Abstract
Chronic lymphocytic leukemia (CLL) cells are thought to have diminished cell-cycling capacity, a view challenged by their phenotypic resemblance to activated human B lymphocytes. The present study addresses the cell-cycling status of CLL cells, focusing on those leukemic cells expressing CD38, a molecule involved in signaling and activation that also serves as a prognostic marker in this disease. CD38+ and CD38− members of individual CLL clones were analyzed for coexpression of molecules associated with cellular activation (CD27, CD62L, and CD69), cell-cycle entry (Ki-67), signaling (ZAP-70), and protection from apoptosis (telomerase and Bcl-2). Regardless of the size of the CD38+ fraction within a CLL clone, CD38+ subclones are markedly enriched for expression of Ki-67, ZAP-70, human telomerase reverse transcriptase, and telomerase activity. Although the percentage of cells (approximately 2%) entering the cell cycle as defined by Ki-67 expression is small, the absolute number within a clone can be sizeable and is contained primarily within the CD38+ fraction. Despite these activation/proliferation differences, both CD38+ and CD38− fractions have similar telomere lengths, suggesting that CD38 expression is dynamic and transient. These findings may help explain why high percentages of CD38+ cells within clones are associated with poor clinical outcome.
Introduction
Chronic lymphocytic leukemia (CLL) results from amplification and accumulation of clonal CD5+ B cells. Although initially thought to be homogenous in manifestations and mechanisms, it is now clear that CLL is quite heterogeneous. Subgroups can be defined by differences in IgVH gene mutations,1 CD382 and ZAP-703,4 expression, presence of chromosomal abnormalities,5 and p53 dysfunction,6 with cases expressing unmutated IgVH genes (U-CLL; Damle et al2 and Hamblin et al7), elevated numbers of CD38+ cells2,8–10 or ZAP-70+ cells,4,11–14 deletions at 17p and 11q,15,16 or impaired p53 activity6 having worse clinical outcomes. Among cell-surface markers, expression of CD38 and its relevance to the pathobiology of CLL17 has been the subject of intense study.18 It is now clear that this molecule binds CD3119, enabling important cell-cell interactions that signal activation and survival pathways20,21 in normal22 and leukemic lymphocytes17 and antigen-presenting cells.23
Despite the heterogeneity of expression of various molecular and cellular markers in CLL, the disease appears relatively homogeneous by gene expression profiling. Only a small number of genes are differentially expressed between U-CLL and CLL patients with mutated IgVH genes (M-CLL),3,24 implying that all CLL clones likely derive from antigen-experienced/memory-like B cells.24 Similarly, different gene expression signatures distinguish instances of CLL defined by CD38 and ZAP-70 expression.25
A paradoxic feature of circulating CLL cells is the expression of multiple features of activated, antigen-experienced B cells by lymphocytes that are mostly arrested in the G0/G1 phase of the cell cycle. Although the majority of CLL cells from most patients express activation-related26–28 and certain cell cycle–related29–32 markers, surprisingly low percentages of Ki-67–expressing cells have been found in the blood of patients with CLL compared with those observed in other lymphoid malignancies.32 Furthermore, a proliferative compartment exists in CLL, although this probably resides in the solid tissues.33 Of note, data derived using tissue microarrays suggest that most CLL cells exist in late G1 phase (cyclin E+), and a surprising number of cells exist in the S (cyclin A+) and G2/M phases (cyclin B1+) of the cell cycle.34 These data are at variance with other studies mentioned, and may support a difference in cell-cycle progression between circulating and tissue-bound CLL cells.
Questions remain as to how many cells bearing evidence for cellular activation actually enter and complete the cell cycle. Since analyses of bulk populations limit the extent to which properties of members of cell populations can be understood, efforts are now focusing on fractionating CLL clones and defining differences in cellular components. In this regard, despite their monoclonal origin, highly purified CD38+ and CD38− subpopulations derived from the same patient with CLL exhibit distinct gene expression signatures.35
In an attempt to address this dilemma and to quantify the percentage of cells that enter the cell cycle, we have studied differences in expression of Ki-67 in relation to ZAP-70, Bcl-2, and surface membrane activation marker expression in CD38+ and CD38− subclones within a series of CLL clones from various patients differing in their overall CD38 status. We have also assessed differences in the replicative history and potential of CD38 subpopulations within individual patients with CLL. These studies identify a close association between CD38 expression and increased Ki-67 and ZAP-70 positivity, suggesting that CD38+ clonal members are more activated and could more frequently enter the cell cycle than their counterpart CD38− cells. Although CD38+ cells exhibited greater telomerase activity than companion CD38− cells, these 2 cell subsets did not differ in telomere length, implying that CD38 expression is a temporal feature of the cells' activation state that can change over time.
Patients, materials, and methods
Patients and healthy donors
The Institutional Review Board of the North Shore–LIJ Health System approved these studies. Following informed consent obtained in accordance with the Declaration of Helsinki, venous blood was collected from 95 randomly chosen patients with CLL for whom IgVH gene DNA sequence data were available. Leukocyte-enriched fractions of blood donated by 20 healthy volunteers matched for age with the patients with CLL (60 years and older) were purchased from Long Island Blood Services (Melville, NY); these samples were negative for HIV and hepatitis B virus (HBV) antigens. Peripheral blood mononuclear cells (PBMCs) were separated from heparinized venous blood and leukocyte-enriched fractions by density gradient centrifugation using Ficoll-Paque (Pharmacia LKB Biotechnology, Piscataway, NJ) and cryopreserved for future use using a programmable cell-freezing machine (Cryomed, Mt Clemens, MI).
Analysis of surface membrane and intracellular antigens by flow cytometry
The following fluorochrome-conjugated mAbs were used: anti-CD5–APC, anti-CD27–FITC, anti–Ki-67–FITC, anti–Bcl-2–FITC, and anti-CD62L–FITC (BD Pharmingen, San Diego, CA); Simultest Leucogate, fluorochrome-conjugated isotype control mAbs, and anti-CD19–perCP, anti-CD38–PE, anti-CD69–FITC (BD Biosciences, San Diego, CA), and anti–ZAP-70–FITC (eBioscience, San Diego, CA). Unconjugated polyclonal anti–telomerase reverse transcriptase (TERT) antibody was purchased from Calbiochem (San Diego, CA). Cryopreserved PBMCs from 50 patients with CLL were subjected to 4-color immunofluorescence staining that included in each set mAbs to CD38, CD5, and CD19 along with either anti-CD27, anti-CD62L, or anti-CD69 mAbs. PBMCs from the entire cohort of 95 patients with CLL were studied for expression of intracellular antigens (Ki-67, ZAP-70, and Bcl-2) by incubating with mAbs reactive with CD38, CD5, and CD19, permeabilizing and fixing with Cytofix/Cytoperm reagent (BD Biosciences), and then incubating with either antibodies to Ki-67, ZAP-70, Bcl-2 or isotype control mAbs for an additional 25 minutes at 4°C. Cells were analyzed with a fluorescence-activated cell sorting (FACS) Calibur flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA).
