Abstract
Human MHC class I-related molecules, MICA and MICB, are stress-induced antigens that are recognized by a subset of γδ T cells expressing the variable region Vδ1. This functional association has been found to be limited to intestinal epithelium, where these T cells are prevalent and where MICA and, presumably, MICB are mainly expressed. However, increased frequencies of Vδ1 γδ T cells have been observed in various epithelial tumors; moreover, MICA/B are expressed on diverse cultured epithelial tumor cells. With freshly isolated tumor specimens, expression of MICA/B was documented in many, but not all, carcinomas of the lung, breast, kidney, ovary, prostate, and colon. In tumors that were positive for MICA/B, the frequencies of Vδ1 γδ T cells were significantly higher than in those that were negative. Vδ1 γδ T cell lines and clones derived from different tumors recognized MICA/B on autologous and heterologous tumor cells. In accord with previous evidence, no constraints were observed in these interactions, such as those imposed by specific peptide ligands. Thus, MICA/B are tumor-associated antigens that can be recognized, in an apparently unconditional manner, by a subset of tumor-infiltrating γδ T cells. These results raise the possibility that an induced expression of MICA/B, by conditions that may be related to tumor homeostasis and growth, could play a role in immune responses against tumors.
Cytolytic T cell responses against tumors require the recognition of specific peptides derived from tumor antigens in association with MHC class I molecules by CD8+ T cells expressing αβ T cell receptors (TCRs). Such responses are limited by antigenic peptides (1), which must be generated by intracellular antigen processing (2), by their highly selective binding to only some of the numerous polymorphic MHC class I molecules (3), and by the often-impaired expression of MHC class I on tumor cells (4). By contrast, T cells expressing γδ TCRs (5, 6) recognize antigens directly, with no requirement for antigen processing or presentation (7–9). Recently identified ligands for human γδ T cells are MICA and MICB, which are distantly related to MHC class I but are functionally distinct. These molecules seem to have no role in the presentation of intracellular peptide antigens; instead, they may function as self-antigens that can be stress-induced (10). MICA and MICB are closely related and functionally indistinguishable (10–12). Both are recognized by cytotoxic γδ T cells expressing the TCR variable region Vδ1. Although the recognition of MICA/B appears to be mediated by TCR engagement, these interactions are unusual because T cells expressing diverse γ and δ chain Vδ1(D)J junctional sequences are capable of recognizing these molecules (10). This common antigen specificity may enable a subset of Vδ1 γδ T cells, which are relatively infrequent and endowed with a limited TCR repertoire, to respond uniformly to expression of MICA/B as a generic tissue distress signal that may be triggered by infection, noxious conditions, or transformation.
Expression of MICA and, presumably, of MICB in normal tissues is restricted mainly to intestinal epithelium (13). This limited distribution correlates with increased frequencies of Vδ1 γδ T cells in this site (14–16), which is opposed to their usually low numbers in lymphoid organs and peripheral blood (17). However, increased frequencies of these T cells also have been found in various epithelial tumors, including lung, colon, and renal cell carcinomas (18–21). Moreover, we have observed that many cell lines of diverse epithelial tumor origins express MICA/B (refs. 10 and 13; V.G. and T.S., unpublished data). Hence, this evidence led us to investigate a potential role of these molecules as tumor-associated antigens. As reported here, expression of MICA/B was documented in many freshly isolated specimens of carcinomas of the lung, breast, kidney, ovary, prostate, and colon. Significantly increased frequencies of Vδ1 γδ T cells were found in tumors that were positive for MICA/B, and Vδ1 γδ T cell lines and clones derived from such tumors recognized these molecules on diverse tumor cell lines and on freshly isolated heterologous and autologous tumor cells. These results provide a basis for further investigation of the biology underlying the tumor-associated expression of MICA/B and of their potential significance as antigens for Vδ1 γδ T cells in an immune surveillance of tumors.
MATERIALS AND METHODS
Tumor and Lymphocyte Cell Suspensions and Flow Cytometry.
Specimens of carcinomas (of breast, lung, ovary, prostate, colon, and kidney) were from resections undertaken for diagnostic or therapeutic purposes at various institutions in the United States and Brazil and were provided by Corixa Corp. (Seattle, WA). The tumor specimens were diagnosed by histopathological and immunohistochemical criteria and contained only malignant epithelium. Tumor samples were minced, enzymatically digested for 12 h [using collagenase type III (100 μg/ml), dispase (550 μg/ml), and DNase I (200 units/ml)] and sieved through wire mesh. Cell suspensions were centrifuged in differential 100% and 75% Ficoll/Hypaque (Pharmacia) gradients. Tumor cell and lymphocyte fractions, collected from separate gradient interfaces, that were of at least 80% purity as judged by microscopy were cryopreserved until use. Thawed and washed tumor cell suspensions were stained with mAb 6D4 (anti-MICA/B) or 2C10 (anti-MICA) or with Ig isotype-matched control antibody and examined by indirect immunofluorescence by using a FACStar flow cytometer (Becton Dickinson).
