Abstract
HIV infects macrophages and microglia in the central nervous system (CNS). Mechanisms that enhance HIV macrophage/microglial tropism are not well understood. Here, we identify an HIV Env variant in the V4 region of gp120, Asp 386 (D386), that eliminates an N-linked glycosylation site at position 386, enhances viral replication in macrophages, and is present at a higher frequency in AIDS patients with HIV-associated dementia (HAD) compared with non-HAD patients. D386 enhances HIV entry and replication in macrophages but not in microglia or peripheral blood mononuclear cells, possibly due to differential glycosylation in these cell types. A D386N mutation in the UK1br Env, which restores the N-linked glycan site, reduced neutralization sensitivity to the IgG1b12 (b12) monoclonal antibody, which recognizes a conserved neutralization epitope that overlaps the CD4 binding site. Molecular modeling suggested that loss of the glycan at position 386 increases exposure of the CD4 and b12 binding sites on gp120. Loss of a glycan at 386 was more frequent in Envs from HAD patients (26%; n=185) compared with non-HAD patients (7%; n=99; p<0.001). The most significant association of these Env variants with HAD was in blood or lymphoid tissue rather than brain. These findings suggest that increased exposure of the b12 epitope overlapping the CD4 binding site via elimination of a glycan at position 386 is associated with enhanced HIV macrophage tropism, and provide evidence that determinants of macrophage and microglia tropism are overlapping but distinct.
Keywords: Human immunodeficiency virus type 1 (HIV), envelope, CD4, macrophage tropism, neurotropism, neutralization, b12 antibody, neuropathogenesis, glycosylation
Introduction
Human immunodeficiency virus type 1 (HIV) infects macrophages and microglia in the central nervous system (CNS) and causes HIV-associated dementia (HAD) or milder forms of neurocognitive impairment (minor cognitive motor disorder (MCMD)) in 10–20% of patients with AIDS (Gonzalez-Scarano and Martin-Garcia, 2005; Kaul et al., 2001; Kaul et al., 2005; McArthur et al., 2003; Sacktor et al., 2002). Highly active anti-retroviral therapy (HAART) has reduced the incidence of HAD (d'Arminio Monforte et al., 2004; Dore et al., 1999; Sacktor et al., 2001; Sacktor et al., 2002), but most current antiviral drugs have relatively poor CNS penetration and the prevalence of MCMD has increased as HIV-infected patients survive for longer times on anti-retroviral therapies (Dore et al., 2003; McArthur et al., 2003; Neuenburg et al., 2002; Valcour et al., 2004). Thus, neurological disease continues to be a significant complication of HIV infection.
HIV variants in brain are genetically distinct from those in lymphoid tissues and other organs, and sequences in the viral envelope glycoprotein (Env) have been associated with brain compartmentalization (Dunfee et al., 2006; Gartner et al., 1997; Korber et al., 1994; Morris et al., 1999; Ohagen et al., 2003; Power et al., 1995; Shapshak et al., 1999; Wang et al., 2001). Furthermore, brain-derived Envs from HAD and non-HAD AIDS patients are genetically and biologically distinct (Power et al., 1994; Power et al., 1998; Shah et al., 2006; Shapshak et al., 1999; Smit et al., 2001). The capacity of HIV or simian immunodeficiency virus (SIV) strains to replicate efficiently in macrophages has been correlated with increased neurotropism (defined here as the ability to replicate in microglia) (Ghorpade et al., 1998; Gorry et al., 2001; Li et al., 1999; Peters et al., 2004; Sharma et al., 1992; Wang et al., 2001), and may also be linked to progression of HIV/SIV disease (Desrosiers et al., 1991; Gray et al., 2005; Mori et al., 1992). However, mechanisms that enhance HIV replication in macrophages in the CNS and other macrophage-rich tissues such as lung, colon, and bone marrow are not well understood.
