Abstract
Endothelial cells secrete prothrombotic ultralarge von Willebrand factor (VWF) multimers, and the metalloprotease ADAMTS13 cleaves them into smaller, less dangerous multimers. This reaction is stimulated by tensile force applied to the VWF substrate, which may occur on cell surfaces or in the circulating blood. The cleavage of soluble VWF by ADAMTS13 was accelerated dramatically by a combination of platelets and fluid shear stress applied in a cone-plate viscometer. Platelet-dependent cleavage of VWF was blocked by an anti-GPIbα monoclonal antibody or by a recombinant soluble fragment of GPIbα that prevents platelet-VWF binding. Multimeric gel analysis showed that shear and platelet-dependent cleavage consumed large VWF multimers. Therefore, ADAMTS13 preferentially acts on platelet-VWF complexes under fluid shear stress. This reaction is likely to account for a majority of VWF proteolysis after secretion and to determine the steady-state size distribution of circulating VWF multimers in vivo.
Introduction
von Willebrand factor (VWF) is a multimeric plasma glycoprotein that plays an essential role in tethering platelets at the site of vascular injury. VWF is synthesized in endothelial cells, where a portion is stored in granules called Weibel-Palade bodies as “unusually large” or “ultralarge” multimers (ULVWF) and secreted upon endothelial stimulation.1,2 Secreted ULVWF multimers bind platelets with relatively high affinity and are thought to be prothrombotic. ULVWF is cleaved into smaller and less dangerous multimers by the metalloprotease ADAMTS13, a member of the A Disintegrin And Metalloprotease with ThromboSpondin type I repeat family.3–5 Inherited or acquired deficiency of ADAMTS13 causes life-threatening microvascular thrombosis that is characteristic of thrombotic thrombocytopenic purpura.4,6,7 Conversely, mutations in von Willebrand disease type 2A cause bleeding by increasing the cleavage of VWF by ADAMTS13 and impairing platelet adhesion.8–10 Therefore, normal hemostasis depends on the precise regulation of VWF proteolysis.
ADAMTS13 cleaves the Tyr1605-Met1606 bond in the A2 domain of VWF, but this bond is buried and relatively inaccessible until the A2 domain is unfolded, presumably by tensile force in vivo.10,11 The shear stress required to apply this force will vary depending on whether VWF is immobilized at the vessel wall or moving with the flowing blood, and whether platelets are bound to it. The rate of VWF cleavage also can be modulated by cofactors that bind to the A1 domain, including platelet GPIb and heparin.12 Thus, ADAMTS13 is presented with VWF multimers in plasma or on endothelial cell surfaces that vary in their susceptibility to cleavage, with or without attached platelets.
The relevance of each of these potential substrates to the catabolism of VWF is unknown. Several studies suggest that VWF strings on endothelial cells must be cleaved to inhibit thrombus growth,13,14 but the role of proteolysis in the fluid phase has not been established. Therefore, the cleavage of VWF by ADAMTS13 was assessed in a cone-plate viscometer to minimize the contribution of surface interactions. The results indicate that proteolysis of fluid phase VWF-platelet complexes is likely to determine the steady state size distribution of circulating VWF multimers in vivo.
Methods
Recombinant ADAMTS13
Full-length human ADAMTS13 with a C-terminal V5 tag was expressed in TRex 293 cells (Invitrogen) as described previously15 and partially purified by anion exchange chromatography. In brief, conditioned medium containing recombinant ADAMTS13 was supplemented with proteinase inhibitors (0.1 μmol/L d-Phe-Pro-Arg-chloromethane and 144 μmol/L phenylmethylsulfonyl fluoride and applied to tandem columns of HiTrap Q Sepharose (2 × 5 mL; GE Healthcare, Chalfont St Giles, United Kingdom). The columns were washed with 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, and developed with a linear gradient of 0 to 50 mM CaCl2 in 20 mM Tris-HCl, pH 8.0, and 100 mM NaCl. Fractions containing ADAMTS13 were combined, dialyzed against 20 mM Tris-HCl, pH 8.0, and 100 mM NaCl, and concentrated by ultrafiltration (YM100; Millipore, Billerica, MA). Protein concentration was determined by bicinchoninic acid assay (Pierce, Rockford, IL). ADAMTS13 antigen concentration was determined with an IMUBIND ADAMTS13 enzyme-linked immunosorbent assay (ELISA) Kit (American Diagnostica, Greenwich, CT), and by ELISA using monoclonal anti-human ADAMTS13 antibody 2G3 provided by Hans Deckmyn (Catholic University of Leuven, Leuven, Belgium)16 for coating and horseradish peroxidase-conjugated anti-V5 antibody (Invitrogen, Carlsbad, CA) for detection. ADAMTS13 activity was assayed based on the cleavage of substrate FRETS-VWF73 (Peptides International, Louisville, KY).17 The recombinant ADAMTS13 stock solution contained approximately 250 units/mL, whereas the concentration of active ADAMTS13 in normal pooled plasma is defined as 1 unit/mL.
Platelets and plasma
Lyophilized formalin-fixed human platelets (Helena Laboratories, Beaumont, TX) were reconstituted in 20 mM Tris-HCl, pH 7.4, and 100 mM NaCl and centrifuged at 6400g for 5 minutes. The pellet was resuspended in the same buffer to a platelet concentration of 2 × 107/μL as determined by counting in a phase hemacytometer.
Washed fresh human platelets were prepared as described previously18 from blood anticoagulated with 4.4 mM K2-ethylenediaminetetraacetic acid (K2EDTA) and 10 ng/mL prostaglandin I2 (Cayman Chemical, Ann Arbor, MI).19 The washed platelets were gently resuspended in platelet wash buffer (6.6 g/L NaCl, 1 g/L dextrose, 0.747 g/L K2HPO4, 1.15 g/L Na2HPO4 · 7H2O, 3.37 g/L NaH2PO4 · H2O, plus 10 ng/mL prostaglandin I2) to a concentration of 2 × 107/μL and used within 24 hours.
