Abstract
The human multidrug-resistance protein (MRP) gene family contains at least six members: MRP1, encoding the multidrug-resistance protein; MRP2 or cMOAT, encoding the canalicular multispecific organic anion transporter; and four homologs, called MRP3, MRP4, MRP5, and MRP6. In this report, we characterize MRP3, the closest homolog of MRP1. Cell lines were retrovirally transduced with MRP3 cDNA, and new monoclonal antibodies specific for MRP3 were generated. We show that MRP3 is an organic anion and multidrug transporter, like the GS-X pumps MRP1 and MRP2. In Madin–Darby canine kidney II cells, MRP3 routes to the basolateral membrane and mediates transport of the organic anion S-(2,4-dinitrophenyl-)glutathione toward the basolateral side of the monolayer. In ovarian carcinoma cells (2008), expression of MRP3 results in low-level resistance to the epipodophyllotoxins etoposide and teniposide. In short-term drug exposure experiments, MRP3 also confers high-level resistance to methotrexate. Neither 2008 cells nor Madin–Darby canine kidney II cells overexpressing MRP3 showed an increase in glutathione export or a decrease in the level of intracellular glutathione, in contrast to cells overexpressing MRP1 or MRP2. We discuss the possible function of MRP3 in (hepatic) physiology and its potential contribution to drug resistance of cancer cells.
Keywords: multidrug resistance, methotrexate, etoposide
Two members of the large family of ABC transporters are known thus far to confer multidrug resistance in human cancer cells. These are the MDR1 P-glycoprotein (1) and the multidrug-resistance protein MRP1 (2). Both membrane proteins transport a wide range of drugs with different cellular targets and confer resistance by decreasing the intracellular concentration of drugs. P-glycoprotein transports these drugs in unmodified form, whereas MRP1 can transport drugs either conjugated to anionic ligands such as glutathione (GSH), glucuronide, or sulfate, or in an unmodified form, possibly together with GSH. Well known substrates for MRP1 are, for example, cysteinyl leukotrienes, glutathione disulfide, S-(2,4-dinitrophenyl-)glutathione, ethacrynic acid S-glutathione, etoposide glucuronide, certain steroid glucuronides, and bile salt derivatives (3–6). Transporters with the characteristics of MRP1 are known as GS-X pumps (7) or multispecific organic anion transporters (8).
Another GS-X pump is MRP2, a homolog of MRP1. Unlike MRP1, which is nearly ubiquitously expressed (9), MRP2 is present mainly in the canalicular membrane of hepatocytes (10), but is also present in other apical domains of polarized cells such as the epithelial cells of the proximal tubules of the kidney (11). Studies with mutant rats (TR−/GY or EHBR), which lack the MRP2 protein in the canalicular membrane of hepatocytes, have shown that the substrate specificity of MRP2 is very similar to that of MRP1 (12, 13). MRP2 also contributes to transport of anti-cancer drugs and some metals. The mutant rats showed a reduced biliary clearance of methotrexate (14), of the topoisomerase I inhibitor CPT-11 and its metabolites (15), and of mercury, cadmium, and arsenite (refs. 16 and 17; R.O.E., unpublished observation). Cells transduced with an MRP2 cDNA construct transport the cytostatic drug vinblastine (18). Moreover, overexpression of the MRP2 gene has been found in several cisplatin-resistant cell lines (19, 20), and transfection of an MRP2-antisense construct into liver cells was reported to confer an increased sensitivity to cytotoxic drugs (21). All these observations strongly suggest that MRP2 may confer multidrug resistance, but whether it does so in cancer patients remains to be established.
Besides MRP1 and MRP2, there are at least four MRP homologs expressed in humans, called MRP3, MRP4, MRP5, and MRP6 (20, 22). Not much is known about the substrate specificity of these putative new transporters. We found MRP3, MRP5, and MRP6, but not MRP4, overexpressed in some drug-resistant cell lines, but no correlation was found thus far between their expression and the resistance of these cells (20, 22). Overexpression of the MRP6 gene in MDR cells is invariably associated with the amplification of the adjacent MRP1 gene and MRP6 probably does not contribute to resistance (22). To examine the role of these putative transporters in further detail, their substrate specificity, cellular location, and physiological functions have to be determined.
Of all the MRP family members, MRP3 is closest to MRP1 (58% amino acid identity). In a previous report, we have shown that MRP3 is mainly expressed in the liver, colon, intestine, and adrenal gland, and to a lower extent in several other tissues (20). With monoclonal antibodies we show here that in liver MRP3 is mainly expressed in bile-duct epithelial cells and to a lower extent also in hepatocytes. To study the properties of the MRP3 protein, we cloned the MRP3 cDNA and retrovirally transduced this into various nonpolarized and polarized cell lines. We show here that MRP3 is, like MRP1 and MRP2, also an organic anion transporter and is able to confer resistance to the anti-cancer drugs methotrexate, etoposide, and teniposide.
MATERIALS AND METHODS
Materials.
1-Chloro-2,4-dinitro[14C]benzene ([14C]CDNB; 10 Ci/mol) was obtained from Amersham. Other chemicals and drugs were obtained from Sigma.
Cell Lines.
All cell lines used in this study and their culture conditions have been described before in refs. 18 and 20 and references therein.
Cloning and Sequencing of MRP3 cDNA.
A human liver 5′ stretch-plus cDNA library (CLONTECH) was screened by using a ±1200-bp EcoRI-SacI fragment from EST clone 84966 as a probe. Several overlapping cDNAs were isolated, but at the 5′ end ±600 bp were lacking. This missing 5′ end-of-coding sequence was isolated by using human liver RNA and a RACE protocol (Life Technologies, Grand Island, NY). MRP3 cDNA was sequenced by using the ABI377 automatic sequencer. The sequence has been deposited in GenBank under accession no. AF009670.
Transfections and Protein Analysis.