Cell-Surface Labeling and Immunoprecipitation.
These procedures were carried out exactly as described, by using biotinylation of cells with Sulfo-NHS-LC-biotin (Pierce) (13). Aliquots of immunocomplexes were treated with N-glycanase (PNGase F; New England Biolabs) as recommended by the manufacturer, separated by SDS/PAGE, electroblotted onto nitrocellulose, and reacted with avidin-horseradish peroxidase (Vector Laboratories) (13).
Immunohistochemistry.
Cryosections of diagnosed solid tumor specimens and of normal tissues embedded in OCT compound (Baxter Scientific Products, McGaw Park, IL) were air-dried, fixed in cold acetone, quenched with H2O2, treated with 5% goat serum in PBS, and incubated with a titered dilution of mAb 6D4 (anti-MICA/B), mAb 35βH11 (anti-cytokeratin; Dako), or isotype-matched control Ig. Antibody binding was visualized by using biotinylated goat anti-mouse Ig (Dako), peroxidase-conjugated streptavidin (Zymed), and diaminobenzidine (Polysciences) or the Envision System (Dako). The images shown in Fig. 2 were generated with the latter procedure. Slides were counterstained with hematoxylin, dehydrated in graded alcohols, and mounted in xylene. Tissue specimens were from Corixa Corp. and the Department of Pathology, Fred Hutchinson Cancer Research Center. Their use was conducted under protocols approved by the institutional review board at the Fred Hutchinson Cancer Research Center. Formalin-fixed and paraffin-embedded specimen sections of breast carcinomas and control tissue from reduction surgery were provided by PhenoPath Laboratories (Seattle) and processed with the Envision System as described by the manufacturer (Dako).
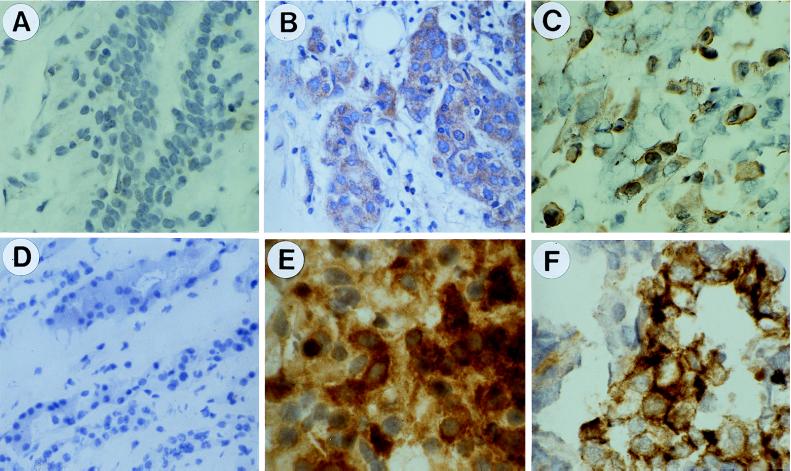
Figure 2.
Immunohistochemical detection of MICA/B in epithelial tumors by staining with mAb 6D4. (A) Normal breast tissue. (B) Breast carcinoma. (C) Lung carcinoma. (D) Normal kidney. (E) Renal cell carcinoma. (F) Ovary carcinoma. Sections in A and B and C–F were from paraffin-embedded and frozen specimens, respectively. Nuclei were counterstained with hematoxylin (see Materials and Methods). These images were representative of stainings of numerous control tissue and tumor specimens. See text for further explanation.
Cell Lines, Transfectants, and Antibodies.