Platelet-rich plasma was prepared from freshly drawn venous blood anticoagulated with 14.4 U/mL heparin, 10 ng/mL prostaglandin I2, and 10 μmol/L d-Phe-Pro-Arg-chloromethane by centrifugation at 480g for 10 minutes. The platelet-rich plasma supernatant was centrifuged at 6400g for 5 minutes to prepare platelet-poor plasma.
Shear-induced proteolytic cleavage of VWF
Reactions were performed in a programmable cone-plate viscometer (HAAKE RheoStress 1; Thermo Fisher Scientific, Waltham, MA) in a total volume of 50 μL (with a 20-mm diameter cone, 0.5° cone angle) or 500 μL (with a 60-mm diameter cone, 0.5° cone angle) of reaction buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 144 μmol/L phenylmethylsulfonyl fluoride, 10 μmol/L d-Phe-Pro-Arg-chloromethane, 0.1 μmol/L ZnCl2, 5 mM CaCl2, and 3 mg/mL bovine serum albumin [BSA]), with various amounts of purified plasma VWF (Haematologic Technologies, Essex Junction, VT), platelets, ADAMTS13, and other components as indicated in the article or figure legends. Complete reaction mixtures were preincubated at 37°C for 10 minutes and then subjected to a constant fluid shear stress for various times. In some experiments, platelets were preincubated for 30 minutes with 30 μg/mL of monoclonal anti-human GPIbα antibody 6D120 provided by Dr Barry Coller (Rockefeller University, New York, NY), a recombinant chimeric protein consisting of human GPIbα residues 1-290 fused to the Fc region of human IgG1 (GPIbα-Ig/2V),21 or mouse IgG1 (BioLegend, San Diego, CA). Reactions were stopped by mixing 15-μL samples with an equal volume of Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA). Platelets were obtained for these studies under a protocol approved by the Washington University institutional review board. Informed consent was obtained in accordance with the Declaration of Helsinki.
A standard cleavage product was prepared in a 500-μL reaction containing 2 × 106 platelets/μL, 30 μg/mL VWF, and 5 U/mL ADAMTS13. The mixture was subjected to 50 dyne/cm2 shear stress for 30 minutes, mixed with 500 μL of Laemmli sample buffer, and stored in aliquots of 20 μL at −70°C. For use, an aliquot was mixed with 180 μL of Laemmli sample buffer containing 3 mg/mL BSA and processed in parallel with experimental reactions. A sample of the diluted standard solution (7.5 μL) was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) in a reference lane for each gel.
Western blotting
Reaction products in Laemmli sample buffer were heated at 90°C for 5 minutes with occasional vortexing and centrifuged at 6400g for 5 minutes. The supernatants were subjected to 4% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Invitrogen). The membranes were incubated (1 hour at room temperature, or overnight at 4°C) in 20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.1% Tween 20 (TBST) containing 0.5% casein, and then incubated (1 hour at room temperature, or overnight at 4°C) with a 1:2500 dilution of horseradish peroxidase-conjugated rabbit anti-VWF antibody (P0226, Dako, Carpinteria, CA) in TBST. Membranes were washed twice for 15 minutes in TBST and developed with the ECL plus Western blotting detection system (GE Healthcare, NJ).
Image analysis
Membranes were exposed directly to film and scanned films were analyzed using NIH ImageJ (rsb.inf.nih.gov/ij). Membranes also were scanned with a Storm or Typhoon scanner (GE Healthcare) for chemifluorescence detection and analysis using ImageQuant software (version 5.1 or TL, GE Healthcare). The intensities of homodimeric 350-kDA product bands7,11 were normalized to the intensity of the standard cleavage product, which was included on each gel.
Multimer gels
Samples were removed from reactions and mixed with an equal volume of 50 mM EDTA. VWF multimers were analyzed as described previously22 by 1.5% SDS-agarose gel electrophoresis and transfer to PVDF membranes. Multimers were visualized with horseradish peroxidase-conjugated rabbit anti-VWF antibody and the ECL chemiluminescence system (GE Healthcare).
Results
VWF multimers encounter many distinct environments that are likely to differ in their importance for the proteolytic remodeling of VWF by ADAMTS13. Upon secretion by endothelial cells, VWF multimers can remain bound to the cell surface or detach and circulate in the blood. Cell-associated VWF binds platelets with high affinity and is cleaved rapidly by ADAMTS13, suggesting that cell surfaces are an important site for VWF proteolysis.13 Although VWF multimers in the blood are relatively resistant to ADAMTS13 under static conditions they are cleaved readily at high levels of fluid shear stress that occur in the microvasculature,23 suggesting that fluid phase cleavage of VWF also may be significant in vivo. In addition, shear stress promotes VWF-dependent platelet aggregation,24 which could influence the recognition of VWF by ADAMTS13. Therefore, the dependence of VWF cleavage in solution on shear stress and platelet count was investigated in a cone-plate viscometer, to minimize the contribution of wall shear stress and focus on fluid phase interactions between VWF and platelets. VWF cleavage by ADAMTS13 was assessed as described previously,7,11 using SDS-PAGE and Western blotting to detect a homodimeric 350 kDa product composed of C-terminal fragments of the VWF subunit.