To make stable MRP3-expressing cell lines, we cloned the MRP3 cDNA behind the cytomegalovirus (CMV) promoter in the retroviral vector pCMV-neo, which contains the bacterial neo gene for selection with G418 (18). pCMV-MRP3 was subsequently transfected into the amphotropic retroviral packaging cell line Phoenix and 48 h after transfection, the level of MRP3 protein was checked with the monoclonal antibody M3II-9. No detectable MRP3 was found. We therefore reduced the number of nucleotides upstream of the ATG start codon to only 13 nucleotides and changed the cytosine at position −3 of the ATG for a more optimal adenine according to the Kozak rule (23). As a control, we also generated 2008 cells overexpressing MRP1. MRP1 cDNA was removed from pJ3Ω-MRP1 as a SalI-NotI fragment (24), blunted, and inserted into the vector pCMV-neo. Retroviral transductions, identification of clones expressing the different constructs, and protein analysis were as described (18, 20).
Monoclonal Antibodies.
For the generation of MRP3-specific monoclonal antibodies, a fusion gene consisting of the gene for the Escherichia coli maltose-binding protein and a part of the MRP3 cDNA corresponding to amino acids 830–949 (the “linker” region) was constructed in pMal-c (20). The fusion protein was produced in E. coli strain DH5α and purified by amylose affinity chromatography. The injection of mice with the fusion protein and the production of monoclonal antibodies were as described (25). As the members of the MRP family show considerable homology (Table 1), we check every antibody for specificity against other MRPs in ELISA with fusion proteins and in cells transduced with cDNA constructs of other MRPs. Monoclonal antibodies M3II-9 and M3II-21 recognized specifically the MRP3 protein and did not crossreact with either human MRP1, human and rat MRP2, or human MRP5 (data not shown).
Table 1.
Sequence comparison of human MRP3 protein (1,527 amino acids) with related ABC transporters
| Protein | Amino acids | Identity, % | Similarity, % | Accession No. |
|---|---|---|---|---|
| Human MRP1 | 1531 | 57.6 | 66.1 | L05628 |
| Human MRP2 (cMOAT) | 1545 | 47.0 | 57.1 | U49248 |
| Human MRP4 (MOAT-B) | 1325 | 35.3 | 47.3 | AF071202 |
| Human MRP5 (MOAT-C) | 1437 | 33.2 | 42.7 | AF104942 |
| Human MRP6 | 1503 | 43.6 | 51.4 | AF076622 |
| Human SUR1 | 1581 | 31.5 | 41.6 | L78207 |
| Human SUR2 | 1549 | 33.5 | 43.9 | AF061289 |
| Human CFTR | 1481 | 27.5 | 39.7 | M28668 |
For this comparison, the gap program of GCG (54) has been used.
Immunocytochemistry.
For immunolocalization of the MRP3 protein in cells, 2008 cells were grown overnight on glass slides and MDCKII cells, for 3 days on microporous polycarbonate filters (3-μm pore size, 24.5-mm diameter, Transwell 3414; Costar, Cambridge, MA). Detection of MRP3 in these cells with monoclonal antibodies M3II-9 or M3II-21 as primary antibody and Alexa 488 labeled goat-anti-mouse IgG (1:50, Molecular Probes) as secondary antibody was as described (18). For detection of MRP3, MRP2, and cytokeratin 7 in human liver sections, the following primary antibodies were used: M3II-9 (IgG1) and M3II-21 (IgG2b) for MRP3, M2III-6 (IgG2a) for MRP2 (26), and a polyclonal antibody for cytokeratin 7 (27). The stainings were carried out as described (27).
Cytotoxicity Assays.
The drug sensitivity of cells was determined in growth inhibition assays with either short-term or continuous exposure to drugs. Eighteen hundred cells per well were seeded in triplicate in 96-well plates and incubated for 24 h at 37°C and 5% CO2. Drug was added and cells were incubated for either 4 h or 4 days at 37°C. In the case of the short-term exposure of drugs, medium with drugs was removed after 4 h, cells were washed four times with fresh medium and subsequently incubated for 4 days in fresh medium at 37°C. After this, medium was removed and cells were frozen at −80°C for 30–45 min. Subsequently the cells were thawed and the total number of cells was determined with fluorescence by using the CyQuant Cell Proliferation Assay Kit (Molecular Probes). Fluorescence was measured with the CytoFluor 4000 fluorescence plate reader (Perkin–Elmer). The relative resistance was calculated as the ratio of inhibitory concentration (IC)50 (where 50% of the cells survive) of the resistant cell line to the IC50 of the parental cell line.
GSH Assays.
GSH assays were carried out as described (20) with the difference that for GSH export measurements, cells were washed with Hepes-buffered saline solution, pH 7.4 (HBS) and subsequently incubated in 1 ml HBS for 2 h at 37°C in the presence of 0.5 mM acivicin to inhibit γ-glutamyl transferase activity. After 2 h, a sample was taken and measured for GSH content according to the recycling method of Tietze (28).
Transport Assays.
Cells were grown for 3–4 days on microporous polycarbonate membrane filters at a density of 2 × 106 cells per well. Export of [14C]DNP-GS from cells was determined by incubating cells with [14C]CDNB as described previously (18).
[3H]MTX Accumulation and Polyglutamylation.
2008 and 2008/MRP3–8 cells were grown as monolayer cells in 80-cm2 flasks until 60% confluency. At this stage, cells were exposed to 1 μM [3H]MTX (0.5 Ci/mmol) for a period of 4 and 24 h. After these time periods, cells were washed twice with 10 ml ice-cold HBS followed by trypsinization, and cells were collected in 10 ml HBS. After centrifugation, the cells were resuspended in 1 ml HBS, of which 10 μl was used for cell counting, 100 μl for radioactivity counting, and 890 μl for [3H]MTX-polyglutamate analysis by HPLC as described by Westerhof et al. (29).
RESULTS
Cloning and Sequencing of the Human MRP3 cDNA.
