-2A and reduction in mFox-2F correlated with the exon 16 splicing switch during MELC differentiation (Figure 4D; ± E16). These results suggest that the erythroid differentiation inducible mFox-2A and the generally expressed mFox-2F play unequal roles in the splicing switch of exon 16 during erythoid differentiation. The up-regulation of mFox-2A most likely plays a critical role in the exon 16 splicing switch in differentiated MELCs.
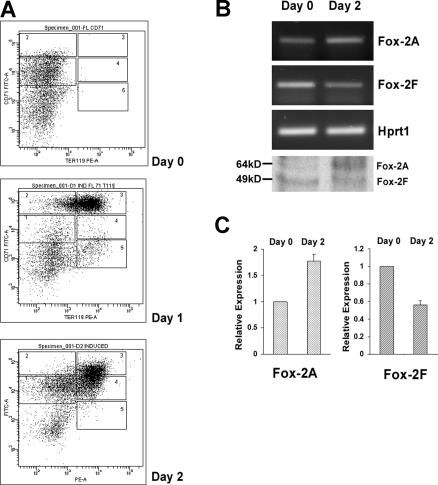
We asked whether mFox-2 isoforms switched in a more physiologic primary cell system. We adopted the mouse fetal liver cells in vitro erythroid differentiation system.32 Cultured fetal liver cells were double labeled for erythroid-specific TER119 and non–erythroid-specific transferrin receptor (CD71) and analyzed by flow cytometry.32 The day-0 cells contained only the TER119− cells (Figure 5A; day 0). Day-1 and -2 cells shifted from a less mature population of CD71highTER119high to a more mature population of CD71lowTER119high (Figure 5A; day 2).
Figure 5.
Analysis of Fox-2 isoforms expression during erythroid differentiation of mouse fetal liver cells. (A) Flow cytometry of cultured fetal liver cells at 0, 1, and 2 days in erythroid-differentiation medium. Cells were double-stained for a PE-conjugated anti-TER119 mAb and an FITC-conjugated anti-CD71 mAb and analyzed by flow cytometry. Axes indicate relative logarithmic fluorescence units for PE (x-axis) and FITC (y-axis). Regions 1 to 5 are defined by characteristic staining pattern of cells, including CD71medTER119low, CD71highTER119low, CD71highTER119high, CD71medTER119high, and CD71lowTER119high, respectively. (B) Fox-2A and -2F isoforms expression analyzed by RT-PCR and Western blot analyses (bottom panel). Hprt1 serves as an internal control for RT-PCR. (C) Relative expression of Fox-2A and Fox-2F in erythroid differentiation of fetal liver cells. Semiquantitative RT-PCR products were scanned and analyzed by the ChemiImager 5500 system (Alpha Innotech Co, San Leandro, CA). Day-0 and day-2 cells were analyzed for expression of mFox-2 isoforms. The bar graph represents the relative levels of mFox-2 mRNA in the day 0 and day 2 cells. Day 2 mFox-2 levels are expressed relative to those in day 0 cells and normalized with Hprt1. Error bars represent standard deviation. Day 0 was taken as 1.
Analysis of these cells revealed a reciprocal expression of the mFox-2A (Figure 5B; Fox-2A, days 0 and 2) and 2F isoforms (Figure 5B; Fox-2F, days 0 and 2). There was an approximately 80% mFox-2A increase in day-2 cells when compared with that of day-0 cells (Figure 5C; Fox-2A) in concert with a 50% to 60% reduction in mFox-2F (Figure 5C; Fox-2F). Reciprocal expression of mFox-2A and mFox-2F at protein levels was also confirmed by a Western blot (Figure 5B bottom panel). Thus, the up-regulation of mFox-2A most likely plays an important role in primary erythroid cell differentiation as well.