For analyses of Ki-67 expression, mAb B56 (BD Pharmingen) was used; this mAb reacts with the same epitope as mAb MIB-1. To define optimum conditions for Ki-67 detection and, in particular, levels of Ki-67 expression in various stages of the cell cycle defined by DNA content, we examined tonsillar mononuclear cells (MNCs) that contain activated cells at different stages of maturation. Cells in S and G2/M phases were selectively enriched in Ki-67 expression compared with cells in the G0/G1 phase of the cell cycle (data not shown).
FACS
Cryopreserved PBMCs from 20 patients with CLL were incubated with mAbs specific for CD19, CD5, and CD38 and with fluorochrome-conjugated isotype control mAbs for 25 minutes at 4°C, and subjected to flow sorting on a FACS-Aria fluorescence activated cell sorter (FACS; Becton Dickinson Immunocytometry Systems). CD19+CD5+CD38− and CD19+CD5+CD38+ subsets from each patient were processed for telomere length and telomerase quantification.
Quantification of telomere length and telomerase activity
A flow–fluorescence in situ hybridization (FISH) protocol detailed elsewhere36,37 was used to quantify mean telomere lengths in purified cell populations. Telomerase activity was assessed in the flow-sorted CD19+CD5+CD38− and CD19+CD5+CD38+ subsets using the TRAPeze telomerase detection kit (Chemicon International, Temecula, CA) as described37; this approach uses the telomere repeat amplification protocol (TRAP; Kim et al38).
Statistical analyses
Expression of Ki-67 and other markers was compared between healthy donors and patients with CLL using the Mann-Whitney test. Comparison across CD38 groupings (negative, low, intermediate, and high) with respect to Ki-67 and ZAP-70 expression was carried out using the Kruskal-Wallis test. Upon finding significant differences across groups, a Bonferroni-like adjusted pairwise comparison was made to determine which groups differed from each another. Differences between percentages of cells expressing the same marker within CD38-based subsets were evaluated using the Wilcoxon signed-rank test. The Spearman rank correlation coefficient was calculated to determine the strength of association between various parameters.
Results
Expression of Ki-67 by CD5+ normal and leukemic B cells
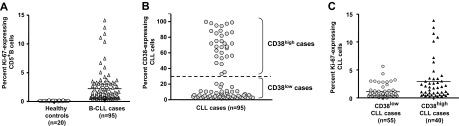
We analyzed Ki-67 expression among PBMCs from a cohort of 95 randomly selected patients with CLL and 20 age-matched healthy donors to identify cells that had traversed the G0/G1 phase (Figure 1A). On average, the CLL clones contained 1.77% Ki-67–expressing cells (range, 0.1%-13.8%), which was a significantly higher percentage than that of CD5+ B cells from healthy donors (average, 0.23%; range, 0.0%-0.98%; P < .001, Kruskal-Wallis test).
Figure 1.
Ki-67 expression in CD5+ B cells from healthy elderly donors and patients with CLL. PBMCs from 20 healthy elderly donors and 95 patients with CLL were analyzed for expression of CD5, CD38, CD19, and Ki-67. (A) Percentage of CD5+CD19+ cells from healthy controls and patients with CLL expressing Ki-67 (P < .001; Mann-Whitney test). (B) Percentage of CD38+ B cells in CLL clones designated CD38low (55 of 95) and CD38high (40 of 95) based on a 30% cut-off. (C) Significant differences in Ki-67 expression between CD38low and CD38high patients with CLL (P < .001; Mann-Whitney test). Horizontal lines in panels A and B indicate averages of values in corresponding groups.
Expression of Ki-67 in CD38high and CD38low CLL clones
Because CD38 is up-regulated with activation and maturation of normal human B lymphocytes,39 and its expression is intimately linked to cellular activation17 and disease course2 in CLL, we determined the relationship between expression of Ki-67 and CD38 within a leukemic clone. The 95 patients with CLL were divided into subgroups based on the percentage of CD5+CD19+ cells expressing CD38 (Figure 1B) by selectively gating and analyzing FACS data obtained after incubation with mAb reactive with CD19, CD5, CD38, and Ki-67. A total of 58% (55 of 95) of the clones contained less than 30% CD38-expressing leukemic cells (CD38low), and 42% (40 of 95) of the clones contained 30% or more CD38-expressing cells (CD38high). As shown in Figure 1C, the average number of Ki-67+ cells was significantly greater in CD38high patients (mean, 2.66%; range, 0.11%-13.80%) than in CD38low patients (mean, 1.06%; range, 0.0%-5.63%; P < .001, Kruskal-Wallis test). Of note, the average number of Ki-67+ cells was also significantly greater in patients with U-CLL (mean, 2.05%; range, 0.0%-13.8%) than in patients with M-CLL (mean, 1.23%; range, 0.0%-3.98%; P < .001, Kruskal-Wallis test; data not shown).
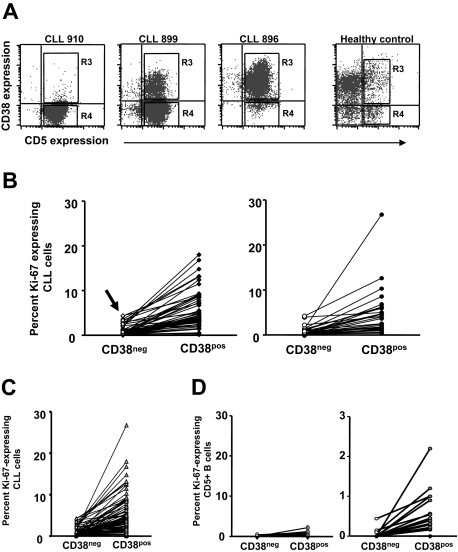
Expression of Ki-67 by CD38+ and CD38− cells within CLL clones
Recent studies suggest that CD38+ and CD38− members of CLL clones differ in gene expression25,35 and certain cellular functions (eg, signaling through the B-cell receptor [BCR]).40 Therefore, Ki-67 expression by CD38+ and CD38− cells of the same 95 CLL clones was analyzed. To accomplish this, 2 regions corresponding to both ends of the CD38 expression spectrum were demarcated by FACS (Figure 2A): region R3, which contains most of the CD19+CD5+CD38+ fraction; and region R4, which contains most of the CD19+CD5+CD38− fraction.