The full-length cDNA encoding MRP3 contains a single ORF of 1,527 amino acids, with a predicted molecular mass of 169.4 kDa. The homology between the MRP3 protein and the other members of the human MRP protein family ranges from 33% with MRP5 to 58% with MRP1 (Table 1). Multiple MRP proteins with substantial homology to the human MRP proteins are also encoded by other organisms, but only in the rat has a clear MRP3 homolog (also called Mlp2) been found, which shows 80% amino acid identity with human MRP3 (30). Our hydrophobicity analysis of the MRP3 amino acid sequence indicates an organization of putative transmembrane domains resembling that suggested for MRP1, MRP2, and MRP6 (not shown; available on request). These proteins probably all span the membrane 17 times with the NH2 terminus being extracellular (31–33). This unusual topology is supported by the demonstration that the Asn19 and Asn23 sites at the NH2 terminus of MRP1 are indeed glycosylated in vivo (31). In MRP2, MRP3, and MRP6, either one or two of these glycosylation sites at the NH2 terminus are conserved. An additional glycosylation site in MRP1 is found at Asn1006. Potential glycosylation sites in this region are also present in MRP2–5 but not in MRP6.
Expression of MRP3 in Human Liver.
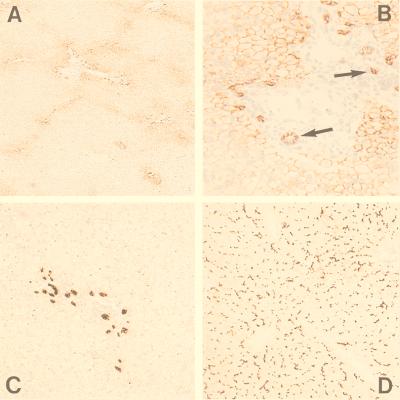
MRP3 RNA is mainly found in liver, colon, small intestine, and adrenal gland, and also, but at lower levels, in pancreas, kidney, and prostate (20, 34–36). To determine where in the liver MRP3 is localized, we used our monoclonal antibodies specific for MRP3 on frozen sections of normal human liver. Monoclonal antibodies M3II-9 and M3II-21 gave similar results. Overall staining for MRP3 was low, and MRP3 staining was detected mainly in the intrahepatic bile-duct epithelial cells (cholangiocytes), and at their lateral side (Fig. 1 A and B; see arrows). The nature of these cells was confirmed with an antibody against cytokeratin 7, which stains cholangiocytes (Fig. 1C). MRP3 staining was also seen in the hepatocytes, but only in those surrounding the portal tracts. The MRP3 staining was at the basolateral or lateral membranes and no staining at all was detected at the canalicular membranes. In contrast, canalicular membranes and no basolateral membranes stained prominently with a control antibody against MRP2 (Fig. 1D), as shown before (10).
Figure 1.
Immunohistochemical staining of human liver. (A and B) Immunohistochemical detection of MRP3 in the lateral membranes of cholangiocytes (indicated by arrows) and (baso)lateral membranes of hepatocytes surrounding the portal tracts by using monoclonal antibody M3II-9. B is an enlargement of A. (C) Immunohistochemical detection of cytokeratin 7 in bile-duct epithelium cells. (D) Immunostaining with M2III-6 against MRP2, showing canalicular staining of the hepatocytes through out the entire liver lobule.
MRP3 Transfectants.
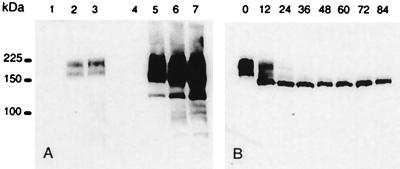
To generate stable cell lines overproducing MRP3, we cloned MRP3 cDNA into a retroviral vector, used previously to overproduce MRP2 (18). Several cell lines were transduced with the retrovirus produced by the transfected Phoenix cells. No MRP3 at all was detectable in G418-resistant cells of transduced pig kidney LLC-PK1 cells, human ovarian carcinoma A2780 cells, and human ovarian carcinoma PXN94 cells, and only low levels of MRP3 were obtained in the human nonsmall cell lung cancer cell line SW1573/S1, in the human colon carcinoma cell line HCT8, and in the human bladder carcinoma cell line T24 (data not shown). Higher levels were obtained in the human ovarian carcinoma cell line 2008 and much higher levels in the canine kidney-derived MDCKII cells (Fig. 2A).
Figure 2.
Western blot analysis of MRP3 protein in parental cells and several clones of transduced 2008 and MDCKII cells. Total cell lysates (10 μg/lane for the 2008 cells and 5 μg for the MDCKII cells) were size fractionated in a 7.5% polyacrylamide gel containing 0.5% SDS. The fractionated proteins were transferred to a nitrocellulose membrane, and MRP3 was detected by incubation with monoclonal antibody M3II-9. (A) 1 = 2008; 2 = 2008/MRP3–4; 3 = 2008/MRP3–8; 4 = MDCKII; 5 = MDCKII/MRP3–17; 6 = MDCKII/MRP3–18; 7 = MDCKII/MRP3–20. (B) Time course in hours of MRP3 expression in 2008/MRP3–8 cells after treatment with tunicamycin (5 μg/ml). Ten μg of total cell lysate was used per lane.
In both the MDCKII/MRP3 and 2008/MRP3 transfectants, the MRP3 protein yields a double band of 170–190 kDa on Western blots (Fig. 2A). This is in agreement with the predicted size of the MRP3 protein. The two bands are reduced to one, after incubation of the cells with tunicamycin (Fig. 2B), and they are therefore attributable to differential glycosylation. Double bands of similar size are also present in membrane preparations of human tissues expressing MRP3 (M.K., unpublished work). The experiments with tunicamycin were also used to determine the half life of MRP3 in 2008 cells. For MRP3 this is only 12–15 h, compared with a t1/2 of MRP1 and MRP2 in the same cells of 20–24 h (not shown).
Immunolocalization of MRP3 Protein in Transduced Cells.
