Abstract
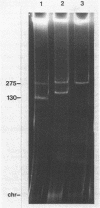
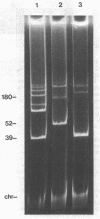
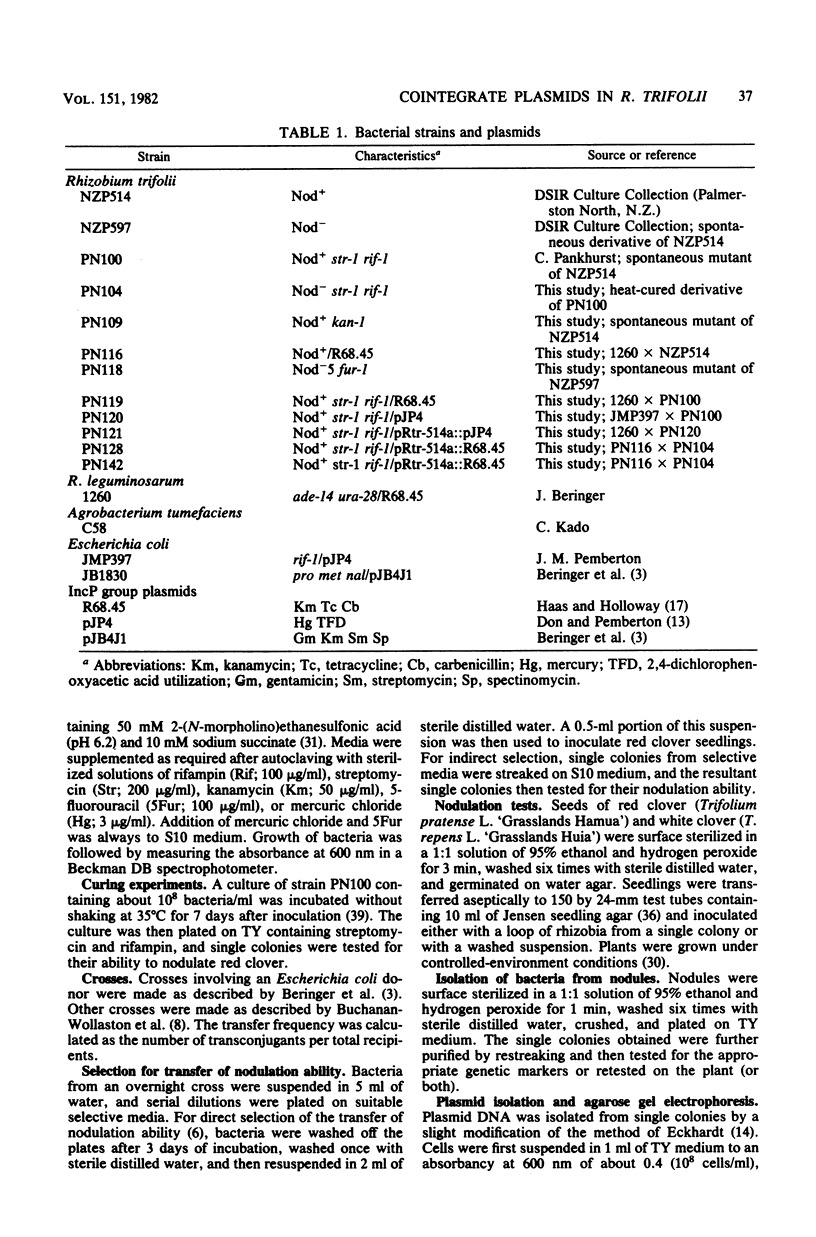
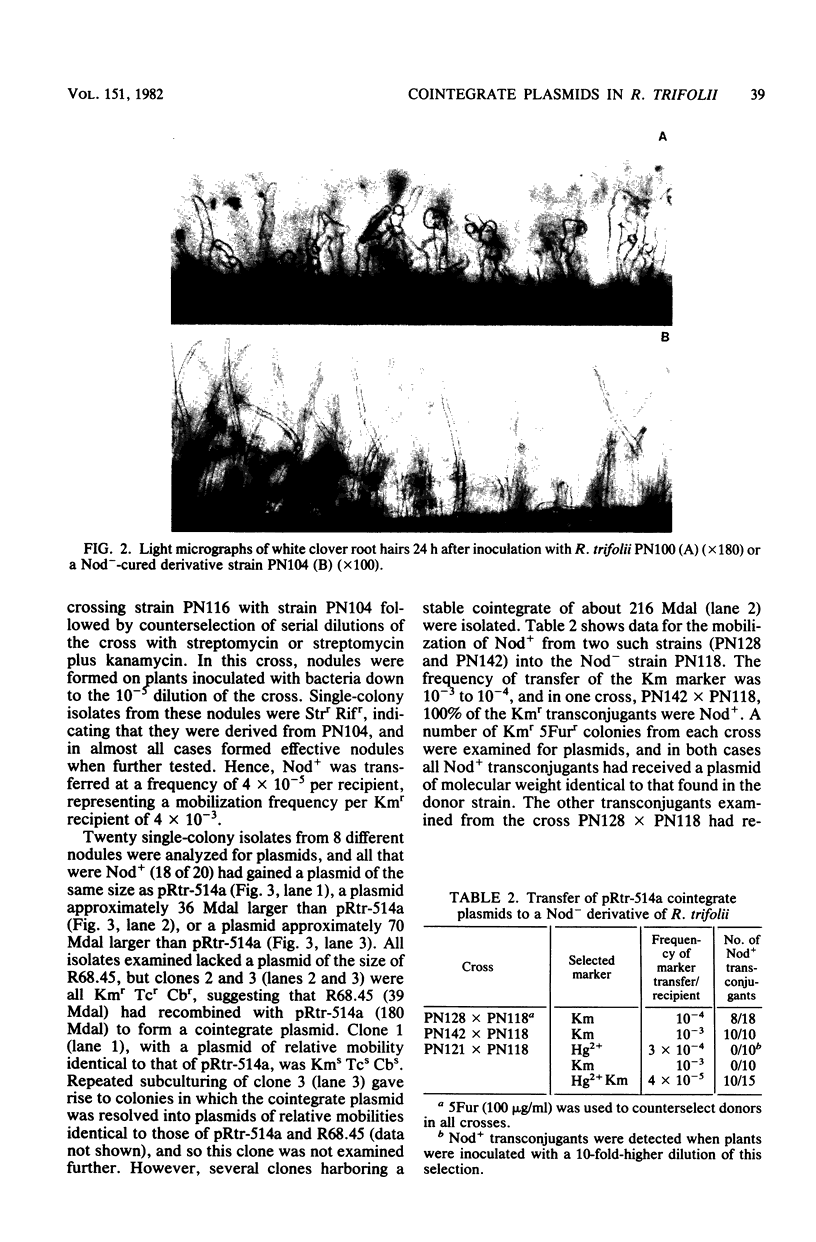
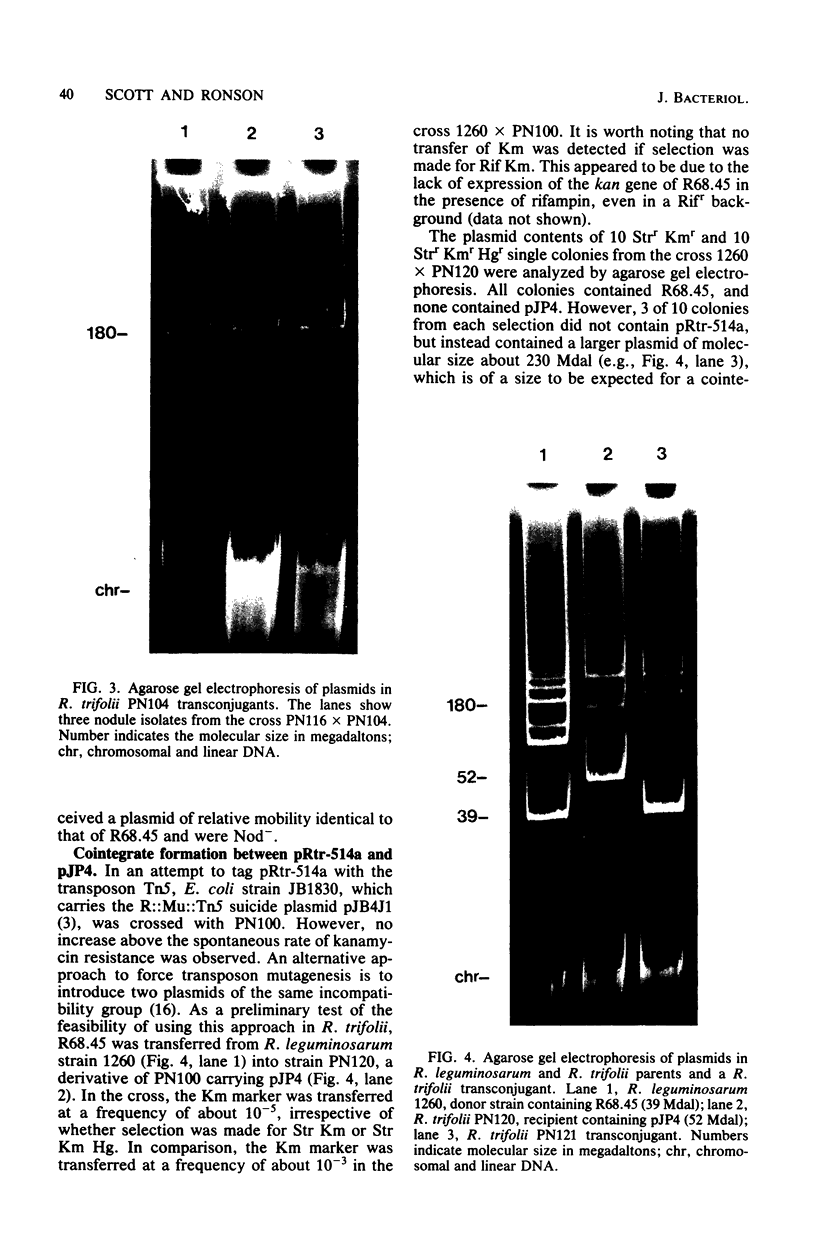
A nodulation plasmid, pRtr-514a, of molecular size 180 megadaltons (Mdal) was identified in Rhizobium trifolii strain NZP514. This plasmid was absent in both spontaneous and heat-cured Nod- derivatives of NZP514, and these strains were unable to induce root hair curling. The ability to nodulate clover was transferred from the wild-type strain to a Nod- derivatives, PN104, with the broad-host-range plasmid R68.45 (39 megadaltons) at a cotransfer frequency of about 4 X 10(-3). Most of the Nod+ transconjugants were resistant to kanamycin, tetracycline, and carbenicillin and had received a plasmid approximately 36 or 70 Mdal larger than pRtr514a but did not contain a plasmid of the size of R68.45, indicating that pRtr-514a was mobilized as a cointegrate plasmid containing either one or possibly two copies of R68.45. Use of these cointegrate-containing strains as donors in further crosses with the Nod- derivative strain PN118 resulted in high-frequency transfer of Nod+ (10(-3) to 10(-4), with cotransfer frequencies with kanamycin of up to 100%. Introduction of R68.45 into a derivative of NZP514 containing the broad-host-range