HIV Env, which is organized into trimers on virions, consists of the gp120 surface and gp41 transmembrane subunits. HIV entry into cells is initiated by interaction between gp120 and CD4, which induces a conformational change in gp120 that exposes the coreceptor binding site (Doms, 2000). The interaction of CD4-bound gp120 with the coreceptor triggers a conformational change in gp120, which leads to structural rearrangements in gp41 that enable fusion and virus entry. CCR5, the primary coreceptor used for infection of macrophages and microglia (Albright et al., 1999; Ghorpade et al., 1998; He et al., 1997; Li et al., 1999), is the coreceptor used by most viruses isolated from brain (Albright et al., 1999; Gorry et al., 2001; He et al., 1997; Li et al., 1999; Smit et al., 2001). However, CCR5 usage is neither necessary nor sufficient for neurotropism (Gorry et al., 2001; Yi et al., 2003). Tissue macrophages and brain microglia express lower cell surface levels of CD4 than CD4+ T cells in peripheral blood (Lewin et al., 1996; Wang et al., 2002). HIV Envs with enhanced tropism for macrophages/microglia have an increased capacity to mediate fusion with cells expressing low levels of CD4 and CCR5, suggesting that reduced dependence on CD4/CCR5 levels may enhance viral replication preferentially in the CNS (Gorry et al., 2002; Gray et al., 2005; Martin et al., 2001; Peters et al., 2004). We previously identified an HIV Env variant associated with brain infection and HAD, N283 in the C2 region of gp120, that increases gp120 affinity for CD4, enhancing the capacity of HIV Envs to use low levels of CD4 for virus entry and increasing viral tropism for macrophages and microglia (Dunfee et al., 2006). However, additional mechanisms by which HIV acquires an enhanced capacity to enter cells when receptor levels are low are unclear.
The gp120 surface subunit of HIV Env is heavily glycosylated (Leonard et al., 1990), resulting in occlusion of receptor binding sites and epitopes for neutralizing antibodies (Kwong et al., 1998; Wyatt et al., 1998). The HIV Clade B consensus gp120 amino acid sequence contains 26 motifs for attachment of N-linked glycans (NXS/T, where X is any amino acid except proline). Amino acid variants that result in the loss of N-linked glycosylation sites in the V1/V2 or V4 regions of HIV or SIV Envs can facilitate infection of cells expressing little or no CD4 (Edwards et al., 2001; Hoffman et al., 1999; Kolchinsky et al., 2001; Puffer et al., 2002). Furthermore, studies of pathogenic SIV and chimeric simian-human immunodeficiency virus (SHIV) infections in rhesus macaques demonstrated that macrophage tropism was associated with loss of conserved N-linked glycosylation sites in Env (Igarashi et al., 2003; Puffer et al., 2002). Whether loss of particular N-linked glycosylation sites in HIV/SIV Envs enhances viral replication in the brain in vivo remains unknown.
To investigate mechanisms by which HIV acquires enhanced tropism for macrophages and microglia, we analyzed sequences of brain-derived HIV Envs from 2 patients with HAD for changes in N-linked glycosylation sites and identified an amino acid variant, Asp 386 (D386), that eliminates an NXS/T motif at position 386 in the V4 variable region of HIV gp120. D386 preferentially enhances viral entry and replication in macrophages but not microglia or peripheral blood mononuclear cells (PBMC), possibly due to differential N-linked glycosylation in these cell types. Restoring the glycan at 386 increases resistance to neutralizing antibody IgG1b12 (b12), which recognizes a conserved epitope that overlaps the CD4 binding site. Molecular modeling suggested that loss of the glycan at position 386 increases exposure of the CD4 and b12 binding sites on gp120. Loss of a glycan at 386 was significantly more frequent in Envs from HAD patients compared with non-HAD patients. These findings suggest that increased exposure of the b12 epitope overlapping the CD4 binding site via elimination of a glycan at position 386 enhances HIV macrophage tropism, and provide evidence that determinants of macrophage and microglia tropism are overlapping but distinct.