The tumor cell lines were from the American Type Culture Collection. Transfectants of C1R cells expressing MICA or MICB have been described (10, 13). The mAbs 2C10 and 6D4 are specific for MICA and MICA/B, respectively, as shown by staining and flow cytometry of various transfectants and of heat-shock-induced and uninduced epithelial cell lines (10, 13); the specificity of mAb 2C10 has been documented further by immunoprecipitation of MICA from lysates of metabolically labeled transfectants (13). Both mAbs inhibit Vδ1 γδ T cell responses against target cells expressing MICA or MICB (10). The epitopes recognized by these mAbs are within the α1α2 domains of MICA/B, as shown with transfectants expressing mouse H-2D or K hybrid molecules in which various domains had been replaced by the corresponding sequences of MICA (ref. 13; V.G. and T.S., unpublished data). Antibodies used for phenotyping or sorting of tumor T cell populations, T cell lines, and clones were biotinylated or FITC-conjugated anti-CD3, anti-TCR-α/β-1, anti-TCR-γ/δ-1 (Becton Dickinson), anti-PanTCRγδ, anti-Vδ2, and mAb δTCS1 (anti-Vδ1) (Endogen, Cambridge, MA). Second-layer reagents were phycoerythrin-streptavidin (Becton Dickinson) or FITC-F(ab′)2 goat anti-mouse Ig conjugates (BioSource International, Camarillo, CA).
T Cell Lines and Clones, Cytotoxicity, and IFN-γ Release Assays.
Frozen lymphocyte preparations from tumors were thawed, washed, and seeded in 96-well round-bottom plates (103 cells per well) and cultured with γ-irradiated C1R-MICA transfectants (2 × 104 cells per well) in RPMI 1640 medium with 8% FBS, 2% pooled human serum, rhIL-2 (2 units/ml; Chiron), rhIL-7 (10 ng/ml; a gift of N. Vita, Sanofi Recherche, France), phytohemagglutinin (0.5 μg/ml; Difco), glutamine, and antibiotics (10). Vδ1 γδ T cell lines were sorted from pooled lymphocytes after 1 week by using mAb δTCS1 and a FACS VANTAGE cell sorter (Becton Dickinson) and grown as above with weekly restimulations. After 3 weeks, T cells were phenotyped by antibody stainings and functionally tested. Long-term T cell cultures were in the additional presence of irradiated allogeneic peripheral blood mononuclear cells. T cell clones were derived after the first week of lymphocyte bulk culture by sorting directly into 96-well plates (0.5 cells per well). The αβ T cell lines and the ovary tumor-derived Vδ2 γδ T cell line and clones were seeded by sorting, by using mAbs anti-TCR-α/β-1 and anti-Vδ2, respectively, and grown under the same conditions. The chromium-release assays and treatment of target cells with mAb 2C10 or mAb 6D4 were as described (10). Secretion of IFN-γ was measured with equal numbers of T cells and irradiated mock C1R cells or stimulator C1R-MICA transfectants, or autologous or allogeneic tumor cells that were positive for MICA/B (each, 105 irradiated cells and T cells per well). Phorbol myristate acetate (1 ng/ml; Sigma) and rhIL-2 (2 units/ml; Chiron) were included. Cytokine levels were determined after 48 h by ELISA with matched antibody in relation to cytokine standard pairs (R & D Systems). For inhibition experiments, stimulator cells were mixed with titered amounts of mAb 6D4 or Ig isotype-matched control mAb shortly before addition of T cells.
Reverse Transcription–PCR and cDNA Sequence Analysis.
RNA was prepared from T cell clones by using STAT-60 reagent (Tel-Test, Friendswood, TX) and cDNA synthesized with oligo(dT) and avian myeloblastosis virus reverse transcriptase (Promega). TCR-δ chain sequences were amplified by PCR by using Taq polymerase (Pharmacia) and specific primers for Vδ1 (5′-CCGTCGACGTCAACTTCAAGAAAGCAGC-3′) and Cδ [biotin-(5′-GTAGAATTCCTTCACCAGAC-3′)] in 30 cycles (30 s at 95°C, 20 s at 56°C, and 60 s at 72°C) (10). PCR products were sequenced directly by using a dye terminator cycle-sequencing kit (Perkin–Elmer). Because of space limitations, the sequences are not shown but will be made available on request.
RESULTS AND DISCUSSION
Tumor-Associated Expression of MICA and MICB.