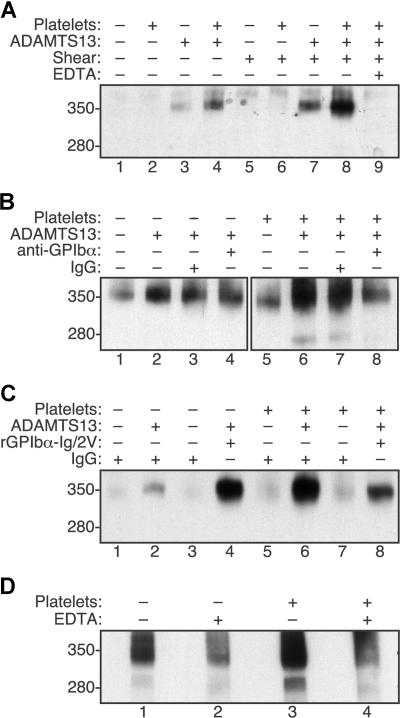
VWF was cleaved very slowly by ADAMTS13 under static conditions in the absence of platelets (Figure 1A lane 3). Cleavage was increased approximately 2-fold by adding platelets (Figure 1A lane 4), which is consistent with the previously described stimulation of ADAMTS13 activity upon binding of a high affinity variant of GPIbα to recombinant VWF substrates12 even though relatively little VWF-platelet binding occurs in the absence of shear stress.25–27 Fluid shear stress alone increased the cleavage of VWF approximately 3-fold (Figure 1A lane 7), whereas the combination of shear stress and platelets increased the cleavage of VWF approximately 12-fold (Figure 1A lane 8). As expected, the addition of EDTA prevented cleavage (Figure 1A lane 9). Thus, platelet-VWF complexes subjected to shear stress appear to be good substrates for ADAMTS13.
Figure 1.
Fluid shear stress and platelet-VWF interactions increase VWF cleavage by ADAMTS13. Reactions containing VWF (30 μg/mL) were treated with (+) or without (−) shear stress and other components as indicated. Samples were analyzed by SDS-PAGE and Western blotting with polyclonal anti-VWF to detect the 350-kDa homodimeric cleavage product containing C-terminal fragments of VWF. The corresponding 280-kDa N-terminal homodimeric product is recognized less efficiently by anti-VWF and was visible in some reactions. (A) Cleavage of VWF by ADAMTS13 (5 U/mL) was increased slightly by fluid shear stress (50 dyne/cm2 for 10 minutes, lane 7) or formalin-fixed platelets (106/μL, lane 4). Maximal cleavage required both shear stress and platelets (lane 8) and was blocked by 5 mM EDTA (lane 9). (B) Monoclonal antibody 6D1 against GPIbα (30 μg/mL) blocked the cleavage of VWF exposed to fluid shear stress (50 dyne/cm2 for 10 minutes) in reactions containing platelets (lane 8) but not in reactions without platelets (lane 4). Control mouse IgG1 had no effect (lanes 3 and 7). (C) Recombinant GPIbα-Ig/2V (30 μg/mL) increased the cleavage of VWF by ADAMTS13 (2.5 U/mL) in reactions exposed to fluid shear stress (16 dyne/cm2 for 5 minutes) without platelets (lane 4) and partially inhibited the cleavage of VWF in reactions with platelets (lane 8). (D) The cleavage of endogenous VWF in fresh platelet-rich plasma subjected to fluid shear stress (20 dyne/cm2 for 5 minutes) was markedly greater than the cleavage of VWF in platelet-poor plasma. Cleavage was prevented by 50 mM EDTA.
The role of direct VWF-platelet interactions was demonstrated by 2 approaches. Monoclonal anti-GPIbα antibody 6D1 prevents the binding of VWF to platelet GPIbα,20 and pretreatment of platelets with antibody 6D1 also blocked the shear stress-induced cleavage of VWF (Figure 1B lane 8 vs lane 7). Antibody 6D1 had no effect on shear stress-induced cleavage of VWF when platelets were omitted (Figure 1B lane 4 vs lane 3).
Similar results were obtained using a recombinant fragment of GPIbα fused to the Fc region of human IgG1 (GPIbα-Ig/2V).21 This construct contains 2 mutations that cause platelet-type or pseudo-von Willebrand disease, G233V and M239V.28,29 These mutations increase the affinity of binding to VWF, and GPIbα-Ig/2V binds immobilized VWF with an IC50 of 0.5 μg/mL.21 At the concentrations of VWF and GPIbα-Ig/2V used in these experiments (30 μg/mL each), almost all GPIbα-Ig/2V binding sites on VWF will be occupied. In the absence of platelets, GPIbα-Ig/2V substantially increased the cleavage of VWF (Figure 1C lane 4 vs lane 2), probably by binding to VWF domain A1 and enhancing the recognition of the adjacent A2 domain by ADAMTS13 as reported for a similar construct.12 The addition of platelets markedly increased the cleavage of VWF (Figure 1C lane 6) and this increase was partially inhibited by GPIbα-Ig/2V (Figure 1C lane 8). These results suggest that GPIbα-Ig/2V is both a cofactor for ADAMTS13 because it binds VWF domain A1,12 and an antagonist of shear-induced VWF cleavage because it inhibits VWF binding to platelets.21
These results demonstrate that platelets increase the shear-dependent cleavage of VWF by ADAMTS13 using purified proteins. To establish that a similar effect occurs under more physiologic conditions, similar studies were performed in platelet-rich and platelet-poor plasma anticoagulated with heparin, prostacyclin and d-Phe-Pro-Arg-chloromethane (Figure 1D). Under these conditions, a normal platelet count markedly increased the shear-dependent cleavage of endogenous plasma VWF by endogenous ADAMTS13.