Results
Loss of a potential N-linked glycosylation site at position 386 in the V4 region in brain Envs from patients with HAD
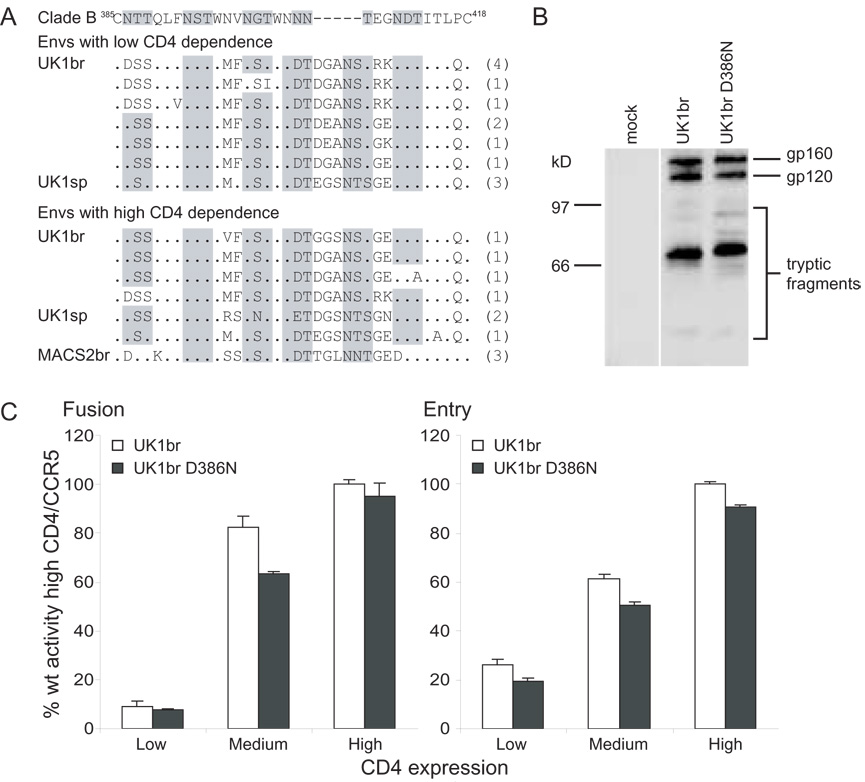
Recently, we cloned and characterized HIV Envs from AIDS patients with HAD and showed that Envs with reduced dependence on CD4 and CCR5 levels are more frequent in brain compared to lymphoid tissues (Thomas et al., 2007), suggesting that viral adaptation for replication in the CNS selects for variants with an enhanced capacity to enter cells expressing low levels of receptors. To investigate mechanisms by which neurotropic Envs acquire the ability to use low CD4, we analyzed 23 full-length Env amino acid sequences from HAD patients MACS2 and UK1 (6 Envs cloned from viral isolates and 17 Envs cloned directly from brain and lymphoid tissues (Gorry et al., 2002; Thomas et al., 2007) for variability in N-linked glycosylation motifs (NXS/T, where X is any amino acid except proline) that might affect exposure of the CD4 binding site. 10/17 brain-derived Envs had an Asp at position 386 in the V4 region, which resulted in elimination of a potential N-linked glycosylation site relative to the Clade B consensus. In contrast, 6/6 lymphoid-derived Envs had an intact N-linked glycosylation site at this position (Fig. 1A). The frequency of D386 was similar in Envs with low versus high CD4 dependence (6/13 and 4/10, respectively). These results suggest that D386 in the V4 region, which results in loss of a potential N-linked glycosylation site, is more frequent in brain compared to lymphoid Envs from two HAD patients.
Fig. 1. D386 is associated with brain-derived Envs from 2 HAD patients and has a minor effect on reduced CD4 dependence.
(A) Amino acid sequences from the V4 region of gp120 from 2 HAD patients were obtained from published studies (Gorry et al., 2002; Thomas et al., 2007) and aligned using Clustal X. Potential N-linked glycosylation sites are highlighted in gray. Sequences are numbered relative to the HXB2 reference sequence. Sequence names are notated as follows: br, brain-derived; sp, spleen-derived. The number in parentheses represents the number of Env clones with each sequence. Dots represent residues identical to the Clade B consensus sequence, and dashes represent gaps. (B) Lysates from 293T cells transfected with UK1br wild-type or D386N mutant Envs were incubated with 0.01 µg/ml trypsin for 2 h at 37°C and analyzed by Western blotting with anti-gp120. (C) Cf2-Luc cells expressing different levels of CD4 and high CCR5 were mixed with 293T cells expressing UK1br wild-type or D386N mutant Envs in cell-cell fusion assays (left) or infected with recombinant viruses expressing wild-type or mutant Envs in entry assays (right). Cells were lysed after 8–12 h incubation or 48 h post-infection, respectively, and analyzed for luciferase activity. Data were normalized to luciferase activity of the wild-type Env on cells expressing high CD4 and are expressed as means of duplicate wells from a single experiment. Error bars represent standard deviations. Data are representative of 3 independent experiments.