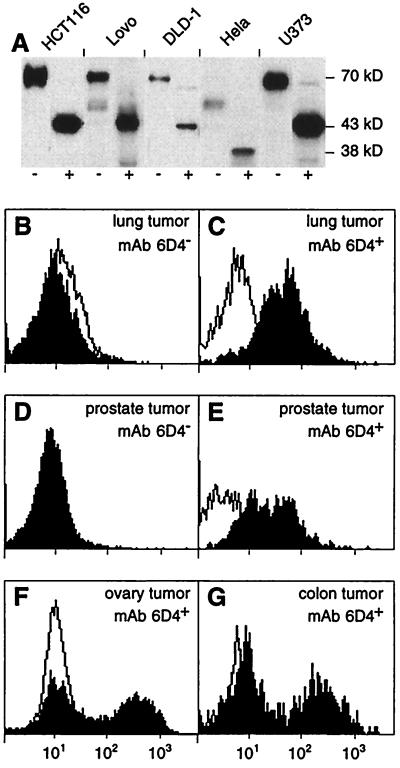
Expression of MICA/B in solid epithelial tumors was tested with cell suspensions prepared from a number of freshly isolated tumor specimens by using flow cytometry and the mAbs 6D4 and 2C10, which bind to epitopes on the α1α2 domains of MICA/B and MICA, respectively. The reactivity of these mAbs has been documented by flow cytometry by using various transfectants and stress-induced or uninduced tumor cell lines (mAbs 6D4 and 2C10) and by immunoprecipitation of metabolically labeled MICA (mAb 2C10) (10, 13). With epithelial tumor cell lines, the specificity of mAb 2C10 was supported further by immunoprecipitation of surface-labeled MICA (Fig. 1A). The tumor specimens examined were diagnosed by standard histopathological and immunohistochemical criteria and contained only malignant epithelium. By cytokeratin staining, the purified tumor suspensions consisted of at least 80% epithelial cells, which, by inference, were tumor cells. Significant proportions of cells positive for binding of mAb 6D4 were seen in some, but not all, samples of breast (2/10), lung (2/6), ovary (5/6), prostate (2/4), renal cell (2/2), and colon (4/5) carcinomas. Representative fluorescence profiles are shown in Fig. 1 C and E–G. The staining intensities were heterogeneous, with about 40–70% of cells showing intermediate to high levels of expression of MICA/B. Similar results were obtained with mAb 2C10, except each one of the colon and renal cell carcinoma suspensions that were positive for binding of mAb 6D4 were not reactive with mAb 2C10. With both mAbs, negative tumor cell populations showed no more staining than with isotype-matched control Ig (Fig. 1 B, D, F, and G).
Figure 1.
Surface expression of MICA on epithelial tumor cell lines and of MICA/B on freshly isolated tumor cells. (A) Immunoprecipitation of MICA with mAb 2C10 from lysates of surface-biotinylated HCT116, Lovo, DLD-1 (colon carcinomas), Hela (cervical carcinoma), and U373 (astrocytoma) tumor cell lines. Immunocomplexes were left untreated (−) or were treated (+) with N-glycanase before SDS/PAGE. Labeled proteins on immunoblots were visualized with avidin-horseradish peroxidase. MICA is heavily glycosylated and has a polypeptide backbone of 43 kDa (11, 13). These results were in accord with previous data obtained with the anti-MICA mAb 56 by using transfectants and epithelial cell lines (13). Hela cells are homozygous for a truncated variant of MICA of 38 kDa that lacks the cytoplasmic tail. This variant corresponds to allele 008, as determined by reverse transcription–PCR and direct cDNA sequencing (unpublished data) (24). (B–G) Tumor cell suspensions prepared from lung (B and C), prostate (D and E), ovary (F), and colon carcinomas (G) were stained with mAb 6D4 (solid profiles) or with isotype-matched Ig (open profiles) and examined by flow cytometry. B and D show examples of tumor cell preparations that were negative for binding of mAb 6D4 whereas C and E–G show positive stainings. These data are representative of the range of staining intensities seen with all of the tumor samples tested. Similar results were obtained with mAb 2C10.
These results were refined by immunohistochemistry by using mAb 6D4 and sections made from frozen or formalin-fixed tumor and control tissue specimens. In an initial, extensive tissue screen, no staining was observed in skin, lung, tonsil, liver, kidney, adrenal gland, ovary, and prostate, whereas intestinal epithelium was consistently positive. Also negative were peripheral blood mononuclear cells examined by flow cytometry (data not shown). With breast tumors, however, sections from 7 of 20 different tumor specimens gave positive stainings of pathologically discernible tumor cells, which was contrasted by the absence of staining of connective tissue (Fig. 2B). By contrast, all of the six paired and seven unpaired normal tissue controls showed no significant staining (Fig. 2A). Moreover, each two of two lung and renal cell, two of three ovary, and two of two prostate carcinomas were positive, whereas the tissue-matched unpaired controls (in numbers of 3, 3, 1, and 2, respectively) were negative (Fig. 2 C–F; data not shown). Thus, together with the flow cytometry data, these results indicated that MICA/B were expressed in substantial numbers of epithelial tumors with no restriction to tumor type.
Relative Frequencies of Tumor-Derived Vδ1 γδ T Cells and Recognition of MICA/B.