Figure 2.
Ki-67 expression by CD38+ and CD38− B cells within individual patients with CLL. PBMCs from 95 patients with CLL and 20 elderly healthy donors were stained as in Figure 1. (A) Dotplots of CD5 and CD38 expression by B cells from 3 patients with CLL and 1 healthy donor. (B) Left panel: Lines connect data points for percentage of Ki-67+ cells in CD38− and CD38+ subsets within each patients with CLL in the CD38low subgroup. Note the detection of Ki-67+ cells even in the CD38− subset of CD38low patients (arrow). Right panel: Lines connect data points for percentage of Ki-67+ cells in CD38− and CD38+ subsets within each CLL clone in the CD38high subgroup. (C) Lines connect data points for percentage of Ki-67+ cells in CD38− and CD38+ subsets of all patients with CLL. (D) Left panel: Lines connect data points for percentage of Ki-67+ cells in CD38− and CD38+ subsets within each healthy participant. Right panel: Same data from left graph drawn to a smaller scale.
The percentage of cells expressing Ki-67 in CD19+CD5+CD38+ and CD19+CD5+CD38− fractions within CD38low and CD38high patients was tabulated (Figure 2B). Significantly higher percentages of CD38+ cells expressed Ki-67 than did CD38− cells (mean Ki-67 expression in CD38+, 4.79%; range, 0.1%-26%; mean Ki-67 expression in CD38−, 0.87%; range, 0%-4.4%; P < .001, Wilcoxon signed-rank test), regardless of the type of patient analyzed (ie, CD38low and CD38high). Surprisingly, even in CD38low patients, a sizable fraction of CD38+CD5+ cells expressed Ki-67 (mean, 5.57% ± 0.6%; n = 55), reflecting entry into the cell cycle (Figure 2B, arrow, left side).
Comparison of Ki-67 expression in CD38+ versus CD38− cells of normal CD5+ B cells and CLL cells
Many normal human B cells express varying levels of CD38, regardless of CD5 coexpression. Therefore, we analyzed expression of Ki-67 by CD38+ and CD38− fractions of normal circulating CD5+ B cells and CLL cells (Figure 2C), and then compared them with each other (Figure 2D). Ki-67+ cells are more often found in CD38+ subsets of both CLL and normal B lymphocytes (Figure 2C; healthy donors: mean, 0.23%; patients with CLL: mean, 4.79%; P < .001, Mann-Whitney test), and there are many more Ki-67+ cells in both CD38− as well as CD38+ fractions of CLL cells (Figure 2C) compared with normal CD5+ B cells (percentage of Ki-67 in CD38−: 0.0%; percentage of Ki-67 in CD38+: mean, 0.23%; P < .001, Mann-Whitney test). Figure 2D (right side) depicts the same data obtained for the normal CD5+ B cells (Figure 2D, left side) but represented on a magnified scale to indicate that significant differences in Ki-67 expression exist even between CD38+ and CD38− subsets of normal B cells. Among both CLL clones and normal B-cell populations, CD38 expression marks a CD5+ population containing more cells that have entered the cell cycle. This is the case even when the CD38+ cells are from CLL clones that have only a small CD38+ fraction (eg, as low as 0.3%; Figure 2B).
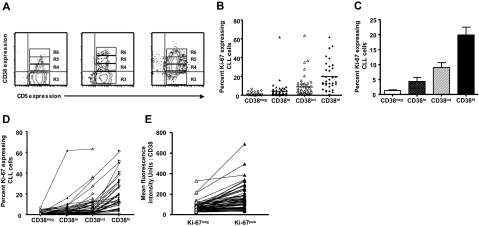
Expression of Ki-67 in CLL cells differing in density of cell-surface CD38
To determine whether Ki-67 expression is related to the density of CD38 expressed on the surfaces of different CD19+CD5+ CLL cells, we further divided FACS region R3 from Figure 2A into 3 arbitrary “slices” based on the intensity of CD38 expression (R4 = CD38low, R5 = CD38int, and R6 = CD38high; Figure 3A for 3 representative patients with CLL). Although all 95 patients with CLL were analyzed for CD38 expression, data on CD38 intensity slices were tabulated only in patients in which more than 50 cells were detected in either R4, R5, or R6; therefore, these analyses include data from 60 of the original 95 patients. The mean fluorescence intensity (MFI) values for CD38 expression in R6 were more than 3.5-fold greater than that of cells in R5 and 10-fold greater than that of R4. The MFI values for R5 were more than 3.5-fold greater than that of cells in R4.
Figure 3.
Ki-67 expression in relation to density of CD38. PBMCs from patients with CLL were stained as described for Figure 1, and contour plots depicting expression of CD5 and CD38 in CD19+ cells of patients with CLL were generated. (A) The CD38+ fraction was divided into 3 regions—R4, R5, and R6—referred to as CD38low, CD38int, and CD38high, respectively. Percentages of Ki-67–expressing cells were scored only if at least 50 cells fell within marked regions. (B) Percentages of Ki-67–expressing cells in CD38low, CD38int, and CD38high subsets from 60 of 95 patients with CLL. Horizontal lines indicate averages of values in corresponding populations. (C) Average (± SE) of values from panel B. (D) Lines connect data points depicting percent Ki-67+ cells in each CD38 subset for each of 60 patients with CLL. (E) In every patient studied, CD5+Ki-67+ cells showed higher density of CD38 compared with CD5+Ki-67− cells from the same case.
The percentages of Ki-67–expressing cells in regions R3, R4, R5, and R6 for each of the 60 patients analyzed were significantly different from each other (Figure 3B,C; P < .001, Kruskal-Wallis test), indicating clearly a direct relationship between Ki-67 positivity and density of CD38 expression on CLL cells. This relationship existed in every patient analyzed (Figure 3D). Similarly, CD38 density defined by MFI was higher in the Ki-67+ subset of the clone compared with the Ki-67− subset in each patient (Figure 3E; P < .01, Mann-Whitney test).