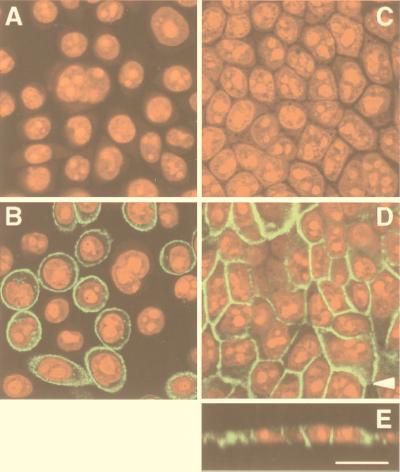
By using confocal laser scanning microscopy, the subcellular localization of MRP3 protein was determined in 2008/MRP3–8 and MDCKII/MRP3–17 cells with monoclonal antibody M3II-9. In the 2008/MRP3–8 cells, the MRP3 protein was predominantly found at the plasma membrane, but some MRP3 was found intracellular (Fig. 3B), similar to what we have seen with MRP1 protein overexpressed in the 2008 cells (data not shown). No MRP3 was detected in the parental 2008 cells (Fig. 3A).
Figure 3.
Immunolocalization of MRP3 in 2008 cells and MDCKII monolayers by confocal laser scanning microscopy. MRP3 is detected by indirect immunofluorescence (green signal) with monoclonal antibody M3II-9. Nucleic acids were detected by counterstaining with propidium iodide (red signal). (A) Parental 2008 cells; (B) 2008/MRP3–8 cells; (C) parental MDCKII cells; (D) MDCKII/MRP3–17 cells; (E) vertical X/Z section of the monolayer shown in D. The arrow in D indicates the position where the X/Z section was made. (Bar = 20 μm.)
MDCKII/MRP3–17 cells were grown to confluency on microporous membrane filters to establish optimal polarization. Fig. 3D shows that the anti-MRP3 antibody stains MRP3 at the plasma membrane of the cells, although some intracellular staining was present as well. No staining was detected in the parental MDCKII cells (Fig. 3C). Examination of the cells at a plane perpendicular to the filter showed that MRP3 is localized in the lateral membrane (Fig. 3E), in agreement with its (baso)lateral localization in cholangiocytes and hepatocytes.
Cytotoxicity Assays.
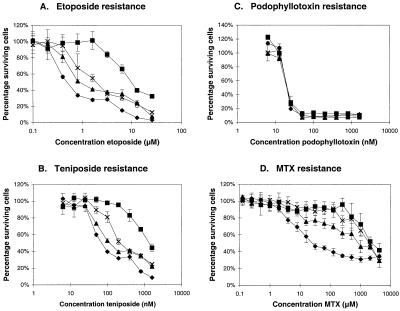
Growth inhibition assays with continuous drug exposure were performed with several independent clones of the 2008/MRP3 cells to test whether the MRP3 protein can act as a drug transporter and confer resistance to the cells. Tested substrates include many cytostatic drugs, such as vinca-alkaloids (vincristine, vinblastine), anthracyclins (doxorubicin, daunorubicin), mitoxantrone, epipodophyllotoxins (etoposide, teniposide), campthotecin analogs (CPT-11, SN-38, topotecan), cisplatin, paclitaxel, methotrexate, heavy metals (arsenite, antimonite, cadmium), antiinflammatory drugs (indo-, ibu-, keto-, and flurbiprofen), and several bile salts in conjugated and unconjugated form. Among all these compounds, we found clear resistance only against the epipodophyllotoxins etoposide [resistance factor (RF) 3.3 ± 0.8 for the 2008/MRP3–8 clone] and teniposide (RF 2.7 ± 0.4 for the 2008/MRP3–8 clone) but not against the analog podophyllotoxin (Fig. 4), suggesting that the sugar moiety in etoposide and teniposide is important for the substrate specificity. These RFs are low in comparison with what we obtained in a parallel experiment with 2008 cells expressing MRP1 (RF 20.4 ± 5.3 for etoposide and 17.1 ± 2.3 for teniposide) (Fig. 4). To test whether the low resistance against epipodophyllotoxins obtained with the MRP3 transfectants is really caused by the overexpression of MRP3 and not by clonal variation, we isolated 16 additional 2008 clones in an independent transduction experiment with MRP3. All clones expressing MRP3 again showed resistance against etoposide and teniposide (RF between 1.6 and 3.5), whereas 16 control clones were not or hardly resistant (three clones showed a RF between 1.6 and 1.8).
Figure 4.
Representative growth inhibition curves for cytotoxicity experiments with (A) etoposide, (B) teniposide, (C) podophyllotoxin, and (D) methotrexate by using 2008 cells (♦), 2008/MRP1–4 cells overexpressing MRP1 (■), and 2008/MRP3–4 (▴) and 2008/MRP3–8 cells (X) both overexpressing MRP3. A–C, experiments with continuous (96-h) drug exposure. D, experiments with short-term (4-h) drug exposure. The vertical bars for each data point represent ± SD of three replicate determinations. The IC50 values of 2008, 2008/MRP1–4, 2008/MRP3–4, and 2008/MRP3–8 cells in this particular experiment were for etoposide: 0.51 nM, 10.29 nM, 0.88 nM, and 1.99 nM; for teniposide: 0.074 μM, 1.388 μM, 0.120 μM, and 0.226 μM; for podophyllotoxin: 21.68 nM, 21.85 nM, 20.54 nM, and 21.20 nM; for MTX: 0.027 mM, 2.504 mM, 0.887 mM, and 2.030 mM.
In long-term continuous drug exposure experiments, we did not observe significant resistance to methotrexate (MTX) (results not shown). Recently, however, we noted that overexpression of MRP1 in 2008 cells results in high resistance to MTX in short-term drug exposure experiments, whereas resistance is low in long-term exposure experiments. We therefore repeated our experiments using a 4-h exposure to high concentrations of MTX. Fig. 4D shows that under these conditions the MRP3 transfectants are also highly resistant to MTX (RF 33 and 75 for the 2008/MRP3–4 and 2008/MRP3–8 clones, respectively), similar to the MRP1 transfectant (RF 93).
Accumulation and Polyglutamylation of [3H]MTX.