mFox-2A localizes to the nucleus and binds to UGCAUG in an electrophoretic mobility shift assay
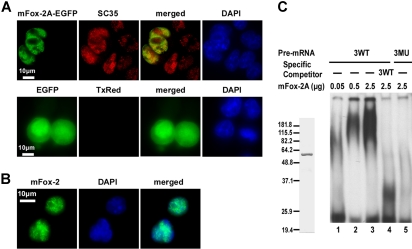
Splicing factors predominantly reside within the nucleus. To examine whether mFox-2A is also localized to the nucleus, we expressed a mFox-2A/EGFP fusion protein in MELCs and analyzed its localization relative to splicing factor SC35. The fusion protein mFox-2A/EGFP localized exclusively in the nucleus of MELCs as a punctate structure (Figure 6A; mFox-2A/EGFP, EGFP). Similarly, SC35 localized to the nucleus with a punctate pattern in both mFox-2A/EGFP–expressing and –nonexpressing cells (Figure 6A; mFox-2A/EGFP, SC35). Superimposition of mFox-2A/EGFP and SC35 revealed that although both proteins are intensely localized in the nucleus, they are not precisely colocalized together (Figure 6A; mFox-2A/EGFP, merged). EGFP was shown to localize throughout the cell when transfected with the vector alone (Figure 6A; EGFP, EGFP). No labeling was apparent when the primary antibody SC35 was omitted (Figure 6A; EGFP, TxRed). Thus, mFox-2A localized to the nucleus of MELCs. The nuclear localization was further confirmed in TER119+ fetal liver cells (Figure 6B). Superimposed images (Figure 6B; merged) of mFox-2 and DNA (Figure 6B; mFox-2, DAPI) suggest that the punctate structure of Fox-2 is localized in the nucleus.
Figure 6.
mFox-2A localizes in the nucleus and specifically binds to UGCAUG sequence in an gel mobility shift assay. (A) MELCs transfected either with mFox-2A/EGFP or EGFP vector alone were fixed and stained for endogenous SC35 and for nucleic acids (DNA), then visualized for the presence of EGFP, SC35, and DAPI. Green represents EGFP epitopes; red, SC35 epitopes; yellow, the colocalization of mFox-2A and SC35; and blue, DNA stained with DAPI. TxRed indicates EGFP-transfected cells were stained with a secondary mouse antibody conjugated with Texas Red but without the primary antibody and served as a negative control. (B) TER119+ fetal liver cells stained for endogenous Fox-2 and DNA. Green indicates Fox-2; blue, DNA. (C) Left panel shows purified mFox-2A. mFox-2A expressed as GST fusion proteins were cleaved with PreScission protease (Amersham Pharmacia) and purified to homogeneity. Right panel shows gel mobility shift assay using 3 copies of sequences between the first and second UGCAUG repeats. 32P-labeled wild-type RNA (3WT; lanes 1-4) or mutant RNA (3MU; lane 5) were incubated with increasing amounts of mFox-2A in the presence of nonspecific competitor tRNA. A 10-fold excess (compared with the 32P-labeled probe) of unlabeled 3WT RNA (3WT; lane 4) was added as a competitor.
To examine whether mFox-2A binds to UGCAUG directly, we performed an electrophoretic mobility shift analysis using UGCAUG repeats and purified mFox-2A protein (Figure 6B). We examined the specific interaction between mFox-2A and UGCAUG using a probe derived from 3WT consisting of 3 copies of the sequences between the first and second repeats (Figure 1C). The mutant 3MU (Figure 1C) served as a negative control. Intensity of the retarded band increased when the WT probe was incubated with increasing amounts of mFox-2A (Figure 6C; 3WT, lanes 0.05, 0.5, and 2.5 μg). The WT UGCAUG-containing RNA competed for the specific binding site and drastically reduced the intensity of the retarded band (Figure 6C; 3WT, lanes WT, 2.5 μg). Retarded bands were absent when large amounts of mFox-2A were incubated with the mutant probe (Figure 6C; 3MU, lane 2.5 μg). These results suggest that mFox-2A directly binds to the UGCAUG sequence.