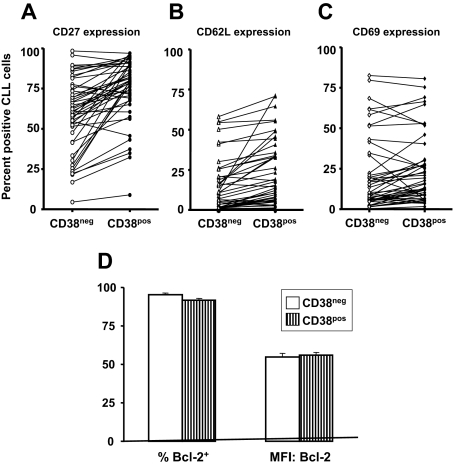
Expression of activation-related cell-surface markers and Bcl-2 in CD38+ and CD38− subsets of CLL clones
In addition to CD38, CLL cells express other activation- and maturation-associated markers27,41,42 (eg, CD69 that is up-regulated rapidly with cellular activation and retains cells in the vicinity of the inductive stimulus,43,44 CD62L that is involved in cell adhesion and eventually lost from the cell surface after cell activation,45 and CD27 that is reflective of a memory and an activated phenotype46,47). These markers were also analyzed in CD38+ and CD38− fractions of individual CLL clones (Figure 4). The percentages of cells within patients expressing these markers were significantly different (CD27:CD38+: mean, 75.1%; CD38−: mean, 58.8%; P < .001; CD62L:CD38+: mean, 21.4%; CD38−: mean, 13.5%; P < .001; and CD69:CD38+: mean, 23.9%; CD38−: mean, 20.8%; P < .01; signed-rank test), although the trend of differences between cells in the CD38+ and CD38− subsets were not as consistent as found for Ki-67 expression.
Figure 4.
Expression of cell activation- and apoptosis- related molecules in CD38+ and CD38− cell subsets within patients with CLL. PBMCs from 50 patients with CLL were analyzed for expression of CD27, CD62L, or CD69 by CD5+, CD19+, and CD38+ cells. (A-C) Significant differences in expression of CD27, CD62L, and CD69, respectively, within CD38+ and CD38− subsets of the clone (CD27 and CD62L, P < .001; CD69, P < .01). (D) Means (± SE) for percentage of Bcl-2+ cells (left) and means (± SE) for MFI of Bcl-2 expression (right).
Expression of ZAP-70 in CD38+ and CD38− subsets of CLL clones
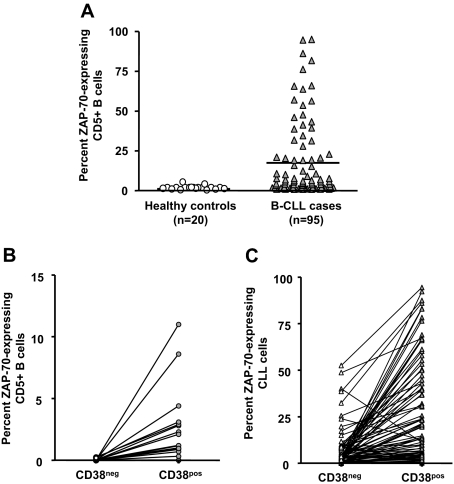
ZAP-70 expression in normal human B lymphocytes is linked to cellular activation48–50 and has an important cell-signaling role in CLL.51–54 We therefore compared expression of ZAP-70 in CLL cells of the same 95 patients and CD19+CD5+ cells from healthy elderly volunteers. Significantly higher percentages of CLL cells expressed ZAP-70 compared with B lymphocytes from control participants (Figure 5A).
Figure 5.
ZAP-70 expression in CD38+ and CD38− cells. PBMCs from 20 elderly healthy donors and 95 patients with CLL were incubated with mAbs to CD5, CD38, CD19, ZAP-70, or appropriate isotype control mAbs. CD19+CD5+ cells exhibiting a ZAP-70 staining in excess of the isotype control mAbs were considered ZAP-70+. (A) Significant differences in percentage of CD5+CD19+ cells expressing ZAP-70 (P < .001; Mann-Whitney test). These data were further analyzed to determine differences in expression of ZAP-70 between the CD38− and CD38+ populations in healthy controls. Horizontal line indicates average of values in corresponding group. (B) and in patients with CLL (C). Significant differences exist between ZAP-70 expression within CD38− and CD38+ subsets from healthy donors and patients with CLL (P < .001; Kruskal-Wallis test).
Furthermore, none of the cells in the CD38− subset from healthy donors expressed ZAP-70 (0 of 20), even though the percentage of ZAP-70–expressing cells in the CD38+ subset ranged from 0.3% to 11.2% (Figure 5B). In contrast, 98% (93 of 95) of patients with CLL had a higher percentage of CD38+ cells expressing ZAP-70 than CD38− cells within the same clones (Figure 5C). In individual patients, ZAP-70 expression exhibited strong positive correlation with both CD38 expression and Ki-67 expression (Table 1; P < .001, Spearman correlation). These findings were true regardless of percentage of the clonal members expressing CD38, further supporting the concept that expression of CD38 and ZAP-70 coevolves.
Table 1.
Correlation of ZAP-70 expression with CD38+ and Ki-67+ cells in individual CLL clones
| CD38 expression | Ki-67 expression | |
|---|---|---|
| r | .244 | .274 |
| P | .017 | .007 |
Percentages of cells CD-expressing ZAP-70 with either CD38 or Ki-67 in individual patients were subjected to Spearman correlation test.
Telomerase activity and telomere length of flow-sorted CLL cells
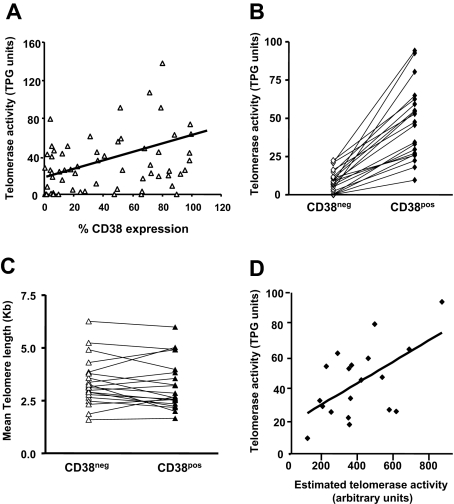
Telomerase activity is enhanced on activation of normal55,56 and leukemic B cells.37 Therefore, telomerase activity was quantified in B cells from CLL clones using the TRAP assay. Figure 6A indicates a modest direct relationship between telomerase activity and percentage of cells in CLL clones expressing CD38 (r = .35; P < .01). The analyses were refined by purifying CD5+CD38+ and CD5+CD38− B cells from 20 patients with CLL using FACS and the same gating approach illustrated in Figure 2A, and quantifying telomerase activity in these 2 subsets. In every instance, flow-sorted CD38+ CLL cells possessed higher telomerase activity than CD38− cells from the same clone (Figure 6B).
Figure 6.