D386 makes a minor contribution to reduced CD4 dependence
To investigate whether D386 influences the capacity of Envs to use low CD4, we constructed a D386N mutant in UK1br15, a neurotropic Env that efficiently uses low levels of CD4 to mediate fusion and entry (Dunfee et al., 2006; Gorry et al., 2002). Hereafter, this Env is referred to as UK1br. To determine if the D386N mutation, which restores a NXS/T motif, resulted in a functional glycosylation site, UK1br wild-type and D386N mutant Envs were analyzed by partial tryptic digestion of 293T cell lysates. Trypsin-digested fragments of UK1br D386N Env migrated more slowly in SDS-PAGE gels compared to wild-type Env fragments, consistent with an additional ~2 kD N-linked glycan relative to wild-type Env (Fig. 1B). The D386N mutation in UK1br Env resulted in a minor reduction in virus entry at all levels of CD4 (Fig. 1C). The magnitude of this effect was less than that of the N283T mutation in UK1br Env, which significantly decreased virus entry into cells expressing low levels of CD4 (Dunfee et al., 2006). These data suggest that loss of a glycan at position 386 of UK1br results in a minor enhancement of virus entry at all CD4 concentrations.
D386 preferentially enhances macrophage tropism
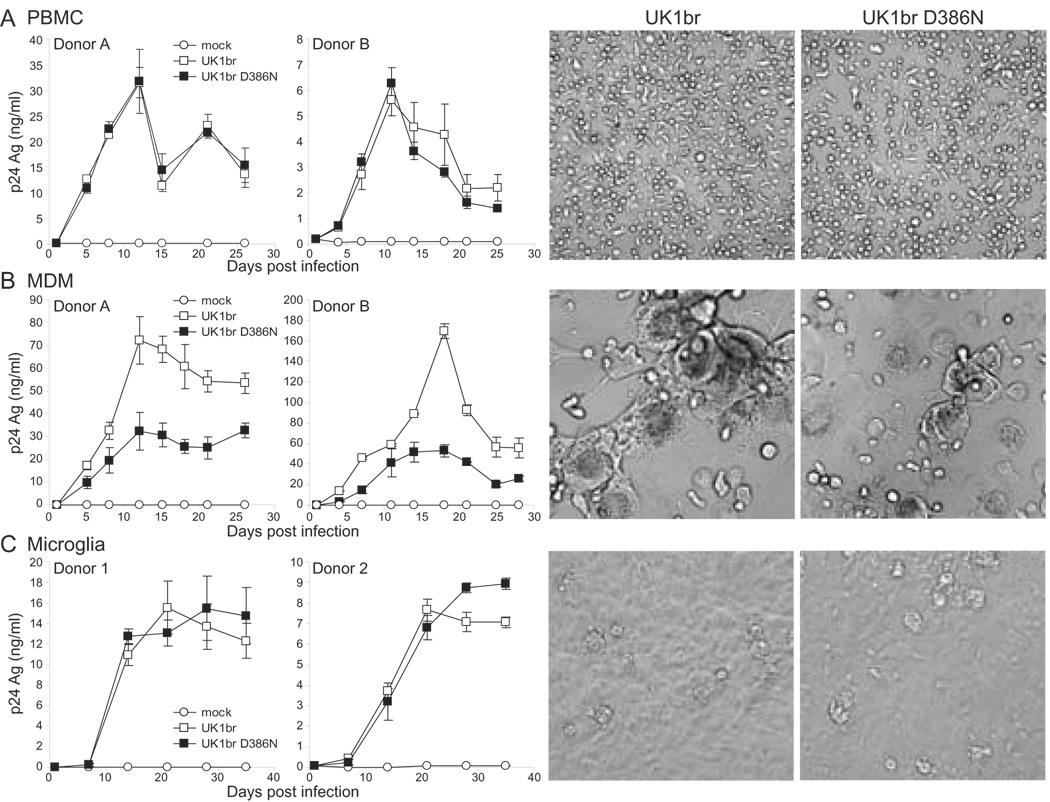
To determine if D386 influences macrophage and/or microglia tropism, we cloned UK1br wild-type or D386N mutant Envs into pNL4-3 and infected PBMC, monocyte-derived macrophages (MDM), or microglia in primary human brain cultures. Viruses expressing wild-type or mutant Envs had similar replication kinetics and cytopathic effects in PBMC (n=4 donors; Fig. 2A) and microglia in primary brain cultures (n=4 of 5 donors; Fig. 2C and data not shown). In contrast, the D386N mutant virus had delayed replication kinetics and reduced syncytia formation in MDM (n=4 donors; Fig. 2B), similar to the phenotype of the UK1br N283T mutant virus that we described previously (Dunfee et al., 2006). These results suggest that D386, or the loss of a glycan at position 386, preferentially enhances UK1br tropism for macrophages.