We investigated relationships between tumor expression of MICA/B and their potential function as T cell antigens. Among the lymphocytes extracted from 13 tumor specimens (2 breast, 2 lung, 3 ovary, 2 prostate, 1 renal cell, and 3 colon) that were positive for binding of mAb 6D4 by cell-suspension staining, the frequencies of Vδ1 γδ T cells (detected with mAb δTCS1) among all γδ T cells (detected with mAb anti-TCR-γ/δ-1) were 35–100% (mean 63%) (Table 1). By contrast, among nine tumor specimens (six breast, two lung, and one prostate) that were negative for staining with mAb 6D4, the frequencies of Vδ1 γδ T cells were 2–40% (mean 17%). With all tumors, the proportions of γδ T cells of CD3+ T cells were 1–16% (mean 4.4%), within the normal range (0.5–15%; mean 5%) defined previously in peripheral blood (17) (Table 1). Thus, the frequencies of Vδ1 γδ T cells in proportion to all γδ T cells correlated positively with tumor expression of MICA/B. These frequencies were statistically significantly higher (P < 0.0001, Wilcoxon rank-sum test) than in the tumors that were negative for MICA/B. Thus, these results provided indirect support for a functional association between tumor-derived Vδ1 γδ T cells and their putative antigens, MICA/B.
Table 1.
Proportions (in %) of Vδ1 γδ T cells of all γδ T cells in tumors positive or negative for MICA/B and of γδT cells (in %) of CD3+ T cells
| Tumor | mAb 6D4 | % γδT cells | % Vδ1 γδ cells |
|---|---|---|---|
| CT-001-97 | + | 3 | 80 |
| CT-046-98 | + | 15 | 46 |
| CT-183-73 | + | 7 | 67 |
| BT-046-99 | − | 2 | 25 |
| BT-086-22 | − | 3 | 40 |
| BT-086-38 | − | 2 | 21 |
| BT-140-73 | − | 3 | 9 |
| BT-183-26 | − | 1 | 10 |
| BT-183-43 | − | 4 | 20 |
| BT-183-76 | + | 4 | 51 |
| BT-238-29 | + | 16 | 100 |
| LT-086-36 | − | 2 | 2 |
| LT-086-49 | − | 2 | 2 |
| LT-140-26 | + | 2 | 60 |
| LT-140-66 | + | 1 | 55 |
| OT-183-83 | + | 2 | 80 |
| OT-183-93 | + | 2 | 51 |
| OT-238-01 | + | 4 | 35 |
| PT-140-63 | + | 6 | 65 |
| PT-183-87 | − | 2 | 20 |
| PT-391-97 | + | 10 | 55 |
| RT-046-29 | + | 4 | 70 |
Representative examples for tumor cell suspensions positive or negative for binding of mAb 6D4 are shown in Fig. 1. The frequencies of Vδ1 γδ T cells of all γδ T cells in MICA/B+ tumors were statistically significantly higher (P < 0.0001, Wilcoxon rank-sum test) as compared with the tumors that were MICA/B−. All samples were positive for binding of mAb W6/32 (anti-HLA-A, -B, and -C). Examined were carcinomas of colon (CT), breast (BT), lung (LT), ovary (OT), prostate (PT), and renal cell (RT).