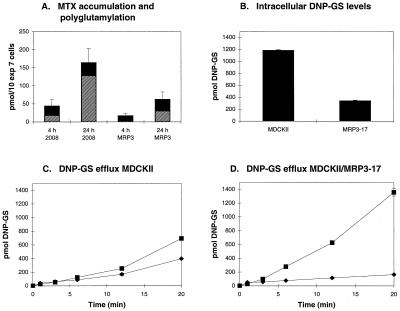
To find out whether the high resistance to MTX in short-term exposures was caused by an altered cellular accumulation and/or polyglutamylation, we measured accumulation of [3H]MTX and its conversion to polyglutamates in 2008 and 2008/MRP3–8 cells after exposure to 1 μM [3H]MTX. After both 4 and 24 h, the total amount of [3H]MTX was 62% lower in the 2008/MRP3–8 cells than in the 2008 cells (Fig. 5A). Analysis of the [3H]MTX-polyglutamate formation after 4 and 24 h showed that the absolute levels of long-chain [3H]MTX-polyglutamates ([3H]MTX-Glu4–6) are 4- to 7-fold lower in the 2008/MRP3–8 cells at both time points compared with the 2008 cells. However, the fraction of total [3H]MTX that is polyglutamylated has increased 2- to 3-fold in both cell lines after a 24-h exposure to [3H]MTX (Fig. 5A).
Figure 5.
(A) Accumulation and polyglutamylation of [3H]MTX. 2008 and 2008/MRP3–8 cells were incubated 1 μM [3H]MTX for 4 and 24 h. Black bars represent the total intracellular pool of [3H]MTX and short-chain [3H]MTX-polyglutamates (MTX-Glu2–3). Hatched bars represent the total intracellular pool of long-chain polyglutamates of [3H]MTX (MTX-Glu4–6). Results presented are the mean ± SD (for total pool of all [3H]MTX) of three to four independent experiments. (B–D) Export of [14C]DNP-GS from MDCKII and MDCKII/MRP3–17 cells grown in monolayers. Cells were incubated at room temperature with 1-chloro-2,4-dinitro[14C]benzene (2 μM) in both the apical and basal compartment. Samples were taken at t = 1, 3, 6, 12, and 20 min and extracted with ethyl acetate. The experiments were done in duplicate. Variation between measurements was in most cases within the size of the symbols. ■ = transport to basal compartment; ♦ = transport to apical compartment. (B) Intracellular [14C]DNP-GS levels at the end of the experiment (t = 20); (C) DNP-GS efflux from MDCKII cells; (D) DNP-GS efflux from MDCKII/MRP3–17 cells.
GSH Efflux and Intracellular GSH Levels of MRP3 Transfectants.
Increased levels of the GS-X pumps MRP1 and MRP2 in transfected cells results in an increased excretion of GSH and often in a decreased cellular GSH content (37–39). This was not observed for the MRP3 transfectants. Neither the 2008 cells nor the MDCKII cells overexpressing MRP3 showed an increase in GSH export or a decrease in the level of intracellular GSH (data not shown).
Transport of Compounds by MDCKII/MRP3 Cells.
We have previously shown that polarized kidney cell monolayers are suitable for studying transport by ABC-transporters that preferentially localize apically or basolaterally (18, 40). The GS-X pumps MRP1 and MRP2 can be studied in these polarized cells by using [14C]CDNB. This hydrophobic compound rapidly diffuses into cells and is conjugated to GSH by intracellular glutathione-S-transferases (41). The resulting hydrophilic [14C]DNP-GS can leave the cell only by carrier-mediated transport. MDCKII cells overexpressing MRP1 preferentially export [14C]DNP-GS toward the basolateral side (40), whereas MDCKII cells overexpressing MRP2 preferentially export [14C]DNP-GS toward the apical side of the cells (18). Using this assay and the MDCKII cells overexpressing MRP3, we found that MRP3 also transports [14C]DNP-GS (Fig. 5 C and D). In comparison with the parental MDCKII cells, there is an increased transport of [14C]DNP-GS toward the basolateral side of the cells overexpressing MRP3, which is similar to what has been found with the MRP1-expressing MDCKII cells (40). Intracellular measurements of [14C]DNP-GS accumulation showed that the MDCKII cells overexpressing MRP3 accumulated less [14C]DNP-GS than the parental cells (Fig. 5B).
DISCUSSION
Our results show that MRP3 is an organic anion transporter and transports DNP-GS, like MRP1 and MRP2. Moreover, MRP3 can cause resistance to the anti-cancer drugs MTX, etoposide, and teniposide, justifying its inclusion in the multidrug resistance protein family.
Among the current members of the MRP family, MRP3 has the highest sequence homology to MRP1 and, like MRP1, it localizes to the (baso)lateral side of polarized cells. Nevertheless, MRP3 is not just a twin brother of MRP1 with a different tissue distribution. The most striking difference found thus far is that 2008 or MDCKII cells overexpressing MRP3 do not detectably excrete more GSH than the parental cells, in contrast to cells transduced with MRP1 or MRP2 constructs [(37); M.K., unpublished work]. We consider the absence of MRP3-catalyzed GSH export important, because several lines of evidence indicate that MRP1-mediated export of some neutral or basic compounds from the cell involves cotransport with GSH [(42); R. Evers, unpublished work]. If the affinity of MRP3 for GSH is lower than that of MRP1 (and MRP2), it might limit the ability of MRP3 to transport nonanionic organic molecules. This might account for the limited resistance spectrum of our MRP3-expressing cells. We cannot rule out, however, that our transduced cells do not contain enough MRP3 to detect resistance against hydrophobic compounds other than etoposide/teniposide. For unknown reasons, we have not succeeded in isolating nonpolarized cells with high MRP3 levels. Even our best 2008 transfectants contain much less MRP3 than the MDCKII/MRP3 cells (Fig. 2A). If the 2008 cells are transduced with MRP1 constructs, RFs up to 20 are obtained. These cells are resistant to many other anti-cancer drugs, but their highest resistance is to etoposide (M.K. et al., unpublished work). Hence the etoposide resistance of the 2008/MRP3 cells could be the tip of the iceberg, and other test systems may be required to determine the whole range of compounds transported by MRP3.