Telomerase activity and telomere length of flow-sorted populations. (A) Telomerase activity was assayed in B cells from 60 patients with CLL using the TRAP assay and percentage of CD38+ CLL cells plotted versus TPG (total product generated) units in the same patients. CD19+CD5+ cells from 20 patients with CLL were sorted into CD38+ and CD38− subpopulations and processed for quantification of telomere length and telomerase activity. (B) Lines connect data points that indicate telomerase activity in the cell subsets of each individual patient. (C) Lines connect data points for mean telomere lengths of flow-sorted CD38+ and CD38− CLL cells. (D) Significant positive correlation (r = .465; P < .045) exists between estimated telomerase activity and observed telomerase activity (TPG units) in CD38+ cells from 20 patients with CLL shown in panels B and C; this correlation does not exist in the CD38− cells. Diagonal lines in panels A and D indicate best fits based on linear regression of data.
Telomerase activity requires functional, human TERT (hTERT) protein.57 To determine if differences in percentage of hTERT+ CLL cells in a bulk population are at the root of the observed differences in enzymatic activity, we analyzed the percentage of CLL cells expressing hTERT by flow cytometry. The product of the percentages of cells expressing hTERT and the intensity of its expression was calculated and called “estimated telomerase activity.” Total product generated (TPG) activity obtained from the TRAP assays showed a positive correlation with these arbitrary units in CD38+ cells but not in CD38− cells (Figure 6D). In addition, there was a positive correlation between percent Ki-67–expressing cells with telomerase activity (r = .515; P = .02) and ZAP-70 expression (r = .26; P < .01).
Notably, despite the differences in hTERT protein and telomerase activity between the CD5+CD38+ and CD5+CD38− subsets, mean telomere lengths of these fractions were comparable within each clone (CD38+: range, 1.66-5.98 kb; mean, 3.34 kb; CD38−: range, 1.59-6.25; mean, 3.36 kb; P = .39; Figure 6C). This finding suggests that CD38+ and CD38− subclones are linked and represent a continuum, distinguished by transient and activation-related expression of CD38.
Discussion
Most circulating CLL cells exhibit features of preactivated cells. Their surface membranes are decorated with molecules found on stimulated B lymphocytes,26–28 with specific antigens displayed differing between U-CLL and M-CLL.27,42,58,59 Based on telomere length, the cells have replicated multiple times and clearly more than B cells from healthy donors matched for age.37,60–62 Finally, in vivo labeling of CLL cells reveals birth of a limited but significant number of new leukemic cells.33
Based on these findings, one would expect that CLL cells are cycling. Surprisingly, few cells in the blood have progressed beyond the G0/G1 phase of the cell cycle, although more cells in solid tissues may have done so.33,34 Although our experiments were carried out on B cells from the peripheral blood since adequate numbers of solid tissue samples were not available, based on the studies mentioned,33,34 the size of the proliferative component in tissues is likely greater than we have estimated for the blood.
Because clonal members can be heterogeneous in expression of genes and markers related to cellular activation and adhesion, in particular CD38,25–27,35,41,42 this intraclonal heterogeneity may translate into differences that determine which cells enter the cell cycle and may help to explain contradictory results. Early hints of such differences came from studies of the pattern of CD38 expression by members of CLL clones, indicating that the presence of a distinct CD38+ population within a leukemic clone, regardless of its apparent size, identified patients who would have more aggressive disease.63
Consequently, we examined how differences in expression of markers of activation by CLL cells correlate with entrance into the cell cycle; in particular, we used expression of Ki-67, a nuclear protein that is up-regulated in the G1, S, G2, and M phases of the cell cycle but is absent from resting cells (G0 phase64). Significantly higher percentages of CD5+ B cells from patients with CLL than CD5+ B cells from controls expressed Ki-67 (Figure 1A). When the patients with CLL were divided into 2 groups based on a cutoff of 30% or more or less than 30% CD38-expressing cells within a leukemic clone, CD38high patients exhibited remarkably more Ki-67+ cells (Figure 1C). Similarly, U-CLL clones contain more Ki-67+ cells than M-CLL clones (not shown).
We further analyzed Ki-67 expression within CD38+ and CD38− subclones of each individual patient with CLL. Whether obtained from a patient with CLL or a control participant, proportionally more Ki-67+ cells were contained in the CD38+ than the CD38− fraction (Figure 2C,D). Even CD38− cells in CD38low patients showed significant numbers of Ki-67–expressing cells (Figure 2B), suggesting that CD38 expression labels cells in an activated state that have crossed the G0/early-G1 boundary of the cell cycle. The finding that the percentages of Ki-67+ cells within the clone increased with the cell-surface density of CD38 (Figure 3C,D) suggests that a cell's level of CD38 expression reflects its extent of activation as well as its proliferative capacity. These findings are consistent with the enhanced transcription of other activation markers (eg, CD18, CD49d, CD20, and subunit 5 of the anaphase-promoting complex/cyclosome) in leukemic B cells from CD38high patients with CLL.65
In addition, we quantified expression of CD69, CD62L, and CD27 in CD38+ and CD38− subsets of the clones from a cohort of 50 patients with CLL. Significant differences were observed in percentages of cells expressing these markers within the subsets, although these paired values did not follow a consistent trend of lower in CD38− and higher in CD38+ in individual patients (Figure 4A-C). It was somewhat surprising to find increased numbers of CD62L+ cells in the CD38+ fraction, since CD62L expression wanes after cellular activation. However, because CD62L and CD69 are involved in retaining lymphocytes at the site of stimulation,43–45 the levels of these 2 molecules on circulating CD38+ cells might be less than those in the solid tissues, and therefore might indicate that CD38+ cells expressing these markers are recent emigrants from such sites. We are currently testing this possibility using in vivo cellular labeling.66
Antiapoptotic proteins such as Bcl-2, Bax, Bak, BAD, and Mcl-1 that might contribute to prolonged survival in vivo are up-regulated in CLL cells.67,68 However, the fact that CD38+ and CD38− cells did not differ with respect to expression of Bcl-2 suggests that this protein does not confer a selective survival advantage to CD38-expressing cells, especially in light of the finding that CD38− cells die faster than CD38+ cells in vitro.35
Since ZAP-70 expression is influenced by activation state and is also one of a few genes differentially expressed between the IgVH gene mutation patient subgroups, we examined its association with CD38 expression. Our observation on elevated percentages of ZAP-70+ cells in the CD38+ subset of the clone compared with those in CD38− CLL cells (Figure 5B,C) within most patients corroborates earlier findings,69 consistent with the suggestion that CD38+ cells might have a better ability to transduce BCR-mediated signals40 with the help of simultaneous ZAP-70 expression.51,52 Although ZAP-70 was not detected in CD38− cells from healthy donors, it was found to a low extent in the CD38+ cells from the same healthy individuals, as reported earlier by others.48–50 It is interesting that in a minority of patients with CLL there were more ZAP-70+ cells in the CD38− fraction of the clone, suggesting that ZAP-70 expression may be retained in a subpopulation of CD38+ cells that have lost CD38 expression.