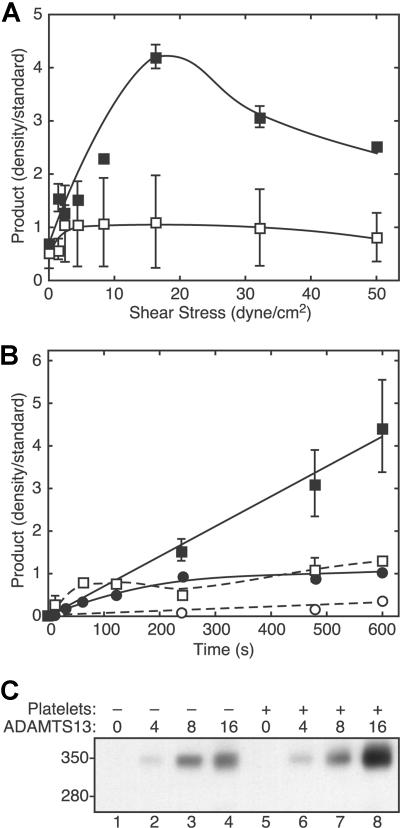
Platelet-dependent cleavage of VWF depended strongly on the level of fluid shear stress (Figure 2A) and exhibited a maximum between 10 and 30 dyne/cm2. In the absence of platelets, the extent of VWF cleavage varied much less as a function of shear stress. With both platelets and shear stress, the extent of VWF cleavage increased approximately linearly with time (Figure 2B), indicating that ADAMTS13 activity was stable during the time course of the experiments. Compared with the very slow rate of product formation without platelets or shear stress, the addition of platelets or exposure to fluid shear stress modestly increased the cleavage of VWF. The extent of VWF cleavage also was proportional to ADAMTS13 concentration (Figure 2C).
Figure 2.
VWF cleavage depends on shear stress, time, platelets, and ADAMTS13. (A) Reactions containing VWF (30 μg/mL) and ADAMTS13 (2.5 U/mL), without platelets (□) or with platelets (1 × 106/μL) (■), were subjected to different levels of fluid shear stress for 3 minutes and analyzed for VWF cleavage by SDS-PAGE and Western blotting with polyclonal anti-VWF. Error bars indicate SD (n = 5, ■) or the range of duplicates (□). (B) Reactions containing VWF (30 μg/mL) and ADAMTS13 (2.5 U/mL), without (●, ○) or with formalin-fixed platelets (106/μL) (■, □) were performed without (□, ○) or with (■, ●) 50 dyne/cm2 shear stress for different times and analyzed for VWF cleavage. ■, 50 dyne/cm2 shear stress and 106 platelets/μL (n = 5); ●, 50 dyne/cm2 and no platelets (n = 2); □, no shear stress and 106 platelets/μL (n = 2); ○, no shear stress and no platelets (n = 1). Error bars indicate SD (■) or the range of duplicates (●, ○). (C) Reactions containing the indicated concentration of ADAMTS13 (U/mL) with or without platelets (106/μL) were exposed to fluid shear stress (16 dyne/cm2 for 5 minutes) and analyzed for VWF cleavage by SDS-PAGE and Western blotting as described in the legend to Figure 1.
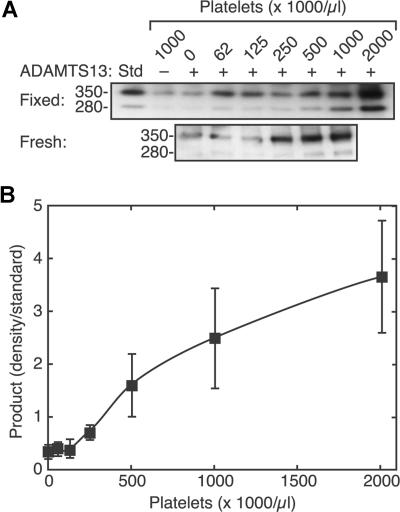
The cleavage of VWF increased markedly with increasing platelet count (Figure 3). Similar results were obtained with formalin fixed platelets and with fresh washed platelets treated with EDTA and prostacyclin to prevent activation and aggregation. Because of the intrinsic variability of the Western blotting assay method, many replicates were required to obtain satisfactory results at low signal intensities associated with lower platelet counts. The results shown represent the average of 9 independent experiments. The maximum VWF cleavage induced at the highest platelet counts varied several-fold among the replicates, which accounts for the relatively large standard errors on the data points at higher platelet counts. A steep increase in VWF cleavage by ADAMTS13 was observed over a physiologically relevant range of platelet counts, between 125 000/μL and 500 000/μL, with little effect at lower counts. This sigmoidal pattern can be fit to a standard binding equation with a Hill coefficient of approximately 2, suggesting that optimal cleavage requires at least 2 platelets to bind simultaneously to a single VWF multimer.
Figure 3.
Sigmoidal dependence of VWF cleavage on platelet count. Reactions containing VWF (30 μg/mL), without (−) or with (+) ADAMTS13 (2.5 U/mL), and various concentrations of platelets were exposed to fluid shear stress (50 dyne/cm2, 3 minutes) and analyzed by SDS-PAGE and Western blotting. (A) Examples of platelet dose response results with formalin-fixed platelets (top) and fresh washed platelets (bottom). (B) Data with fixed platelets were combined by normalizing the density of the 350-kDa product band to the density of a cleavage product standard (Std). Error bars indicate SE (n = 9).