mFox-2 isoforms have differential regulatory effects on exon 16 alternative splicing
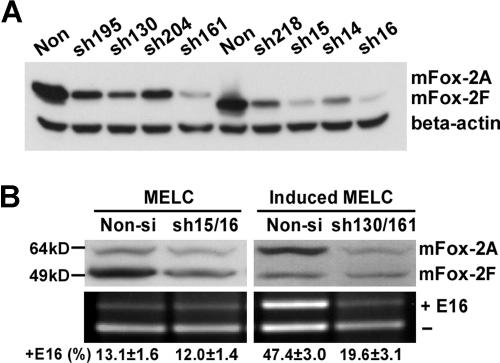
Since the behaviors of mFox-2A and -2F are different during erythroid differentiation, we next examined whether reduction in each endogenous mFox-2 isoform expression would block the exon 16 splicing switch in induced MELCs. Several shRNA constructs specifically targeted to mFox-2A or mFox-2F forms were tested for their potency in MELCs (Figure 7A). The most potent shRNA constructs—sh130 and sh161 for mFox-2A and sh15 and sh16 for mFox-2F—were selected for further studies.
Figure 7.
Isoform-specific mFox-2A reduction inhibits exon 16 splicing switch in differentiated MELCs. (A) Screening of mFox-2A or mFox-2F isoform-specific shRNAs. mFox-2 isoform-specific shRNAs were tested in cells stably transfected with mFox-2A (lanes 1-5) or mFox-2F (lanes 6-10). The reduction of mFox-2 isoforms was analyzed by Western blot using anti-Flag antibody. β-actin was used as a loading control. (B) The reduction of endogenous mFox-2 isoform expression by isoform-specific shRNAs and the effects on exon 16 splicing. Reduction of mFox-2F isoform expression did not affect exon 16 splicing pattern in uninduced MELCs. Reduction of mFox-2A isoform expression in induced MELCs inhibited the exon 16 splicing switch.
We stably expressed mFox-2-shRNAs or its nonfunctional control nonsilencing shRNA in MELCs, and analyzed exon 16 expression. In uninduced cells, the expression of endogenous mFox-2A is low (Figure 7B; MELC, top panel, Non-si lane) and mFox-2F is high (Figure 7B; Induced MELC, top panel, Non-si lane). Treatment of mFox-2F–shRNA reduced mFox-2F expression (Figure 7B; MELC, top panel, Non-si and sh15/16 lanes). Interestingly, the reduction of mFox-2F had no effect on exon 16 splicing when compared with nonsilencing control shRNA (Figure 7B; MELC, bottom panel, Non-si and sh15/16 lanes). In induced MELCs, mFox-2A is the major mFox-2 isoform. Induced cells treated with mFox-2A–shRNA reduced mFox-2A expression (Figure 7B; induced MELC, top panel, Non-si and sh130/160 lanes). Exon 16 inclusion was reduced by more than 2.5-fold in these cells (Figure 7B; induced MELC, bottom panel, Non-si and sh-130/161 lanes). These results further suggest that the exon 16 splicing switch is favored by a differentiation-inducible mFox-2A isoform.
Discussion
The present study identifies a novel mFox-2A isoform that activates exon 16 splicing via interactions with the intronic splicing enhancer UGCAUG. It also demonstrates that up-regulation of mFox-2A mediates the exon 16 splicing switch in differentiated cells. Our finding suggests for the first time that regulated expression of a differentiation-inducible isoform plays an important role in controlling erythroid differentiation stage–specific alternative RNA processing of protein 4.1R exon 16.
The UGCAUG hexanucleotide has been shown to be a splicing regulator of many alternative exons.24–28 Its location relative to the exon determines its positive or negative effect; UGCAUG represses splicing when upstream of the exon,41,42 but activates when located downstream.25,27,28 Recent studies have begun to reveal how the upstream location of UGCAUG determines its activity as a silencer. Zhou and colleagues40 showed that Fox-1 and Fox-2 proteins interact with the upstream UGCAUG elements in a manner that blocks U2AF65 binding to the 3′ splice site upstream of exon 4 of calcitonin/CGRP pre-mRNA. How the downstream location of UGCAUG acts as an enhancer remains to be determined.