It has long been known through the work of Henderson and coworkers that mammalian cells contain several GS-X pumps able to transport MTX (43, 44). By comparing wild-type and mutant rats lacking MRP2 in the canalicular membrane, Masuda et al. (14) showed that MTX is transported by MRP2 into bile. Increased uptake of MTX was found in membrane vesicles isolated from GLC4/ADR cells, which overexpress MRP1 (45). GLC4/ADR cells also show a decreased accumulation of MTX compared with the parental GLC4 cells. Nevertheless, little effect on MTX sensitivity was found in GLC4/ADR cells or cells transfected with MRP1 or MRP2 cDNA constructs (46). This apparent discrepancy was recently solved when we showed that 2008/MRP1 cells are highly resistant to short-term MTX exposure but not to long-term exposure (46). We obtain the same result for the 2008 cells transduced with MRP3. In 4-h exposure experiments, we find up to 75-fold resistance. The absolute RF is somewhat arbitrary because it is highly dependent on how many times the cells are washed after the 4-h incubation with MTX to remove excess drug. No significant resistance against MTX was found in continuous drug exposure (96-h) experiments. The difference can be explained by the fact that MTX is polyglutamylated after entering the cell. The polyglutamylated form is an effective inhibitor of dihydrofolate reductase, and we assume that it is not transported by MRP3. Polyglutamylation of MTX is slow and after short-term exposures to MTX, cells with a good MTX efflux pump can pump out the monoglutamate MTX. These cells will accumulate much less long-chain polylutamylates of MTX that are retained in the cell. In continuous exposures to MTX, a good efflux pump for MTX makes no major difference, because these cells will eventually accumulate enough polyglutamylated MTX to block the DHFR efficiently, resulting in cell death.
Whether MRPs contribute to clinical resistance to MTX remains to be seen. Several mechanisms for resistance to MTX are known, but these do not involve MRPs (47, 48). These mechanisms were elucidated by isolating cells resistant to continuous exposure to MTX. It is possible, however, that MTX concentrations in patients fluctuate sufficiently to allow MRPs to contribute to clinical resistance, and a more directed search for such a contribution would seem of interest.
With the completion of the MRP3 sequence by us and others (34–36) and the sequences of MRP4 (49), MRP5 [(36), J. Wijnholds, unpublished work], and MRP6 (22), the sequences of six members of the human MRP family are already known. Each member is clearly more homologous to MRP1 than to any other ABC-transporter, justifying our previous classification of these genes into one subfamily of ABC-transporter genes, called MRP1–6 (20). Based on the complete sequences, two subgroups can be recognized. One group with MRP1, MRP2, MRP3, and MRP6, sharing 45–58% amino acid identity, is characterized by the presence of an NH2-terminal membrane-bound extension of about 200 amino acids, which adds five putative transmembrane segments to an MDR1-like core (50). This extra domain is also present in the yeast MRP1 homologue YCF1 and the SUR proteins SUR1 and SUR2, but not in the other MRP homologs, MRP4 and MRP5. MRP4 and MRP5 have less homology to MRP1 (34–39% amino acid identity), and their predicted structure is more similar to CFTR and MDR1. The function of the NH2-terminal extension in MRP1 is unknown, but recent data suggest that the part with the extra five transmembrane domains is not required for catalytic function or routing to the plasma membrane (40).
We and others have shown that MRP3 RNA is high in human liver (20, 34–36). However, we show now with immunohistochemistry of frozen sections of normal human liver that there are only modest amounts of MRP3 protein in normal human liver. We see prominent staining of the lateral membranes of intrahepatic bile-duct epithelial cells (cholangiocytes) and basolateral MRP3 staining in the hepatocytes surrounding the portal tracts. The discrepancy between high-MRP3 RNA and low protein in human liver may seem puzzling, but may be explained by a massive induction of MRP3 expression in diseased liver. Hirohashi et al. (30) reported that Mrp3 RNA is low or undetectable in normal rat liver, but that the level is increased in rats made cholestatic by bile-duct ligation. We have also found much higher levels of MRP3 in cholestatic human liver than in normal liver (M.K., unpublished work). It seems therefore likely that the high-MRP3 RNA levels reported for human liver were obtained with RNA from cholestatic liver rather than normal liver. More work is required, however, to assess what determines liver MRP3 levels in humans.
The physiological function of MRP3 in the cholangiocytes and hepatocytes is still unknown. Although the cholangiocytes account for only 3–5% of the hepatic cell population, they play an important role in determining the bile flow and pH of hepatic bile through absorbtive and/or secretory processes (51). Obstructive cholestasis is characterized by a proliferation of the bile-duct epithelium, which may explain the increased expression of Mrp3 in cholestatic rats. The epithelial bile-duct proliferation is believed to be a result of the continuous exposure of these cells to increased levels of toxic bile salts. These bile salts have to be removed from the bile, and it is possible that this is done by the proliferated epithelial cells of the bile duct. It is known that the ileum-bile-salt-transporter IBST is localized at the apical side of these cells and that this transporter is able to take up bile salts (52). The transporter that secretes the bile salts at the basolateral side of the cells into the blood is not known, however, and this might be MRP3. This could also explain why in normal livers the expression of MRP3 is very low (human) or even undetectable (rat). Under normal healthy conditions the bile-duct epithelial cells do not have to take up bile salts from the bile.
We do not find transport of bile salts with our transduced 2008 and MDCKII cells, but these are not suitable for investigating whether bile salts are substrates for MRP3, because bile salts are not taken up by these cells. Good vesicular systems suitable for transport studies will be required to determine the substrate specificity of MRP3.
A possible role for MRP3 in the basolateral transport of bile salts is also compatible with the expression of MRP3 in the small intestine (20). Bile salts, after secretion into the duodenum, are actively taken up by the IBST transporter expressed at the apical side of the epithelial cells in the ileum (53). Also here it is still unknown which transporter is responsible for the resecretion of bile salts into blood. This could be MRP3, because we (20) and others (34–36) have shown that MRP3 is also highly expressed in the small intestine.
Acknowledgments
We thank our colleagues Raymond Evers, Zsolt Hóllo, Jan Wijnholds, and Noam Zelcer for critical reading of the manuscript and Lauran Oomen (Department of Biophysics, Netherlands Cancer Institute) for assistance with the confocal laser scanning microscopy. This work was supported by the Dutch Cancer Society (NKI 94-775).