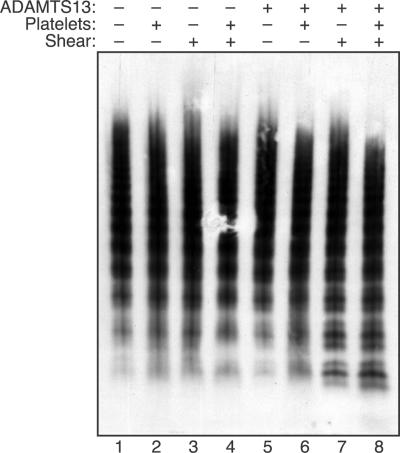
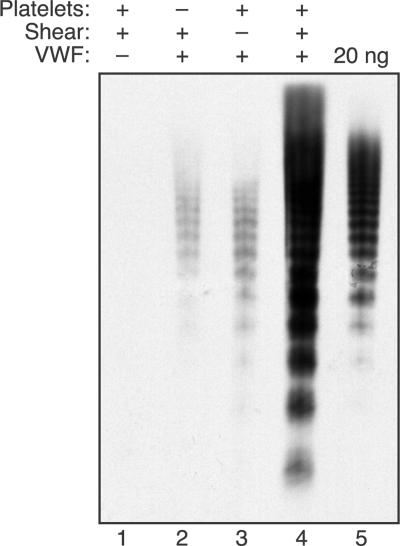
ULVWF multimers bind to platelets more tightly than smaller multimers.30 In addition, the likelihood that a multimer will bind several platelets should increase with multimer length, which correlates with the number of potential platelet binding sites. Therefore, platelets may bind to and promote the cleavage of larger multimers, and the results of multimer gel electrophoresis are consistent with this expectation (Figure 4). When plasma VWF multimers were exposed to fluid shear stress, no change in multimer distribution was observed if either platelets or ADAMTS13 was omitted. A recent study described an increase in VWF multimer size upon exposure of concentrated VWF (0.1 mg/mL) to approximately 65 dyne/cm2 shear stress.31 In the present studies, this effect may have been prevented by the use of a lower VWF concentration (30 μg/mL), lower shear stress (16 dyne/cm2), and the presence of BSA (3 mg/mL). When both ADAMTS13 and platelets were included, larger VWF multimers disappeared and smaller multimers increased. So-called satellite bands flanking the smaller multimers are caused by proteolysis, and these bands increased markedly when VWF was subjected to shear stress with both platelets and ADAMTS13 present.
Figure 4.
Platelets increase the cleavage of large VWF multimers under fluid shear stress. Reactions containing VWF (30 μg/mL) were performed as indicated with (+) or without (−) ADAMTS13 (2.5 U/mL), platelets (2 × 106/μL), and fluid shear stress (16 dyne/cm2, 10 minutes). The distribution of VWF multimers was assessed by 1.5% SDS-agarose gel electrophoresis.
Shear-induced binding of VWF multimers to platelets was demonstrated directly by analyzing the VWF eluted from platelets after exposure to fluid shear stress (Figure 5). No VWF was detected in reactions containing only formalin-fixed platelets, indicating that little or no VWF can be extracted from them. When plasma VWF was included, a small amount of VWF was sedimented in reactions that were subjected to shear stress in the absence of platelets (Figure 5 lane 2). In reactions prepared with platelets but not subjected to shear stress (Figure 5 lane 3), trace amounts of relatively small VWF multimers sedimented with the platelets. In contrast, combining shear stress with platelets markedly increased the recovery of larger VWF multimers in the platelet pellet (Figure 5 lane 4). Comparison with a VWF standard on the same gel suggests that fluid shear stress caused approximately 5% to 10% of the input VWF to bind to platelets. Thus, fluid shear stress induces the binding of large VWF multimers to platelets, and platelet-VWF complexes appear to be preferred substrates for ADAMTS13.
Figure 5.
Shear stress induces large VWF multimers to bind platelets. Reactions were performed as indicated with (+) or without (−) VWF (30 μg/mL), formalin-fixed platelets (106/μL), and fluid shear stress (16 dyne/cm2, 10 minutes). Samples were diluted with an equal volume of reaction buffer and centrifuged for 5 minutes at 6400g. The platelet pellets (or any other pelleted material for reactions without platelets) were washed 3 times by resuspension in reaction buffer and centrifugation. The washed platelets or other contents were dispersed in Laemmli sample buffer, heated at 80°C for 5 minutes, and centrifuged, and supernatant corresponding to 20 μL of the initial reaction volume was analyzed by Western blotting after 1.5% SDS-agarose gel electrophoresis. The 20 ng of input VWF analyzed for reference corresponds to approximately 3% of the VWF initially present in the samples of reactions analyzed in lanes 1 to 4.
Discussion
In the absence of fluid shear stress, VWF binds very weakly to platelet GPIbα with a Kd of 3 to 5 μmol/L.25–27 Therefore, at reference blood concentrations of VWF (∼10 μg/mL, ∼250 000 Da/subunit, ∼40 nM A1 domains) and platelets (∼250 000 platelets/μL, ∼28 000 GPIbα/platelet,26 ∼12 nM GPIbα), only aproximately 0.3% of VWF subunits will be bound to a platelet. Such low occupancy is consistent with the common observation that VWF-platelet binding is usually undetectable in blood under static conditions. This small amount of platelet binding slightly increases the cleavage of VWF by ADAMTS13 (Figures 1A and 2), probably by increasing the accessibility of VWF domain A2.12 A variant of GPIbα that binds VWF more tightly causes a correspondingly larger increase in the cleavage of VWF (Figure 1C).
By itself, shear stress also does not account for the cleavage of VWF by ADAMTS13 in vivo, because physiologic levels of fluid shear stress can exert little force on VWF in solution. For example, at 20 dyne/cm2, a shear stress characteristic of the microvasculature, the peak forces on fluid phase VWF are in the range of 0.2 to 0.8 pN.32 These values are less than the force needed to break a typical hydrogen bond, which suggests that fluid shear stress should have a very small effect on the tertiary structure of VWF. Nevertheless, weak forces in this range are sufficient to cause large scale changes in the conformation of VWF multimers.33 Under static conditions, VWF multimers adopt a collapsed, globular conformation that appears to depend on weak interactions among the subunits. Above a threshold shear force, the multimer unfurls and extends reversibly to expose binding sites for collagen and platelets33 and also to increase the accessibility of VWF domain A2 to ADAMTS13. These considerations are consistent with the modest increase in VWF cleavage by ADAMTS13 as a function of increasing fluid shear stress, even in the absence of platelets (Figure 2).
Although each alone has a relatively small effect, the combination of shear stress and platelets dramatically increases the cleavage of VWF (Figure 2), and this synergism can be explained by the effect of platelets on the force experienced by VWF. Binding to the surface of a single platelet should not significantly increase the tension on VWF,32 but the peak force on 2 platelets held together by VWF is predicted to exceed 390 pN at a shear stress of 20 dyne/cm2.32 This force is enough to unfold many structural proteins and probably unfolds VWF domain A2, facilitating the cleavage of its Tyr1605-Met1606 bond by ADAMTS13.