Fox-2β binds to UGCAUG elements in intron 16.22 mFox-2A, which possesses a distinct N- and C-terminus from that of Fox-2β but an identical RRM, also binds to the same sequence motif (Figure 6B); this RRM is most likely responsible for RNA binding activity. The RRMs for Fox-1 and Fox-2 are identical over their entire lengths and are present in nearly all isoforms in both human and mouse tissue.30 The fact that we detected no Fox-1 mRNA in MELCs suggests that Fox-1 is not involved in exon 16 splicing in differentiating erythroid cells, even though Fox-1 has been shown to enhance exon 16 splicing in HeLa cells.22 Given the ability of Fox-1 to bind to UGCAUG elements, it is possible that Fox-1 also contributed to the inclusion of E2 when these sequences were tested with DUP system in HeLa cells (Figure 1D).
A complex 5′ structure and multiple alternative exons are characteristic of the Fox-2 gene, generating Fox-2 isoforms with diverse amino-terminal domains and variations in the internal and/or C-terminal domain. Tissue-specific isoforms of Fox-1 and Fox-2 exhibit differential splicing activities and play a role in determining tissue specificity of UGCAUG-mediated alternative N30 splicing.29 Our study has identified 2 isoforms of mFox-2 in MELCs, each possessing different amino-terminal peptides. The N-terminal domain of mFox-2A is significantly dissimilar to any protein motifs in current databases. mFox-2A exerts a significantly stronger exon 16 splicing activity than mFox-2F. We attempted to identify the functional differences between mFox-2A and -2F using deletion constructs lacking the RRM domain. Overexpression of mutant mFox-2F did not affect exon 16 splicing. However, cell death occurred when mutant mFox-2A was overexpressed (our unpublished data, May 2007). Whether these peptides complex with different proteins in the splicing machinery and/or in other cellular processing pathways to differentiate their activity remains to be determined.
The reciprocal expression of mFox-2A and -2F isoforms in differentiating MELCs was recapitulated in mouse primary erythroblasts cultured in vitro that closely mimics the in vivo terminal proliferation and maturation of erythroid cells (Figure 5). Ponthier et al22 reported an approximately 48-kDa Fox-2 whose expression was unchanged during erythoid differentiation in erythroblasts from mice treated with the anemia-inducing strain of Friend virus. Whether different antibodies and cell systems (Friend virus infected mice versus normal mice) used in these studies contribute to these discrepancies remains to be determined.
Our studies20 and those of Hou and colleagues21 imply that an antagonistic effect between positive and negative factors may regulate exon 16 splicing, with SF2/ASF possibly functioning through an exonic splicing enhancer and hnRNPA1 possibly functioning through an exonic splicing silencer. Factor expression analysis during erythroid differentiation shows that, compared with nondifferentiated cells, several SR proteins (ASF/SF2,20 m9G8, mU2AF65, and mSC35) are up-regulated, while the negative regulators hnRNP A/B and mPTB are down-regulated in differentiated MELCs (our unpublished data, June 2002). A correlation between the reduction of hnRNPA1 expression and increase in exon 16 inclusion during erythoid differentiation has been reported.21 Even though mFox-2F was abundantly expressed in uninduced MELCs, it exhibited only a slight enhancing effect on exon 16 splicing. It is possible that the effect of mFox-2F is antagonized by the abundantly expressed negative regulators in undifferentiated cells, and the effect of mFox-2A was unmasked by the reduction of the same negative factors. However, the importance of mFox-2A in exon 16 splicing is evident by the fact that nonsilencing control cells exhibit increased exon 16 inclusion, while silenced mFox-2A dramatically reduced exon 16 inclusion in the same low hnRNP A1 background. These results suggest that mFox-2A also plays an important role in exon 16 inclusion regardless of the presence of other factors.