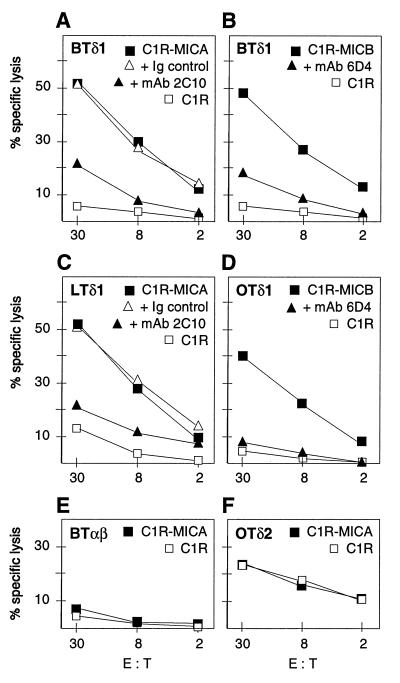
For functional testing, T cell lines were established from carcinomas of the breast, lung, ovary, and colon. The tumor specimens selected were positive for MICA/B by cell-suspension stainings with the mAbs 6D4 and 2C10. Extracted and purified bulk lymphocytes were cultured in the presence of cytokines and irradiated C1R transfectants expressing MICA. Vδ1 γδ T cells were isolated after 1 week of culture by cell sorting and expanded. In parallel, CD8+ αβ T cell lines and a Vδ2 γδ T cell line were established from the same lymphocyte preparations derived from the four tumors and from the ovary carcinoma, respectively. The surface-marker phenotypes of the T cell lines were confirmed after 3 weeks of expansion (data not shown). In functional assays, all of the four Vδ1 γδ T cell lines (BTδ1, LTδ1, OTδ1, and CTδ1) were cytotoxic against C1R-MICA and C1R-MICB transfectants but not against untransfected C1R cells. Selected data representative of multiple experiments with these T cell lines are shown in Fig. 3 A–D. These responses were inhibited when target cells were treated with mAbs 2C10 or 6D4, whereas addition of isotype-matched control Ig or of the anti-HLA mAb W6/32 as a cell-binding control mAb had no effect (Fig. 3 A–D; data not shown) (22). By contrast, none of the CD8+ αβ T cell lines were cytotoxic against the C1R-MICA or C1R-MICB targets. Fig. 3E shows representative data obtained with the breast tumor-derived T cell line (BTαβ). The Vδ2 γδ T cell line was cytotoxic against C1R cells, as has been noted previously with Daudi cells (23), but there was no enhanced activity against the C1R-MICA transfectants (Fig. 3F). No unspecific T cell inhibition was observed in the presence of mAb 6D4 (data not shown). Thus, these results supported previous evidence indicating that T cell recognition of MICA/B was confined to the Vδ1 subset of γδ T cells and showed that T cells with this reactivity could be derived from tumors in which MICA/B were expressed.
Figure 3.
Specific recognition of MICA/B on C1R transfectants by the Vδ1 γδ T cell lines BTδ1, LTδ1, and OTδ1 derived from carcinomas of breast, lung, and ovary, respectively (A–D). The data from chromium release assays shown were representative of numerous experiments with these T cell lines and others derived from prostate and colon carcinomas, which all were tested against the C1R-MICA and C1R-MICB targets. Cytotoxicity was inhibited in the presence of mAb 2C10 (anti-MICA) or mAb 6D4 (anti-MICA and -MICB) but not with control Ig. (E) No responses were seen with the tumor-derived CD8+ αβ T cell lines; as one example, data obtained with the breast tumor-derived BTαβ line are shown. (F) The ovary tumor-derived Vδ2 γδ T cell line (OTδ2) showed no increased response against C1R-MICA targets. E:T, effector-to-target cell ratio. See text for further explanation.
Recognition of MICA and MICB on Heterologous and Autologous Tumor Cells.
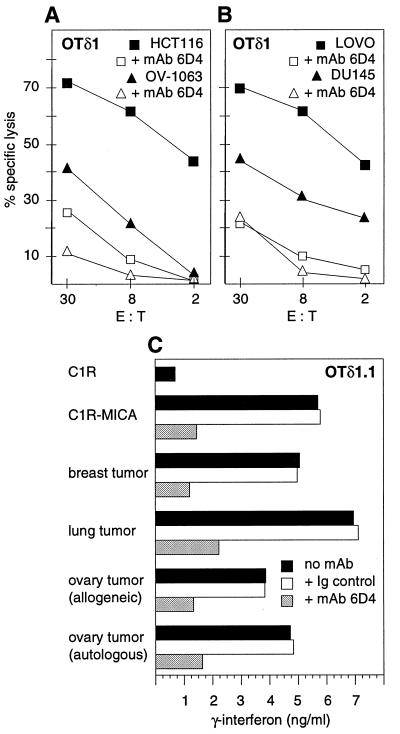
The potential relevance and limitations of these T cell responses were tested by using tumor cell lines and freshly isolated tumor cells. The ovary tumor-derived OTδ1 T cell line was cytolytic against a panel of unrelated tumor cell lines derived from ovary (OV-1063 and SW626), prostate (DU145 and PC-3), colon (HCT116, Lovo, DLD-1, and SW480), liver (HepG2), and cervical (Hela) carcinomas that were positive for expression of MICA/B by staining with mAbs 6D4 and 2C10 and/or by immunoprecipitation of MICA with mAb 2C10 from lysates of surface-labeled cells (Figs. 1A and 4 A and B; data not shown). In all cases, the T cell responses were inhibited by mAb 6D4 but not by isotype-matched control Ig. The selected data shown in Fig. 4 A and B also were representative of additional experiments in which the BTδ1 and LTδ1 T cells derived from the breast and lung tumors, respectively, were tested. Corresponding results were obtained with two Vδ1 γδ T cell clones from different ovary tumors, which were assayed for IFN-γ release after 48 h of stimulation with breast, lung, or autologous or allogeneic ovary tumor cells. These tumor cells were from cell-suspension preparations that were stored frozen and were thawed and used directly, without prior exposure to culture medium; they were selected for positive staining with mAb 6D4. The measured responses were similar to those that were seen with the C1R-MICA transfectants but not with C1R cells and were all inhibited in the presence of mAb 6D4 (Fig. 4C). Thus, MICA/B were recognized on autologous and diverse heterologous tumor cells with no apparent specificity constraints associated with any of the many different tumor cells tested.