The dependence of VWF cleavage on shear stress exhibits a maximum between 10 and 30 dyne/cm2 (Figure 2A), and the decline at higher shear stress may have several explanations. Increasing shear rate will increase the frequency of platelet-VWF collisions, which should increase the rate of cleavage as well. However, this effect will be limited because increased shear stress applies more force to the VWF-GPIbα bond, which will accelerate dissociation from the platelet surface34 and reduce the time during which a VWF multimer is stretched and exposed for cleavage by ADAMTS13. Alternatively, ADAMTS13 may prefer a partially unfolded substrate, and the application of increasing force may overextend the VWF A2 domain so that ADAMTS13 cannot recognize it efficiently. This mechanism would be consistent with the finding that pretreatment with 1.25 mol/L guanidine-HCl increases the susceptibility of VWF to ADAMTS13, but a higher concentration causes VWF to become resistant.9,15
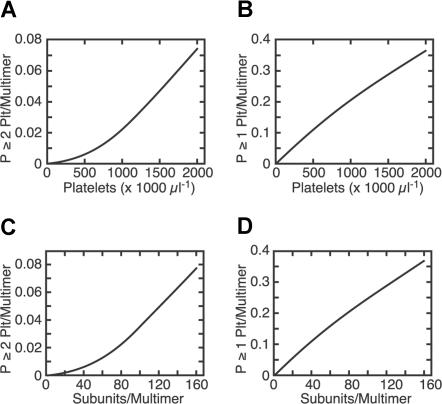
A requirement to bind 2 platelets per multimer implies that cleavage by ADAMTS13 should increase exponentially rather than linearly with platelet count (Figure 6), and it does (Figure 3). Shear-dependent binding of VWF to platelets35 and shear-induced platelet aggregation36 also show a similar nonlinear dependence on platelet count, indicating that these processes require multiple platelets to interact with a single VWF multimer.35
Figure 6.
Binding of platelets to VWF modeled as a function of platelet count and multimer length. The fractional occupancy of platelet binding sites on VWF multimers was calculated from the Kd for platelet GPIbα-A1 domain binding (∼4 μmol/L),25–27 plasma VWF concentration (∼10 μg/mL), VWF subunit mass (∼250 kDa), and number of GPIbα per platelet (∼28 000).26 The probability that a multimer has bound a particular number x of platelets is given by the binomial formula for P(x,n), where n is the number of VWF A1 domains (or subunits) per multimer and y is the fractional occupancy of VWF domain A1 sites by GPIbα ([GPIbα-A1]/[A1]total): P(0,n) = (1 − y)n and P(1,n) = ny(1 − y)n−1. Therefore, the probability of having at least 1 platelet bound per multimer is 1 − (1 − y)n, and the probability of finding at least 2 platelets bound per multimer is 1 − {(1 − y)n + ny(1 − y)n−1}. At a fixed multimer length (n = 20 subunits), P is greater than or equal to 2 platelets/multimer increases exponentially with increasing platelet count (A), whereas P is greater than or equal to 1 platelet/multimer increases approximately linearly with increasing platelet count (B). At a fixed platelet count (250 000/μL), P is greater than or equal to 2 platelets/multimer increases exponentially with increasing multimer length (C), whereas P is greater than or equal to 1 platelet/multimer increases approximately linearly with increasing multimer length (D).
The probability that at least 2 platelets will bind to a multimer is also expected to increase exponentially with multimer length (Figure 6). This prediction is consistent with the finding that shear-induced, platelet dependent cleavage of VWF consumes large multimers (Figure 4) and that shear-induced binding of VWF to platelets preferentially removes the largest multimers (Figure 5). An exponential relationship between multimer length and cleavage by ADAMTS13 can account for the selective proteolysis of large VWF multimers associated with thrombocytosis37,38 or with increased shear stress caused by aortic stenosis.39 Conversely, small multimers are unlikely to bind 2 platelets and therefore rarely become stretched enough to present a susceptible cleavage site for ADAMTS13. Therefore, this relationship also can explain the striking lack of VWF proteolysis associated with von Willebrand disease variants in which only small multimers are secreted.40
Platelets and fluid shear stress cooperate to promote VWF cleavage at several stages in the life history of a VWF multimer. Immediately after secretion, some ULVWF forms long strings that bind to the endothelial cell surface.13 In flowing blood, platelets bind and transmit force to the cell-associated ULVWF strings, which results in prompt cleavage by ADAMTS13 and release of VWF into the circulation.14 A similar mechanism prevents the microvascular thrombosis that characterizes thrombotic thrombocytopenic purpura. However, substantial proteolysis of VWF multimers also occurs in the fluid phase of circulating blood. For example, administration of DDAVP induces an acute 3-fold increase in plasma VWF level that includes a substantial amount of ULVWF. The ULVWF multimers disappear within 2 to 4 hours, accompanied by increased VWF subunit proteolysis. In contrast, the plasma concentration of VWF returns to baseline over a longer time course of 24 hours.41 The rapid evolution of VWF multimers after DDAVP, with disappearance of “ultralarge” species over 2 to 4 hours, indicates that the shear-induced cleavage of VWF in solution is a major pathway for the catabolism of VWF in vivo. This process is sensitive to changes in platelet count, to the size distribution of VWF multimers, and to normal or pathologic variations in fluid shear stress.
Acknowledgments
We thank Professor Barry Coller (Rockefeller University, New York, NY) for monoclonal anti-GPIbα antibody 6D1 and Professor Hans Deckmyn (Catholic University, Leuven, Belgium) for monoclonal anti-ADAMTS13 antibodies.
This work was supported in part by National Institutes of Health grant HL72917.