ABBREVIATIONS
- GSH
glutathione
- [14C]CDNB
1-chloro-2,4-dinitro[14C]benzene
- CMV
cytomegalovirus
- HBS
Hepes-buffered solution
- RF
resistance factor
- MTX
methotrexate
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF009670).
References
- 1.Gottesman M M, Pastan I, Ambudkar S V. Curr Opin Genet Dev. 1996;6:610–617. doi: 10.1016/s0959-437x(96)80091-8. [DOI] [PubMed] [Google Scholar]
- 2.Cole S P C, Deeley R G. BioEssays. 1998;20:931–940. doi: 10.1002/(SICI)1521-1878(199811)20:11<931::AID-BIES8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 3.Leier I, Jedlitschky G, Buchholz U, Cole S P C, Deeley R G, Keppler D. J Biol Chem. 1994;269:27807–27810. [PubMed] [Google Scholar]
- 4.Jedlitschky G, Leier I, Buchholz U, Barnouin K, Kurz G, Keppler D. Cancer Res. 1996;56:988–994. [PubMed] [Google Scholar]
- 5.Zaman G J R, Cnubben N H P, van Bladeren P J, Evers R, Borst P. FEBS Lett. 1996;391:126–130. doi: 10.1016/0014-5793(96)00718-1. [DOI] [PubMed] [Google Scholar]
- 6.Loe D W, Deeley R G, Cole S P C. Eur J Cancer. 1996;32A:945–957. doi: 10.1016/0959-8049(96)00046-9. [DOI] [PubMed] [Google Scholar]
- 7.Ishikawa T. Trends Biochem Sci. 1992;17:463–468. doi: 10.1016/0968-0004(92)90489-v. [DOI] [PubMed] [Google Scholar]
- 8.Jansen P L M, Oude Elferink R P J. In: Defective Hepatic Anion Secretion in Mutant TR-Rats. Tavaloni N, Berk P D, editors. New York: Raven; 1993. pp. 721–731. [Google Scholar]
- 9.Zaman G J R, Versantvoort C H M, Smit J J M, Eijdems E W H M, de Haas M, Smith A J, Broxterman H J, Mulder N H, de Vries E G E, Baas F, et al. Cancer Res. 1993;53:1747–1750. [PubMed] [Google Scholar]
- 10.Paulusma C C, Kool M, Bosma P J, Scheffer G L, ter Borg F, Scheper R J, Tytgat G N J, Borst P, Baas F, Oude Elferink R P J. Hepatology. 1997;25:1539–1542. doi: 10.1002/hep.510250635. [DOI] [PubMed] [Google Scholar]
- 11.Schauab T B, Kartenbeck J, Konig J, Vogel O, Witzgall R, Kriz W, Keppler D. J Am Soc Nephrol. 1997;8:1213–1221. doi: 10.1681/ASN.V881213. [DOI] [PubMed] [Google Scholar]
- 12.Oude Elferink R P J, Meijer D K F, Kuipers F, Jansen P L M, Groen A K, Groothuis G M M. Biochim Biophys Acta. 1995;1241:215–268. doi: 10.1016/0304-4157(95)00006-d. [DOI] [PubMed] [Google Scholar]
- 13.Keppler D, Leier I, Jedlitschky G. Biol Chem. 1997;378:787–791. [PubMed] [Google Scholar]
- 14.Masuda M, I’izuka Y, Yamazaki M, Nishigaki R, Kato Y, Ni’inuma K, Suzuki H, Sugiyama Y. Cancer Res. 1997;57:3506–3510. [PubMed] [Google Scholar]
- 15.Sugiyama, Y., Kato, Y. & Chu, X. (1998) Cancer Chemother. Pharmacol.42, Suppl., 44–49. [DOI] [PubMed]
- 16.Sugawara N, Lai Y R, Sugaware C, Arizono K. Toxicology. 1998;126:23–31. doi: 10.1016/s0300-483x(97)00170-4. [DOI] [PubMed] [Google Scholar]
- 17.Dijkstra M, Havinga R, Vonk R J, Kuipers F. Life Sci. 1996;59:1237–1246. doi: 10.1016/0024-3205(96)00447-x. [DOI] [PubMed] [Google Scholar]
- 18.Evers R, Kool M, van Deemter L, Janssen H, Calafat J, Oomen L C J M, Paulusma C C, Oude Elferink R P J, Baas F, Schinkel A H, et al. J Clin Invest. 1998;101:1310–1319. doi: 10.1172/JCI119886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniguchi K, Wada M, Kohno K, Nakamura T, Kawabe T, Kawakami M, Kagotani K, Okumura K, Akiyama S-i, Kuwano M. Cancer Res. 1996;56:4124–4129. [PubMed] [Google Scholar]
- 20.Kool M, de Haas M, Scheffer G L, Scheper R J, Van Eijk M J T, Juijn J A, Baas F, Borst P. Cancer Res. 1997;57:3537–3547. [PubMed] [Google Scholar]
- 21.Koike K, Kawabe T, Tanaka T, Toh S, Uchiumi T, Wada M, Akiyama S, Ono M, Kuwano M. Cancer Res. 1997;57:5475–5479. [PubMed] [Google Scholar]
- 22.Kool M, van der Linden M, de Haas M, Baas F, Borst P. Cancer Res. 1999;59:175–182. [PubMed] [Google Scholar]
- 23.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaman G J R, Flens M J, van Leusden M R, de Haas M, Mulder H S, Lankelma J, Pinedo H M, Scheper R J, Baas F, Broxterman H J, et al. Proc Natl Acad Sci USA. 1994;91:8822–8826. doi: 10.1073/pnas.91.19.8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flens M J, Izquierdo M A, Scheffer G L, Fritz J M, Meijer C J L M, Scheper R J, Zaman G J R. Cancer Res. 1994;54:4557–4563. [PubMed] [Google Scholar]
- 26.Paulusma C C, Bosma P J, Zaman G J R, Bakker C T M, Otter M, Scheffer G L, Scheper R J, Borst P, Oude Elferink R P J. Science. 1996;271:1126–1128. doi: 10.1126/science.271.5252.1126. [DOI] [PubMed] [Google Scholar]