Figure 4.
Vδ1 γδ T cells recognized MICA/B on epithelial tumor cell lines and on heterologous and autologous tumor cells. (A and B) Cytotoxicity of the ovary tumor-derived OTδ1 T cell line against HCT116 and Lovo (colon carcinoma cell lines), OV-1063 (ovary carcinoma cell line), and DU145 (prostate carcinoma line) was inhibited by mAb 6D4. Addition of isotype-matched control Ig had no effect (data not shown). The target cell lines expressed MICA/B by staining with mAb 6D4 (ref. 10; data not shown). Similar results were obtained with other tumor-derived Vδ1 γδ T cell lines and additional tumor cell line targets (see text). E:T, effector-to-target cell ratio. (C) MICA/B-induced release of IFN-γ by the OTδ1.1 T cell clone was inhibited in the presence of mAb 6D4 but not of isotype-matched control Ig. Release of IFN-γ (ng/ml) after 48 h of T cell stimulation was quantitated by ELISA (see Materials and Methods). Stimulator cells were C1R-MICA transfectants or tumor cell preparations from breast, lung, or heterologous or autologous ovary carcinomas. Similar results were obtained with another Vδ1 γδ T cell clone derived from a different ovary tumor.
The diversity of the Vδ1 γδ T cells capable of recognizing MICA/B was examined after direct seeding of clones by sorting from the tumor-derived bulk lymphocytes and expansion. The clones selected for δ-chain cDNA sequencing showed significant cytotoxic activities against C1R-MICA targets. The 38 sequences derived from the breast, lung, ovary, and colon carcinomas included 12 different δ-chains that were allocated in numbers of 6 of 10, 2 of 11, 3 of 7, and 1 of 10, respectively, and showed no apparent junctional region similarities (data not shown; see Materials and Methods). Because paired peripheral blood samples were not available for analysis, it was not possible to examine whether these oligoclonal sequences were associated with selective expansion or retention of the Vδ1 γδ T cells in tumor sites. However, in accord with previous data (10), these results indicated that MICA/B can be recognized by a substantial subset of Vδ1 γδ T cells that remains to be defined.
CONCLUSIONS
The data presented show that MICA/B, which, under normal conditions, have a limited tissue distribution in intestinal epithelium, are expressed in many epithelial tumors independent of tumor type. This raises the question of whether a similar regulation may underlie this shared expression. Cultured epithelial tumor cells express intermediate to large amounts of MICA/B, which is associated with proliferation and is independent of cellular stress (10, 13). With nonproliferating cells grown at high confluency, however, the expression levels are low but can be increased strongly by heat shock and, presumably, other types of stress (10). Thus, tumor-associated expression of MICA/B could be induced by conditions related to tumor homeostasis and growth. It will be of interest to identify tumor-biology parameters and molecular markers that may be associated positively or negatively with expression of MICA/B.
The results confirm and extend previous evidence that indicated a specific functional association between T cells in the Vδ1 γδ T cell subset and MICA/B. This is supported further by the increased numbers of these T cells in tumors expressing these molecules. The recognition of MICA/B on all of the diverse tumor cells tested argues against an association of these molecules with self-antigen-derived peptide or nonpeptide ligands, which is in agreement with the previous model suggesting a function as self-antigens (10). It is apparent that the Vδ1 γδ T cell responses share similarities with those of natural killer cells. However, although the latter usually are inhibited by binding of inhibitory receptors to MHC class I molecules on target cells, we have not observed similar effects with the Vδ1 γδ T cells. Further studies will be required to determine the in vivo activation phenotypes of these T cells and to define the signaling and costimulatory molecules that are necessary for triggering the cytolytic response. In addition, a link between TCR-γ and -δ chain sequences and the recognition of MICA/B remains to be firmly established.
The physiological role and in vivo effector functions of Vδ1 γδ T cells are unknown, and no antigens except MICA/B have been identified. However, the tumor-associated expression of MICA/B and their apparently unconditional recognition by these T cells, independent of specific processing and ligand association requirements, suggest potential functions as components of an innate immune surveillance of tumors.