In HeLa cells, exon 16 splicing has been shown to be regulated by the antagonistic activities of Fox-2 and hnRNP A1; Fox-2 reduced hnRNP A1 binding to the enhancer element in a concentration-dependent manner in an in vitro competitive binding experiment43. These results further support the involvement of Fox-2 in exon 16 splicing. However, the precise mechanism by which mFox-2A promotes exon 16 splicing activation and its interaction with other splicing regulators are unknown. How Fox-2 isoforms are regulated during erythroid differentiation also remains to be determined. Nevertheless, our data show that an erythroid differentiation-inducible mFox-2A protein is up-regulated and mediates an important exon 16 splicing switch through an interaction with UGCAUG repeats in late erythroid cells. Elucidation of the mechanisms of splicing switch regulation is critical to a better understanding of the regulated expression of other key erythroid-specific forms during late erythropoiesis.
Acknowledgments
We thank Dr D. L. Black (University of California, Los Angeles, CA) for the DUP4-1 minigene.
This work was supported by National Institutes of Health grant no. HL24385 (E.J.B.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: G.Y. designed experiments, performed research, analyzed data, and wrote the manuscript. S.C.H. designed experiments, analyzed data, and wrote the manuscript. J.Y.W. analyzed data and edited the manuscript. E.J.B. designed experiments, analyzed data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shu-Ching Huang, Department of Medical Oncology, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: shu-ching_huang@dfci.harvard.edu.
References
- 1.Parra MK, Gee SL, Koury MJ, Mohandas N, Conboy JG. Alternative 5′ exons and differential splicing regulate expression of protein 4.1R isoforms with distinct N-termini. Blood. 2003;101:4164–4171. doi: 10.1182/blood-2002-06-1796. [DOI] [PubMed] [Google Scholar]
- 2.Conboy JG, Chan J, Mohandas N, Kan YW. Multiple protein 4.1 isoforms produced by alternative splicing in human erythroid cells. Proc Natl Acad Sci U S A. 1988;85:9062–9065. doi: 10.1073/pnas.85.23.9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang TK, Qin Z, Leto T, Marchesi VT, Benz EJ., Jr Heterogeneity of mRNA and protein products arising from the protein 4.1 gene in erythroid and nonerythroid tissues. J Cell Biol. 1990;110:617–624. doi: 10.1083/jcb.110.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang SC, Baklouti F, Tang TK, Benz EJ., Jr Differential utilization of translation initiation sites in alternatively spliced mRNAs arising from the protein 4.1 gene. Trans Assoc Am Physicians. 1992;105:165–171. [PubMed] [Google Scholar]
- 5.Chasis JA, Coulombel L, McGee S, et al. Differential use of protein 4.1 translation initiation sites during erythropoiesis: implications for a mutation-induced stage-specific deficiency of protein 4.1 during erythroid development. Blood. 1996;87:5324–5331. [PubMed] [Google Scholar]
- 6.Subrahmanyam G, Bertics PJ, Anderson RA. Phosphorylation of protein 4.1 on tyrosine-418 modulates its function in vitro. Proc Natl Acad Sci U S A. 1991;88:5222–5226. doi: 10.1073/pnas.88.12.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang SC, Liu ES, Chan SH, et al. Mitotic regulation of protein 4.1R involves phosphorylation by cdc2 kinase. Mol Biol Cell. 2005;16:117–127. doi: 10.1091/mbc.E04-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chasis JA, Coulombel L, Conboy J, et al. Differentiation-associated switches in protein 4.1 expression: synthesis of multiple structural isoforms during normal human erythropoiesis. J Clin Invest. 1993;91:329–338. doi: 10.1172/JCI116189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baklouti F, Huang SC, Tang TK, Delaunay J, Marchesi VT, Benz EJ., Jr Asynchronous regulation of splicing events within protein 4.1 pre-mRNA during erythroid differentiation. Blood. 1996;87:3934–3941. [PubMed] [Google Scholar]