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.S. designed and performed research, analyzed and interpreted data, and wrote the manuscript. P.J.A. performed research, contributed vital new reagents, and analyzed and interpreted data. E.A.T. performed research. E.W. contributed vital new reagents. J.E.S. designed research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: J.E.S. is a consultant for Baxter BioSciences. E.W. is an employee of Wyeth Biotech. The remaining authors declare no competing financial interests.
Correspondence: J. Evan Sadler, MD, PhD, Howard Hughes Medical Institute, Washington University School of Medicine, 660 S Euclid Ave, Box 8022, St Louis, MO 63110; e-mail: esadler@im.wustl.edu.
References
- 1.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 2.Ruggeri ZM. Von Willebrand factor, platelets and endothelial cell interactions. J Thromb Haemost. 2003;1:1335–1342. doi: 10.1046/j.1538-7836.2003.00260.x. [DOI] [PubMed] [Google Scholar]
- 3.Zheng ×, Chung D, Takayama TK, Majerus EM, Sadler JE, Fujikawa K. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J Biol Chem. 2001;276:41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 4.Levy GG, Nichols WC, Lian EC, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413:488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 5.Soejima K, Mimura N, Hirashima M, et al. A novel human metalloprotease synthesized in the liver and secreted into the blood: possibly, the von Willebrand factor-cleaving protease? J Biochem. 2001;130:475–480. doi: 10.1093/oxfordjournals.jbchem.a003009. [DOI] [PubMed] [Google Scholar]
- 6.Furlan M, Robles R, Galbusera M, et al. von Willebrand factor-cleaving protease in thrombotic thrombocytopenic purpura and the hemolytic-uremic syndrome. N Engl J Med. 1998;339:1578–1584. doi: 10.1056/NEJM199811263392202. [DOI] [PubMed] [Google Scholar]
- 7.Tsai HM, Lian EC. Antibodies to von Willebrand factor-cleaving protease in acute thrombotic thrombocytopenic purpura. N Engl J Med. 1998;339:1585–1594. doi: 10.1056/NEJM199811263392203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dent JA, Berkowitz SD, Ware J, Kasper CK, Ruggeri ZM. Identification of a cleavage site directing the immunochemical detection of molecular abnormalities in type IIA von Willebrand factor. Proc Natl Acad Sci U S A. 1990;87:6306–6310. doi: 10.1073/pnas.87.16.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai HM, Sussman II, Ginsburg D, Lankhof H, Sixma JJ, Nagel RL. Proteolytic cleavage of recombinant type 2A von Willebrand factor mutants R834W and R834Q: inhibition by doxycycline and by monoclonal antibody VP-1. Blood. 1997;89:1954–1962. [PubMed] [Google Scholar]
- 10.Sutherland JJ, O'Brien LA, Lillicrap D, Weaver DF. Molecular modeling of the von Willebrand factor A2 Domain and the effects of associated type 2A von Willebrand disease mutations. J Mol Model. 2004;10:259–270. doi: 10.1007/s00894-004-0194-9. [DOI] [PubMed] [Google Scholar]
- 11.Tsai H-M. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87:4235–4244. [PubMed] [Google Scholar]
- 12.Nishio K, Anderson PJ, Zheng XL, Sadler JE. Binding of platelet glycoprotein Ibα to von Willebrand factor domain A1 stimulates the cleavage of the adjacent domain A2 by ADAMTS13. Proc Natl Acad Sci U S A. 2004;101:10578–10583. doi: 10.1073/pnas.0402041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong JF, Moake JL, Nolasco L, et al. ADAMTS-13 rapidly cleaves newly secreted ultra-large von Willebrand factor multimers on the endothelial surface under flowing conditions. Blood. 2002;100:4033–4039. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 14.Chauhan AK, Motto DG, Lamb CB, et al. Systemic antithrombotic effects of ADAMTS13. J Exp Med. 2006;203:767–776. doi: 10.1084/jem.20051732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson PJ, Kokame K, Sadler JE. Zinc and calcium ions cooperatively modulate ADAMTS13 activity. J Biol Chem. 2006;281:850–857. doi: 10.1074/jbc.M504540200. [DOI] [PubMed] [Google Scholar]
- 16.Feys HB, Liu F, Dong N, et al. ADAMTS-13 plasma level determination uncovers antigen absence in acquired thrombotic thrombocytopenic purpura and ethnic differences. J Thromb Haemost. 2006;4:955–962. doi: 10.1111/j.1538-7836.2006.01833.x. [DOI] [PubMed] [Google Scholar]
- 17.Kokame K, Nobe Y, Kokubo Y, Okayama A, Miyata T. FRETS-VWF73, a first fluorogenic substrate for ADAMTS13 assay. Br J Haematol. 2005;129:93–100. doi: 10.1111/j.1365-2141.2005.05420.x. [DOI] [PubMed] [Google Scholar]
- 18.Baenziger NL, Majerus PW. Isolation of human platelets and platelet surface membranes. Methods Enzymol. 1974;31:149–155. doi: 10.1016/0076-6879(74)31015-4. [DOI] [PubMed] [Google Scholar]
- 19.Wu YP, van Breugel HH, Lankhof H, et al. Platelet adhesion to multimeric and dimeric von Willebrand factor and to collagen type III preincubated with von Willebrand factor. Arterioscler Thromb Vasc Biol. 1996;16:611–620. doi: 10.1161/01.atv.16.5.611. [DOI] [PubMed] [Google Scholar]
- 20.Coller BS, Peerschke EI, Scudder LE, Sullivan CA. Studies with a murine monoclonal antibody that abolishes ristocetin-induced binding of von Willebrand factor to platelets: additional evidence in support of GPIb as a platelet receptor for von Willebrand factor. Blood. 1983;61:99–110. [PubMed] [Google Scholar]