In this regard, the finding that highly purified CD38+ and CD38− subfractions of CLL clones showed no differences in mean telomere length (Figure 6C) suggests that the replicative histories of these 2 fractions are not different, and supports the notion that these fractions may represent a continuum distinguished by transient, activation-related expression of CD38.
Thus, regardless of the percentage of CD38+ cells in a patient's CLL clone, CD38 expression is linked to cell activation and labels a proliferative component defined by Ki-67 expression. On average, 1.77% of the leukemic cells in the patients studied expressed Ki-67, and most of these cells were within the CD38+ fraction (4.8% of CD38+ CLL cells coexpressed Ki-67 vs 0.88% of CD38− CLL cells). We need to emphasize, however, that the circulating CD38+Ki-67+ component is small in relation to the percentage of cells within a clone (approximately 2%). Nevertheless, it may represent a sizeable number of cells in toto. Since the size of a CLL clone in vivo ranges from 1012 to 1014 cells, this percentage then represents approximately 1010 to 1012 cells. However, ongoing cell death appears to balance out this continuing proliferation in most patients,33 thereby maintaining absolute cell numbers and explaining the often slow changes in lymphocyte counts in vivo. Since primarily CD38+ CLL cells coexpressed Ki-67, one would anticipate that this fraction would be enriched in proliferating cells; indeed, our preliminary data suggest that the CD38 marks the proliferative compartment in CLL, based on incorporation of 2H label.67
Collectively, our findings may help to explain why the presence of high percentages of CD38+ leukemic cells within a CLL clone is associated with aggressive disease and poor clinical outcome,2,64 since presumably it is from this fraction that new, more dangerous chromosomal abnormalities evolve.70 Moreover, combining quantification of Ki-67+ cells with CD38+ cells may provide even greater prognostic effectiveness.
Acknowledgments
This work was supported in part by RO1 grant CA 87956 from the National Cancer Institute and M01 General Clinical Research Center Grant (RR018535) from the National Center for Research Resources. The Karches Family Foundation, the Peter Jay Sharp Foundation, the Prince Family Foundation, the Marks Family Foundation, the Jean Walton Fund for Lymphoma & Myeloma Research, and the Joseph Eletto Leukemia Research Fund also provided support for these studies.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.N.D. designed and performed research, analyzed data, and wrote the paper; S.T. and T.B. performed research; C.C. and S.Y. wrote the paper; C.S. analyzed data; S.L.A. and K.R.R. analyzed data and wrote the paper; and N.C. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicholas Chiorazzi, The Feinstein Institute for Medical Research, 350 Community Drive, Manhasset, NY, 11030; e-mail: nchizzi@nshs.edu.
References
- 1.Fais F, Ghiotto F, Hashimoto S, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102:1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 3.Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiestner A, Rosenwald A, Barry TS, et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood. 2003;101:4944–4951. doi: 10.1182/blood-2002-10-3306. [DOI] [PubMed] [Google Scholar]
- 5.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 6.Pettitt AR, Sherrington PD, Stewart G, Cawley JC, Taylor AM, Stankovic T. p53 dysfunction in B-cell chronic lymphocytic leukemia: inactivation of ATM as an alternative to TP53 mutation. Blood. 2001;98:814–822. doi: 10.1182/blood.v98.3.814. [DOI] [PubMed] [Google Scholar]
- 7.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig VH genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 8.Jelinek DF, Tschumper RC, Geyer SM, et al. Analysis of clonal B-cell CD38 and immunoglobulin variable region sequence status in relation to clinical outcome for B-chronic lymphocytic leukaemia. Br J Haematol. 2001;115:854–861. doi: 10.1046/j.1365-2141.2001.03149.x. [DOI] [PubMed] [Google Scholar]
- 9.Del Poeta G, Maurillo L, Venditti A, et al. Clinical significance of CD38 expression in chronic lymphocytic leukemia. Blood. 2001;98:2633–2639. doi: 10.1182/blood.v98.9.2633. [DOI] [PubMed] [Google Scholar]
- 10.Marasca R, Maffei R, Morselli M, et al. Immunoglobulin mutational status detected through single-round amplification of partial V(H) region represents a good prognostic marker for clinical outcome in chronic lymphocytic leukemia. J Mol Diagn. 2005;7:566–574. doi: 10.1016/S1525-1578(10)60589-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–1775. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 12.Durig J, Nuckel H, Cremer M, et al. ZAP-70 expression is a prognostic factor in chronic lymphocytic leukemia. Leukemia. 2003;17:2426–2434. doi: 10.1038/sj.leu.2403147. [DOI] [PubMed] [Google Scholar]
- 13.Rassenti LZ, Hunynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 14.Schroers R, Griesinger F, Trumper L, et al. Combined analysis of ZAP-70 and CD38 expression as a predictor of disease progression in B-cell chronic lymphocytic leukemia. Leukemia. 2005 doi: 10.1038/sj.leu.2403707. [DOI] [PubMed] [Google Scholar]
- 15.Krober A, Seiler T, Benner A, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- 16.Stilgenbauer S, Bullinger L, Lichter P, Dohner H. Genetics of chronic lymphocytic leukemia: genomic aberrations and V(H) gene mutation status in pathogenesis and clinical course. Leukemia. 2002;16:993–1007. doi: 10.1038/sj.leu.2402537. [DOI] [PubMed] [Google Scholar]
- 17.Deaglio S, Capobianco A, Bergui L, et al. CD38 is a signaling molecule in B-cell chronic lymphocytic leukemia cells. Blood. 2003;102:2146–2155. doi: 10.1182/blood-2003-03-0989. [DOI] [PubMed] [Google Scholar]