- 10.Horowitz DS, Krainer AR. Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet. 1994;10:100–106. doi: 10.1016/0168-9525(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 11.Norton PA. Alternative pre-mRNA splicing: factors involved in splice site selection. J Cell Sci. 1994;107:1–7. doi: 10.1242/jcs.107.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 13.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 14.Lopez AJ. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 15.Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayeda A, Screaton GR, Chandler SD, Fu XD, Krainer AR. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 19.Deguillien M, Huang SC, Moriniere M, Dreumont N, Benz EJ, Jr, Baklouti F. Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood. 2001;98:3809–3816. doi: 10.1182/blood.v98.13.3809. [DOI] [PubMed] [Google Scholar]
- 20.Yang G, Huang SC, Wu JY, Benz EJ., Jr An erythroid differentiation-specific splicing switch in protein 4.1R mediated by the interaction of SF2/ASF with an exonic splicing enhancer. Blood. 2005;105:2146–2153. doi: 10.1182/blood-2004-05-1757. [DOI] [PubMed] [Google Scholar]
- 21.Hou VC, Lersch R, Gee SL, et al. Decrease in hnRNP A/B expression during erythropoiesis mediates a pre-mRNA splicing switch. EMBO J. 2002;21:6195–6204. doi: 10.1093/emboj/cdf625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponthier JL, Schluepen C, Chen W, et al. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- 23.Brudno M, Gelfand MS, Spengler S, Zorn M, Dubchak I, Conboy JG. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29:2338–2348. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baraniak AP, Lasda EL, Wagner EJ, Garcia-Blanco MA. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol Cell Biol. 2003;23:9327–9337. doi: 10.1128/MCB.23.24.9327-9337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedjran F, Yeakley JM, Huh GS, Hynes RO, Rosenfeld MG. Control of alternative pre-mRNA splicing by distributed pentameric repeats. Proc Natl Acad Sci U S A. 1997;94:12343–12347. doi: 10.1073/pnas.94.23.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawamoto S. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J Biol Chem. 1996;271:17613–17616. [PubMed] [Google Scholar]
- 27.Lim LP, Sharp PA. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol Cell Biol. 1998;18:3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modafferi EF, Black DL. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paddison PJ, Cleary M, Silva JM, et al. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat Methods. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- 33.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Ding L, Sandford AJ. Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol Biol. 2005;6:4. doi: 10.1186/1471-2199-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattagajasingh SN, Huang SC, Hartenstein JS, Snyder M, Marchesi VT, Benz EJ. A nonerythroid isoform of protein 4.1R interacts with the nuclear mitotic apparatus (NuMA) protein. J Cell Biol. 1999;145:29–43. doi: 10.1083/jcb.145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman AP, Friedlich DL, Harmison G, et al. Androgens regulate the mammalian homologues of invertebrate sex determination genes tra-2 and fox-1. Biochem Biophys Res Commun. 2001;282:499–506. doi: 10.1006/bbrc.2001.4617. [DOI] [PubMed] [Google Scholar]
- 37.Norris JD, Fan D, Sherk A, McDonnell DP. A negative coregulator for the human ER. Mol Endocrinol. 2002;16:459–468. doi: 10.1210/mend.16.3.0787. [DOI] [PubMed] [Google Scholar]
- 38.Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.National Center for Biotechnology Information. [Accessed January 6, 2007]; Entrez: http://www.ncbi.nlm.nih.gov/sites/gquery.
- 40.UCSC Genome Bioinformatics. [Accessed January 6, 2007];Gene Sorter: Mouse, Assembly. 2006 Feb; http://genome.ucsc.edu/cgi-bin/hgNear.
- 41.Jin Y, Suzuki H, Maegawa S, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou HL, Baraniak AP, Lou H. A role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/CGRP alternative RNA processing. Mol Cell Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schluepen C, Lersch R, Gee SL, Conboy JG. Protein 4.1R exon 16 splicing regulation by antagonistic activities of Fox-2 and hnRNP A1 splicing factors. Blood. 2005;106 Abstract 804. [Google Scholar]