plasmid pJP4 (52 Mdal) resulted in a high frequency of transconjugants carrying a cointegrate plasmid composed of pRtr-514a and pJP4. When used as donors to Nod- derivatives, such strains cotransferred Nod+ with kanamycin plus mercury at a frequency of 67%. The identification of stable cointegrates between pRtr-514a and the broad-host-range plasmids R68.45 and pJP4 should enable several genetic manipulations to be carried out with this nodulation plasmid, including the transfer of the plasmid to most gram-negative bacterial genera.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Crisona N. J., Nowak J. A., Nagaishi H., Clark A. J. Transposon-mediated conjugational transmission of nonconjugative plasmids. J Bacteriol. 1980 May;142(2):701–713. doi: 10.1128/jb.142.2.701-713.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Pemberton J. M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981 Feb;145(2):681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- FAHRAEUS G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol. 1957 Apr;16(2):374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Howe T. G., Richmond K. M. Translocation of the tetracycline resistance determinant from R100-1 to the Escherichia coli K-12 chromosome. J Bacteriol. 1975 Dec;124(3):1153–1158. doi: 10.1128/jb.124.3.1153-1158.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D., Holloway B. W. R factor variants with enhanced sex factor activity in Pseudomonas aeruginosa. Mol Gen Genet. 1976 Mar 30;144(3):243–251. doi: 10.1007/BF00341722. [DOI] [PubMed] [Google Scholar]
- Holloway B. W. Plasmids that mobilize bacterial chromosome. Plasmid. 1979 Jan;2(1):1–19. doi: 10.1016/0147-619x(79)90002-7. [DOI] [PubMed] [Google Scholar]
- Holsters M., Silva B., Genetello C., Engler G., van Vliet F., de Block M., Villarroel R., van Montagu M., Schell J. Spontaneous formation of cointegrates of the oncogenic Ti-plasmid and the wide-host-range P-plasmid RP4. Plasmid. 1978 Sep;1(4):456–467. doi: 10.1016/0147-619x(78)90004-5. [DOI] [PubMed] [Google Scholar]
- Hooykaas P. J., den Dulk-Ras H., Schilperoort R. A. Molecular mechanism of Ti plasmid mobilization by R plasmids: isolation of Ti plasmids with transposon-insertions in Agrobacterium tumefaciens. Plasmid. 1980 Jul;4(1):64–75. doi: 10.1016/0147-619x(80)90083-9. [DOI] [PubMed] [Google Scholar]
- Meyers J. A., Sanchez D., Elwell L. P., Falkow S. Simple agarose gel electrophoretic method for the identification and characterization of plasmid deoxyribonucleic acid. J Bacteriol. 1976 Sep;127(3):1529–1537. doi: 10.1128/jb.127.3.1529-1537.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. R. Resolution of cointegrates between transposons gamma delta and Tn3 defines the recombination site. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3428–3432. doi: 10.1073/pnas.78.6.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riess G., Holloway B. W., Pühler A. R68.45, a plasmid with chromosome mobilizing ability (Cma) carries a tandem duplication. Genet Res. 1980 Aug;36(1):99–109. doi: 10.1017/s0016672300019704. [DOI] [PubMed] [Google Scholar]
- Robertson J. G., Lyttleton P., Bullivant S., Grayston G. F. Membranes in lupin root nodules. I. The role of Golgi bodies in the biogenesis of infection threads and peribacteroid membranes. J Cell Sci. 1978 Apr;30:129–149. doi: 10.1242/jcs.30.1.129. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Lyttleton P., Robertson J. G. C(4)-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4284–4288. doi: 10.1073/pnas.78.7.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt D., Arthur A., Burke M. Transposon-specified, site-specific recombination systems. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):275–281. doi: 10.1101/sqb.1981.045.01.040. [DOI] [PubMed] [Google Scholar]
- Willetts N. S., Crowther C., Holloway B. W. The insertion sequence IS21 of R68.45 and the molecular basis for mobilization of the bacterial chromosome. Plasmid. 1981 Jul;6(1):30–52. doi: 10.1016/0147-619x(81)90052-4. [DOI] [PubMed] [Google Scholar]