- 27.de Vree J M L, Jacquemin E, Sturm E, Cresteil D, Bosma P J, Aten J, Deleuze J F, Desrochers M, Burdelski M, Bernard O, et al. Proc Natl Acad Sci USA. 1998;95:282–287. doi: 10.1073/pnas.95.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tietze F. Anal Biochem. 1969;27:502–522. doi: 10.1016/0003-2697(69)90064-5. [DOI] [PubMed] [Google Scholar]
- 29.Westerhof G R, Rijnboutt S, Schornagel J H, Pinedo H M, Peters G J, Jansen G. Cancer Res. 1995;55:3795–3802. [PubMed] [Google Scholar]
- 30.Hirohashi T, Suzuki H, Ito K, Ogawa K, Kume K, Shimizu T, Sugiyama Y. Mol Pharmacol. 1998;53:1068–1075. [PubMed] [Google Scholar]
- 31.Hipfner D R, Almquist K C, Leslie E M, Gerlach J H, Grant C E, Deeley R G, Cole S P C. J Biol Chem. 1997;272:23623–23630. doi: 10.1074/jbc.272.38.23623. [DOI] [PubMed] [Google Scholar]
- 32.Kast C, Gros P. Science. 1997;272:26479–26487. doi: 10.1074/jbc.272.42.26479. [DOI] [PubMed] [Google Scholar]
- 33.Kast C, Gros P. Biochemistry. 1998;37:2305–2313. doi: 10.1021/bi972332v. [DOI] [PubMed] [Google Scholar]
- 34.Kiuchi Y, Suzuki H, Hirohashi T, Tyson C A, Sugiyama Y. FEBS Lett. 1998;433:149–152. doi: 10.1016/s0014-5793(98)00899-0. [DOI] [PubMed] [Google Scholar]
- 35.Uchiumi T, Hinoshita E, Haga S, Nakamura T, Tanaka T, Toh S, Furukawa M, Kawabe T, Wada M, Kagotani K, et al. Biochem Biophys Res Commun. 1998;252:103–110. doi: 10.1006/bbrc.1998.9546. [DOI] [PubMed] [Google Scholar]
- 36.Belinsky M G, Bain L J, Balsara B B, Testa J R, Kruh G D. J Natl Cancer Inst. 1998;90:1735–1741. doi: 10.1093/jnci/90.22.1735. [DOI] [PubMed] [Google Scholar]
- 37.Paulusma C C, van Geer M A, Evers R, Heijn M, Ottenhof R, Borst P, Oude Elferink R P J. Biochem J. 1999;338:393–401. [PMC free article] [PubMed] [Google Scholar]
- 38.Rappa G, Lorico A, Flavell R A, Sartorelli A C. Cancer Res. 1997;57:5232–5237. [PubMed] [Google Scholar]
- 39.Zaman G J R, Lankelma J, van Tellingen O, Beijnen J H, Dekker H, Paulusma C C, Oude Elferink R P J, Baas F, Borst P. Proc Natl Acad Sci USA. 1995;92:7690–7694. doi: 10.1073/pnas.92.17.7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakos E, Evers R, Szakacs G, Tusnady G E, Welker E, Szabo K, de Haas M, van Deemter L, Borst P, Varadi A, et al. J Biol Chem. 1998;273:32167–32175. doi: 10.1074/jbc.273.48.32167. [DOI] [PubMed] [Google Scholar]
- 41.Oude Elferink R P J, Bakker C T M, Jansen P L M. Biochem J. 1993;290:759–764. doi: 10.1042/bj2900759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loe D W, Deeley R G, Cole S P C. Cancer Res. 1998;58:5130–5136. [PubMed] [Google Scholar]
- 43.Saxena M, Henderson G B. Biochem Pharmacol. 1996;51:975–982. doi: 10.1016/0006-2952(96)00051-2. [DOI] [PubMed] [Google Scholar]
- 44.Saxena M, Henderson G B. Biochem J. 1996;320:273–281. doi: 10.1042/bj3200273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heijn M, Hooijberg J H, Scheffer G L, Szabo G, Westerhoff H V, Lankelma J. Biochim Biophys Acta. 1997;1326:12–22. doi: 10.1016/s0005-2736(97)00003-5. [DOI] [PubMed] [Google Scholar]
- 46.Hooijberg, J. H., Broxterman, M. J., Kool, M., Assaraf, Y. G., Peters, G. J., Noordhuis, P., Scheper, R. J., Borst, P., Pinedo, N. M. & Jansen, G. (1999) Cancer Res., in press. [PubMed]
- 47.Jansen G, Pieters R. Drug Res Updates. 1998;1:211–218. doi: 10.1016/s1368-7646(98)80042-3. [DOI] [PubMed] [Google Scholar]
- 48.Moscow J A. Leuk Lymphoma. 1998;30:215–224. doi: 10.3109/10428199809057535. [DOI] [PubMed] [Google Scholar]
- 49.Lee K, Belinsky M G, Bell D W, Testa J R, Kruh G D. Cancer Res. 1998;58:2741–2747. [PubMed] [Google Scholar]
- 50.Tusnady G E, Bakos E, Varadi A, Sarkadi B. FEBS Lett. 1997;402:1–3. doi: 10.1016/s0014-5793(96)01478-0. [DOI] [PubMed] [Google Scholar]
- 51.Roberts S K, Ludwig J, Larusso N F. Gastroenterology. 1997;112:269–279. doi: 10.1016/s0016-5085(97)70244-0. [DOI] [PubMed] [Google Scholar]
- 52.Lazaridis K N, Pham L, Tietz P, Marinelli R A, deGroen P C, Levine S, Dawson P A, Larusso N F. J Clin Invest. 1997;100:2714–2721. doi: 10.1172/JCI119816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong M H, Oelkers P, Craddock A L, Dawson P A. J Biol Chem. 1994;269:1340–1347. [PubMed] [Google Scholar]
- 54.Devereux J, Haeberli P, Smithies O A. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]