- 21.Wang Q, Shorten D, Xu X, et al. Effect of von Willebrand factor on the pharmacokinetics of recombinant human platelet glycoprotein Ibα-immunoglobulin G1 chimeric proteins. Pharm Res. 2006;23:1743–1749. doi: 10.1007/s11095-006-9018-1. [DOI] [PubMed] [Google Scholar]
- 22.Raines G, Aumann H, Sykes S, Street A. Multimeric analysis of von Willebrand factor by molecular sieving electrophoresis in sodium dodecyl sulphate agarose gel. Thromb Res. 1990;60:201–212. doi: 10.1016/0049-3848(90)90181-b. [DOI] [PubMed] [Google Scholar]
- 23.Tsai HM, Sussman II, Nagel RL. Shear stress enhances the proteolysis of von Willebrand factor in normal plasma. Blood. 1994;83:2171–2179. [PubMed] [Google Scholar]
- 24.Kroll MH, Hellums JD, McIntire LV, Schafer AI, Moake JL. Platelets and shear stress. Blood. 1996;88:1525–1541. [PubMed] [Google Scholar]
- 25.Sugimoto M, Dent J, McClintock R, Ware J, Ruggeri ZM. Analysis of structure-function relationships in the platelet membrane glycoprotein Ib-binding domain of von Willebrand's factor by expression of deletion mutants. J Biol Chem. 1993;268:12185–12192. [PubMed] [Google Scholar]
- 26.Cruz MA, Handin RI, Wise RJ. The interaction of the von Willebrand factor-A1 domain with platelet glycoprotein Ib/IX. The role of glycosylation and disulfide bonding in a monomeric recombinant A1 domain fragment. J Biol Chem. 1993;268:21238–21245. [PubMed] [Google Scholar]
- 27.Miura S, Li CQ, Cao Z, Wang H, Wardell MR, Sadler JE. Interaction of von Willebrand factor domain A1 with platelet glycoprotein Ibα-(1-289). Slow intrinsic binding kinetics mediate rapid platelet adhesion. J Biol Chem. 2000;275:7539–7546. doi: 10.1074/jbc.275.11.7539. [DOI] [PubMed] [Google Scholar]
- 28.Miller JL, Cunningham D, Lyle VA, Finch CN. Mutation in the gene encoding the 〈 chain of platelet glycoprotein Ib in platelet-type von Willebrand disease. Proc Natl Acad Sci U S A. 1991;88:4761–4765. doi: 10.1073/pnas.88.11.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell SD, Roth GJ. Pseudo-von Willebrand disease: a mutation in the platelet glycoprotein Ib〈 gene associated with a hyperactive surface receptor. Blood. 1993;81:1787–1791. [PubMed] [Google Scholar]
- 30.Arya M, Anvari B, Romo GM, et al. Ultralarge multimers of von Willebrand factor form spontaneous high-strength bonds with the platelet glycoprotein Ib-IX complex: studies using optical tweezers. Blood. 2002;99:3971–3917. doi: 10.1182/blood-2001-11-0060. [DOI] [PubMed] [Google Scholar]
- 31.Shankaran H, Alexandridis P, Neelamegham S. Aspects of hydrodynamic shear regulating shear-induced platelet activation and self-association of von Willebrand factor in suspension. Blood. 2003;101:2637–2645. doi: 10.1182/blood-2002-05-1550. [DOI] [PubMed] [Google Scholar]
- 32.Shankaran H, Neelamegham S. Hydrodynamic forces applied on intercellular bonds, soluble molecules, and cell-surface receptors. Biophys J. 2004;86:576–588. doi: 10.1016/S0006-3495(04)74136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider SW, Nuschele S, Wixforth A, et al. Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proc Natl Acad Sci U S A. 2007;104:7899–7903. doi: 10.1073/pnas.0608422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doggett TA, Girdhar G, Lawshe A, et al. Selectin-like kinetics and biomechanics promote rapid platelet adhesion in flow: the GPIbα-VWF tether bond. Biophys J. 2002;83:194–205. doi: 10.1016/S0006-3495(02)75161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goto S, Salomon DR, Ikeda Y, Ruggeri ZM. Characterization of the unique mechanism mediating the shear-dependent binding of soluble von Willebrand factor to platelets. J Biol Chem. 1995;270:23352–23361. doi: 10.1074/jbc.270.40.23352. [DOI] [PubMed] [Google Scholar]
- 36.Goto S, Kasahara H, Sakai H, et al. Functional compensation of the low platelet count by increased individual platelet size in a patient with May-Hegglin anomaly presenting with acute myocardial infarction. Int J Cardiol. 1998;64:171–177. doi: 10.1016/s0167-5273(98)00043-6. [DOI] [PubMed] [Google Scholar]
- 37.Budde U, Dent JA, Berkowitz SD, Ruggeri ZM, Zimmerman TS. Subunit composition of plasma von Willebrand factor in patients with the myeloproliferative syndrome. Blood. 1986;68:1213–1217. [PubMed] [Google Scholar]
- 38.Budde U, Scharf RE, Franke P, Hartmann-Budde K, Dent J, Ruggeri ZM. Elevated platelet count as a cause of abnormal von Willebrand factor multimer distribution in plasma. Blood. 1993;82:1749–1757. [PubMed] [Google Scholar]
- 39.Vincentelli A, Susen S, Le Tourneau T, et al. Acquired von Willebrand syndrome in aortic stenosis. N Engl J Med. 2003;349:343–349. doi: 10.1056/NEJMoa022831. [DOI] [PubMed] [Google Scholar]
- 40.Zimmerman TS, Dent JA, Ruggeri ZM, Nannini LH. Subunit composition of plasma von Willebrand factor. Cleavage is present in normal individuals, increased in IIA and IIB von Willebrand disease, but minimal in variants with aberrant structure of individual oligomers (types IIC, IID, and IIE). J Clin Invest. 1986;77:947–951. doi: 10.1172/JCI112394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Batlle J, Lopez-Fernandez MF, Lopez-Borrasca A, et al. Proteolytic degradation of von Willebrand factor after DDAVP administration in normal individuals. Blood. 1987;70:173–176. [PubMed] [Google Scholar]