Acknowledgments
We thank M. Beauchamp, A. Gown, and A. Steinle for experimental help, T. Gooley for the statistical analysis, and L. Lanier for helpful comments. This work was supported by National Institutes of Health Grants AI30581 and CA18221.
ABBREVIATIONS
- MIC
MHC class I-related chain
- TCR
T cell receptor
References
- 1.Van den Eynde B J, van der Bruggen P. Curr Opin Immunol. 1997;9:684–693. doi: 10.1016/s0952-7915(97)80050-7. [DOI] [PubMed] [Google Scholar]
- 2.Pamer E, Cresswell P. Annu Rev Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 3.Rammensee H-G, Falk K, Ràtzschke O. Annu Rev Immunol. 1993;11:213–244. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- 4.Ferrone S, Marincola F M. Immunol Today. 1995;16:487–494. doi: 10.1016/0167-5699(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 5.Allison J P, Raulet D H. Semin Immunol. 1990;2:59–65. [PubMed] [Google Scholar]
- 6.Porcelli S, Brenner M B, Band H. Immunol Rev. 1991;120:137–183. doi: 10.1111/j.1600-065x.1991.tb00591.x. [DOI] [PubMed] [Google Scholar]
- 7.Rock E P, Sibbald P R, Davis M M, Chien Y-h. J Exp Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schild H, Mavaddat N, Litzenberger C, Ehrich E W, Davis M M, Bluestone J A, Matis L, Draper R K, Chien Y H. Cell. 1994;76:29–37. doi: 10.1016/0092-8674(94)90170-8. [DOI] [PubMed] [Google Scholar]
- 9.Morita C T, Beckman E M, Bukowski J F, Tanaka Y, Band H, Bloom B R, Golan D E, Brenner M B. Immunity. 1995;3:495–507. doi: 10.1016/1074-7613(95)90178-7. [DOI] [PubMed] [Google Scholar]
- 10.Groh V, Steinle A, Bauer S, Spies T. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 11.Bahram S, Bresnahan M, Geraghty D E, Spies T. Proc Natl Acad Sci USA. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahram S, Spies T. Immunogenetics. 1996;43:230–233. doi: 10.1007/BF00587305. [DOI] [PubMed] [Google Scholar]
- 13.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spencer J, Isaacson P G, Diss T C, MacDonald T T. Eur J Immunol. 1989;19:1335–1338. doi: 10.1002/eji.1830190728. [DOI] [PubMed] [Google Scholar]
- 15.Deusch K, Luling F, Reich K, Classen M, Wagner H, Pfeffer K. Eur J Immunol. 1991;21:1053–1059. doi: 10.1002/eji.1830210429. [DOI] [PubMed] [Google Scholar]
- 16.Chowers Y, Holtmeier W, Harwood J, Morzycka-Wroblewska E, Kagnoff M F. J Exp Med. 1994;180:183–190. doi: 10.1084/jem.180.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker C M, Groh V, Band H, Porcelli S A, Morita C, Fabbi M, Glass D, Strominger J L, Brenner M B. J Exp Med. 1990;171:1597–1612. doi: 10.1084/jem.171.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zocchi M R, Ferrarini M, Rugarli C. Eur J Immunol. 1990;20:2685–2689. doi: 10.1002/eji.1830201224. [DOI] [PubMed] [Google Scholar]
- 19.Zocchi M R, Ferrarini M, Migone N, Casorati G. Immunology. 1994;81:234–239. [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhary A F, Davodeau F, Moreau A, Peyrat M-A, Bonneville M, Jotereau F. J Immunol. 1995;154:3932–3940. [PubMed] [Google Scholar]
- 21.Maeurer M J, Martin D, Walter W, Liu K, Zitvogel L, Halusczcak K, Rabinovich H, Duquesnoy R, Storkus W, Lotze M T. J Exp Med. 1996;183:1681–1696. doi: 10.1084/jem.183.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parham P, Barnstable C J, Bodmer W F. J Immunol. 1979;123:342–349. [PubMed] [Google Scholar]
- 23.Fisch P, Oettel K, Fudim N, Surfus J E, Malkovsky M, Sondel P M. J Immunol. 1992;148:2315–2323. [PubMed] [Google Scholar]
- 24.Fodil N, Laloux L, Wanner V, Pellet P, Hauptmann G, Mizuki N, Inoko H, Spies T, Theodorou I, Bahram S. Immunogenetics. 1996;44:351–357. doi: 10.1007/BF02602779. [DOI] [PubMed] [Google Scholar]