- 18.Deaglio S, Vaisitti T, Aydin S, Ferrero E, Malavasi F. In-tandem insight from basic science combined with clinical research: CD38 as both marker and key component of the pathogenetic network underlying chronic lymphocytic leukemia. Blood. 2006;108:1135–1144. doi: 10.1182/blood-2006-01-013003. [DOI] [PubMed] [Google Scholar]
- 19.Deaglio S, Morra M, Mallone R, et al. Human CD38 (ADP-ribosyl cyclase) is a counter-receptor of CD31, an Ig superfamily member. J Immunol. 1998;160:395–402. [PubMed] [Google Scholar]
- 20.Deaglio S, Vaisitti T, Bergui L, et al. CD38 and CD100 lead a network of surface receptors relaying positive signals for B-CLL growth and survival. Blood. 2005;105:3042–3050. doi: 10.1182/blood-2004-10-3873. [DOI] [PubMed] [Google Scholar]
- 21.Deaglio S, Vaisitti T, Billington R, et al. CD38/CD19: a lipid raft-dependent signaling complex in human B cells. Blood. 2007;109:5390–5398. doi: 10.1182/blood-2006-12-061812. [DOI] [PubMed] [Google Scholar]
- 22.Funaro A, Morra M, Calosso L, Zini MG, Ausiello CM, Malavasi F. Role of the human CD38 molecule in B cell activation and proliferation. Tissue Antigens. 1997;49:7–15. doi: 10.1111/j.1399-0039.1997.tb02703.x. [DOI] [PubMed] [Google Scholar]
- 23.Frasca L, Fedele G, Deaglio S, et al. CD38 orchestrates migration, survival, and Th1 immune response of human mature dendritic cells. Blood. 2006;107:2392–2399. doi: 10.1182/blood-2005-07-2913. [DOI] [PubMed] [Google Scholar]
- 24.Klein U, Tu Y, Stolovitzky GA, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194:1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huttmann A, Klein-Hitpass L, Thomale J, et al. Gene expression signatures separate B-cell chronic lymphocytic leukaemia prognostic subgroups defined by ZAP-70 and CD38 expression status. Leukemia. 2006;20:1774–1782. doi: 10.1038/sj.leu.2404363. [DOI] [PubMed] [Google Scholar]
- 26.Sembries S, Pahl H, Stilgenbauer S, Dohner H, Schriever F. Reduced expression of adhesion molecules and cell signaling receptors by chronic lymphocytic leukemia cells with 11q deletion. Blood. 1999;93:624–631. [PubMed] [Google Scholar]
- 27.Damle RN, Ghiotto F, Valetto A, et al. B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes. Blood. 2002;99:4087–4093. doi: 10.1182/blood.v99.11.4087. [DOI] [PubMed] [Google Scholar]
- 28.Trentin L, Zambello R, Sancetta R, et al. B lymphocytes from patients with chronic lymphoproliferative disorders are equipped with different co stimulatory molecules. Cancer Res. 1997;57:4940–4947. [PubMed] [Google Scholar]
- 29.Delmer A, Ajchenbaum-Cymbalista F, Tang R, et al. Overexpression of cyclin D2 in chronic B-cell malignancies. Blood. 1995;85:2870–2876. [PubMed] [Google Scholar]
- 30.Vrhovac R, Delmer A, Tang R, Marie JP, Zittoun R, Ajchenbaum-Cymbalista F. Prognostic significance of the cell cycle inhibitor p27Kip1 in chronic B-cell lymphocytic leukemia. Blood. 1998;91:4694–4700. [PubMed] [Google Scholar]
- 31.Wolowiec D, Ciszak L, Kosmaczewska A, et al. Cell cycle regulatory proteins and apoptosis in B-cell chronic lymphocytic leukemia. Haematologica. 2001;86:1296–1304. [PubMed] [Google Scholar]
- 32.Jaroslav P, Martina H, Jiri S, et al. Expression of cyclins D1, D2, and D3 and Ki-67 in leukemia. Leuk Lymphoma. 2005;46:1605–1612. doi: 10.1080/10428190500215100. [DOI] [PubMed] [Google Scholar]
- 33.Messmer BT, Messmer D, Allen SL, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest. 2005;115:755–764. doi: 10.1172/JCI23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obermann EC, Went P, Tzankov A, et al. Cell cycle phase distribution analysis in chronic lymphocytic leukaemia: a significant number of cells reside in early G1-phase. J Clin Pathol. 2006;60:794–797. doi: 10.1136/jcp.2006.040956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pepper C, Ward R, Lin TT, et al. Highly purified CD38(+) and CD38(-) sub-clones derived from the same chronic lymphocytic leukemia patient have distinct gene expression signatures despite their monoclonal origin. Leukemia. 2007;21:687–696. doi: 10.1038/sj.leu.2404587. [DOI] [PubMed] [Google Scholar]
- 36.Rufer N, Dragowska W, Thornbury G, Roosnek E, Lansdorp PM. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat Biotechnol. 1998;16:743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- 37.Damle RN, Batliwalla FM, Ghiotto F, et al. Telomere length and telomerase activity delineate distinctive replicative features of the B-CLL subgroups defined by immunoglobulin V gene mutations. Blood. 2004;103:375–382. doi: 10.1182/blood-2003-04-1345. [DOI] [PubMed] [Google Scholar]
- 38.Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 39.Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. J Exp Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cutrona G, Colombo M, Matis S, et al. Clonal heterogeneity in CLL cells: response to surface IgM crosslinking is more efficient in CD38, ZAP-70 positive cells. Haematologica. 2007 doi: 10.3324/haematol.11646. In press. [DOI] [PubMed] [Google Scholar]
- 41.Hulkkonen J, Vilpo L, Hurme M, Vilpo J. Surface antigen expression in chronic lymphocytic leukemia: clustering analysis, interrelationships and effects of chromosomal abnormalities. Leukemia. 2002;16:178–185. doi: 10.1038/sj.leu.2402363. [DOI] [PubMed] [Google Scholar]
- 42.Vilpo J, Tobin G, Hulkkonen J, et al. Mitogen induced activation, proliferation and surface antigen expression patterns in unmutated and hypermutated chronic lymphocytic leukemia cells. Eur J Haematol. 2005;75:34–40. doi: 10.1111/j.1600-0609.2005.00443.x. [DOI] [PubMed] [Google Scholar]
- 43.Simms PE, Ellis TM. Utility of flow cytometric detection of CD69 expression as a rapid method for determining poly- and oligoclonal lymphocyte activation. Clin Diagn Lab Immunol. 1996;3:301–304. doi: 10.1128/cdli.3.3.301-304.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiow LR, Rosen DB, Brdickova N, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 45.Kishimoto TK, Jutila MA, Butcher EC. Identification of a human peripheral lymph node homing receptor: a rapidly down-regulated adhesion molecule. Proc Natl Acad Sci U S A. 1990;87:2244–2248. doi: 10.1073/pnas.87.6.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y, Agematsu K, Ochs HD, Sugane K. Functional analysis of human memory B-cell subpopulations: IgD+CD27+ B cells are crucial in secondary immune response by producing high affinity IgM. Clin Immunol. 2003;108:128–137. doi: 10.1016/s1521-6616(03)00092-5. [DOI] [PubMed] [Google Scholar]
