To the editor:
We read with great interest the report by Tran and colleagues in which the authors studied the induction of FOXP3 in naive human T cells.1 The authors state that, in the absence of exogenous TGF-β, only a minor subset of activated human T cells express FOXP3. In contrast, several prior reports including ours2–4 have shown FOXP3 expression on a substantial proportion of activated T cells (reviewed in Pillai and Karandikar5). The authors attribute these findings to the “nonspecific” staining of human T cells by clone PCH101 (eBioscience, San Diego, CA), used in multiple prior studies. The authors rely on clone 259D (Biolegend, San Diego, CA), claiming that this was similar to other clones such as 206D (Biolegend) and 236A/E7 (eBioscience), distinct from PCH101 (data mentioned but not demonstrated in the publication). If true, these observations would call into question many prior conclusions.
However, we believe that the authors' conclusions are based on somewhat erroneous interpretation of flow cytometric data. In Figure 3 of their publication,1 where PCH101 staining is demonstrated alongside 259D, the authors' “negative gates” were set solely on the basis of isotypes. When evaluating intracellular staining on activated cells (or sometimes even simple surface staining), isotypic antibodies may be insufficient and misleading controls.6,7 Experience shows that multiple flow cytometric antibodies can cause negative populations to significantly shift beyond isotypic cutoffs. Rather than interpreting this as “nonspecific staining,” it is important to include other controls during the validation of staining, such as antibody titrations and cells known to be negative for the marker. Use of unstimulated, FOXP3− cells would have undoubtedly resulted in a higher cutoff and their “2 peaks” of FOXP3 expression would have been correctly interpreted as negative and positive peaks.2 In fact, the authors' siRNA inhibition data demonstrated the disappearance of the positive FOXP3 peak, confirming the specificity of the “second peak.” The first peak remained intact because it was not “FOXP3+” to begin with.
In addition, basing conclusions exclusively on 259D staining may also be problematic. We have performed parallel staining for FOXP3 with multiple clones of antibodies (Figures 1, 2). In these experiments, inclusion of carboxyfluorescein diacetate succinimidyl ester (CFSE) staining allowed us to clearly differentiate stimulated and proliferating cells from those that did not enter cell cycle. Of note, even with “pan T-cell stimulation” such as anti-CD3, not all T cells undergo complete activation, nor do all cells express FOXP3 (providing excellent internal controls). However, almost all of the fully activated cells truly express FOXP3, based on carefully picked cutoffs and titrated antibodies. This is also corroborated by prior PCR analysis of sorted populations.2
Figure 1.
Clone 259D is a less sensitive detector of total FOXP3 expression. Human CD4+CD25− T cells were stained with CFSE and cultured in the presence of no antigen (top row), irradiated, allogeneic T-cell–depleted peripheral blood mononuclear cells (PBMC, middle row) or irradiated, autologous T-cell–depleted PBMC with anti-CD3. Cells were harvested and stained at various time points for CD4, CD25, and FOXP3 and analyzed on a BD LSR II (BD Biosciences, San Jose, CA), as described before2; this is similar to the authors' protocols for permeabilization (eBioscience reagents). The data represent day 6 of in vitro stimulation and demonstrate CFSE on the x-axis and phycoerythrin (PE) staining on the y-axis, with the indicated PE-conjugated antibodies (FMO indicates “fluorescence minus one”; ie, no stain in the PE channel). Thus, clones PCH101 and 236A/E7 showed similar results, with FOXP3 expression robustly detected in virtually all activated, dividing cells (panels I, K, O, Q). Nondividing cells at this late time point showed very few FOXP3+ cells. In contrast, clone 259D could not robustly detect total FOXP3 expression (panels L,R). PCH101 (used here in titrated amounts) did not show appreciable high background staining of unstimulated cells (compare panels A-C). However, clone 236A/E7 showed high background with unstimulated cells (panel E; ┈ represents cutoff with isotypic control from panel D). Thus, panel E (not D) provided the correct cutoff for the analysis of panels K and Q. Numbers on plots are percentages of total CD4+ T cells.
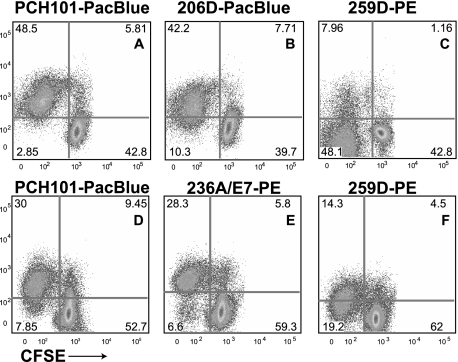
Figure 2.
Clones PCH101, 206D, and 236A/E7 show similar detection of FOXP3, with 259D being the outlier with low sensitivity. Two separate experiments (panels A-C and panels D-F, respectively) were set up similar to Figure 1. Clones PCH101-PacBlue (A,D), 206D-PacBlue (B) and 236A/E7-PE (E) showed robust FOXP3 detection on activated T cells. In contrast, clone 259D was unable to detect total expression on cells from the same culture at the same time point (C,F). Numbers on plots are percentages of total CD4+ T cells.
Clones PCH101, 206D, and 236A/E7 provide essentially similar results, distinct from those of 259D. In fact, clone 259D would miss FOXP3 expression in some cases (Figures 1L, 2C), when all other clones robustly detect such expression (Figures 1I, K, 2B). The differences may arise from the specific epitopes recognized by the antibodies. Regardless, it can be conclusively stated that 259D is the outlier among all these clones and a less sensitive detector of total FOXP3.
Our data reaffirm the validity of previous results using PCH101, provided the correct cutoffs were used. Clearly, CD4+CD25−FOXP3− T cells that undergo full activation express robust amounts of FOXP3. Importantly, these data expose an important pitfall of relying solely on isotypic controls in flow cytometric evaluation, which should help other investigators in their evaluation of this and other markers.
Footnotes
This work was supported by grants (to N.J.K.) from the National Institutes of Health and National Multiple Sclerosis Society (NMSS). V.P. is a postdoctoral fellow of the National Multiple Sclerosis Society and N.J.K. is a Harry Weaver Neuroscience Scholar of the National Multiple Sclerosis Society.
Authorship
Contribution: V.P. designed and performed experiments and wrote the manuscript. N.J.K. designed experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nitin Karandikar, MD, PhD, UT Southwestern Medical Center, Department of Pathology, 6000 Harry Hines Blvd., Dallas, TX 75390-9072; e-mail: nitin.karandikar@utsouthwestern.edu.
References
- 1.Tran DQ, Ramsey H, Shevach EM. Induction of FOXP3 expression in naive human CD4+FOXP3− T cells by T-cell receptor stimulation is TGF{beta}-dependent but does not confer a regulatory phenotype. Blood. 2007;110:2983–2990. doi: 10.1182/blood-2007-06-094656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pillai V, Ortega SB, Wang CK, Karandikar NJ. Transient regulatory T-cells: a state attained by all activated human T-cells. Clin Immunol. 2007;123:18–29. doi: 10.1016/j.clim.2006.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4(+) T cells. Eur J Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- 4.Allan SE, Crome SQ, Crellin NK, et al. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int Immunol. 2007;19:345–354. doi: 10.1093/intimm/dxm014. [DOI] [PubMed] [Google Scholar]
- 5.Pillai V, Karandikar NJ. Human regulatory T cells: a unique, stable thymic subset or a reversible peripheral state of differentiation? Immunol Lett. 2007;114:9–15. doi: 10.1016/j.imlet.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 7.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]