- 47.Borst J, Hendriks J, Xiao Y. CD27 and CD70 in T cell and B cell activation. Curr Opin Immunol. 2005;17:275–281. doi: 10.1016/j.coi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 48.Nolz JC, Tschumper RC, Pittner BT, Darce JR, Kay NE, Jelinek DF. ZAP-70 is expressed by a subset of normal human B-lymphocytes displaying an activated phenotype. Leukemia. 2005;19:1018–1024. doi: 10.1038/sj.leu.2403726. [DOI] [PubMed] [Google Scholar]
- 49.Cutrona G, Colombo M, Matis S, et al. B lymphocytes in humans express ZAP-70 when activated in vivo. Eur J Immunol. 2006;36:558–569. doi: 10.1002/eji.200526355. [DOI] [PubMed] [Google Scholar]
- 50.Scielzo C, Camporeale A, Geuna M, et al. ZAP-70 is expressed by normal and malignant human B-cell subsets of different maturational stage. Leukemia. 2006;20:689–695. doi: 10.1038/sj.leu.2404138. [DOI] [PubMed] [Google Scholar]
- 51.Chen L, Widhopf G, Huynh L, et al. Expression of ZAP-70 is associated with increased B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2002;100:4609–4614. doi: 10.1182/blood-2002-06-1683. [DOI] [PubMed] [Google Scholar]
- 52.Chen L, Apgar J, Huynh L, et al. ZAP-70 directly enhances IgM signaling in chronic lymphocytic leukemia. Blood. 2005;105:2036–2041. doi: 10.1182/blood-2004-05-1715. [DOI] [PubMed] [Google Scholar]
- 53.Richardson SJ, Matthews C, Catherwood MA, et al. ZAP-70 expression is associated with enhanced ability to respond to migratory and survival signals in B-cell chronic lymphocytic leukemia (B-CLL). Blood. 2006;107:3584–3592. doi: 10.1182/blood-2005-04-1718. [DOI] [PubMed] [Google Scholar]
- 54.Gobessi S, Laurenti L, Longo PG, Sica S, Leone G, Efremov DG. ZAP-70 enhances B-cell-receptor signaling despite absent or inefficient tyrosine kinase activation in chronic lymphocytic leukemia and lymphoma B cells. Blood. 2007;109:2032–2039. doi: 10.1182/blood-2006-03-011759. [DOI] [PubMed] [Google Scholar]
- 55.Weng NP, Palmer LD, Levine BL, Lane HC, June CH, Hodes RJ. Tales of tails: regulation of telomere length and telomerase activity during lymphocyte development, differentiation, activation, and aging. Immunol Rev. 1997;160:43–54. doi: 10.1111/j.1600-065x.1997.tb01026.x. [DOI] [PubMed] [Google Scholar]
- 56.Igarashi H, Sakaguchi N. Telomerase activity is induced in human peripheral B lymphocytes by the stimulation to antigen receptor. Blood. 1997;89:1299–1307. [PubMed] [Google Scholar]
- 57.Blackburn EH. The end of the (DNA) line. Nat Struct Biol. 2000;7:847–850. doi: 10.1038/79594. [DOI] [PubMed] [Google Scholar]
- 58.Porakishvili N, Kulikova N, Jewell AP, et al. Differential expression of CD180 and IgM by B-cell chronic lymphocytic leukaemia cells using mutated and unmutated immunoglobulin VH genes. Br J Haematol. 2005;131:313–319. doi: 10.1111/j.1365-2141.2005.05775.x. [DOI] [PubMed] [Google Scholar]
- 59.Zucchetto A, Bomben R, Dal Bo M, et al. A scoring system based on the expression of six surface molecules allows the identification of three prognostic risk groups in B-cell chronic lymphocytic leukemia. J Cell Physiol. 2006;207:354–363. doi: 10.1002/jcp.20570. [DOI] [PubMed] [Google Scholar]
- 60.Hultdin M, Rosenquist R, Thunberg U, et al. Association between telomere length and V(H) gene mutation status in chronic lymphocytic leukaemia: clinical and biological implications. Br J Cancer. 2003;88:593–598. doi: 10.1038/sj.bjc.6600763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grabowski P, Hultdin M, Karlsson K, et al. Telomere length as a prognostic parameter in chronic lymphocytic leukemia with special reference to VH gene mutation status. Blood. 2005;105:4807–4812. doi: 10.1182/blood-2004-11-4394. [DOI] [PubMed] [Google Scholar]
- 62.Ricca I, Rocci A, Drandi D, et al. Telomere length identifies two different prognostic subgroups among VH-unmutated B-cell chronic lymphocytic leukemia patients. Leukemia. 2007;21:697–705. doi: 10.1038/sj.leu.2404544. [DOI] [PubMed] [Google Scholar]
- 63.Ghia P, Guida G, Stella S, et al. The pattern of CD38 expression defines a distinct subset of chronic lymphocytic leukemia (CLL) patients at risk of disease progression. Blood. 2003;101:1262–1269. doi: 10.1182/blood-2002-06-1801. [DOI] [PubMed] [Google Scholar]
- 64.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 65.Pittner BT, Shanafelt TD, Kay NE, Jelinek DF. CD38 expression levels in chronic lymphocytic leukemia B cells are associated with activation marker expression and differential responses to interferon stimulation. Leukemia. 2005;19:2264–2272. doi: 10.1038/sj.leu.2403975. [DOI] [PubMed] [Google Scholar]
- 66.Calissano C, Damle R, Banapour T, et al. In vivo labeling of newly synthesized DNA suggests that the CD38+ fraction is enriched in proliferating cells within a clone of chronic lymphocytic leukemia B cells [abstract]. Blood. 2006;108:12a–13a. [Google Scholar]
- 67.Kitada S, Andersen J, Akar S, et al. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with in vitro and in vivo chemoresponses. Blood. 1998;91:3379–3389. [PubMed] [Google Scholar]
- 68.Granziero L, Ghia P, Circosta P, et al. Survivin is expressed on CD40 stimulation and interfaces proliferation and apoptosis in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2777–2783. doi: 10.1182/blood.v97.9.2777. [DOI] [PubMed] [Google Scholar]
- 69.Pepper C, Brennan P, Alghazal S, et al. CD38+ chronic lymphocytic leukaemia cells co-express high levels of ZAP-70 and are functionally distinct from their CD38- counter-parts. Leukemia. 2006;20:743–744. doi: 10.1038/sj.leu.2404133. [DOI] [PubMed] [Google Scholar]
- 70.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]