Fig. 2. Replication kinetics and cytopathic effects of viruses expressing UK1br wild-type and D386N mutant Envs in PBMC, MDM, and microglia.
PBMC (A), MDM (B), and microglia in primary human brain cultures (C) were infected as described in Materials and Methods. Supernatants were collected every 3–7 days and replication was monitored by p24 ELISA. Results shown are from triplicate samples. Error bars represent standard deviations. Data are representative of independent experiments in 4 different donors per cell type. Photographs were taken at 14 (PBMC and MDM) or 28 (microglia) days post-infection at a 200X magnification.
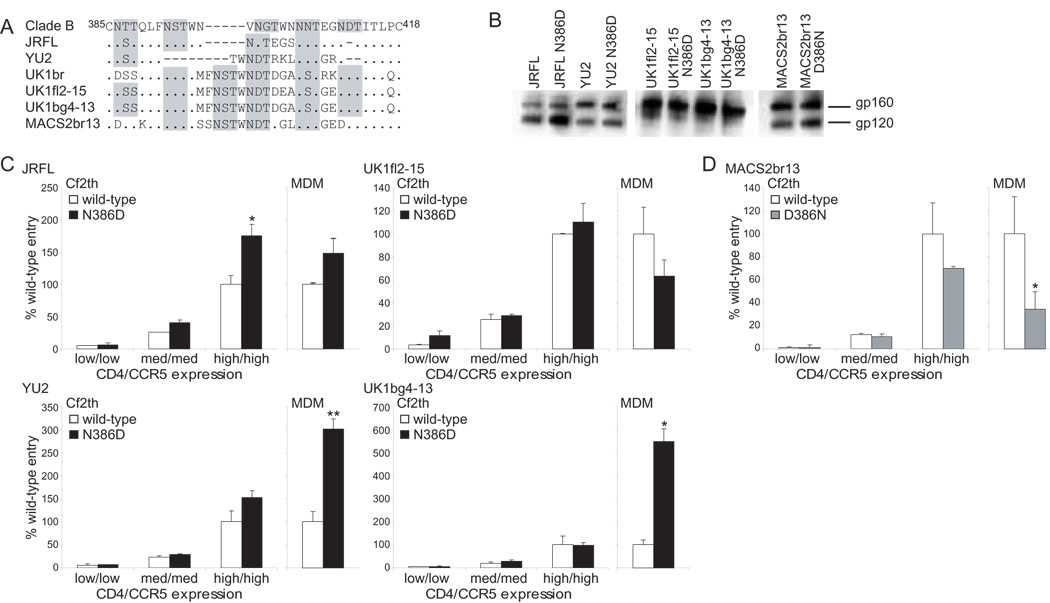
To investigate whether D386 enhances macrophage tropism in other brain-derived Envs, we used site-directed mutagenesis to introduce N386D mutations in JRFL and YU2, well-characterized, macrophage-tropic Envs from brain tissues of HAD patients (Li et al., 1991; O'Brien et al., 1990), and in UK1fl2-15 and UK1bg4-13, Envs cloned directly from the brain of patient UK1 that enter MDM at high and intermediate levels, respectively, compared with other UK1 Envs (Thomas et al., 2007). We also introduced a D386N mutation in MACS2br13, a brain-derived Env from patient MACS2 with high CD4 dependence (Gorry et al., 2002) (Fig. 3A). Wild-type and mutant Envs were expressed and processed to gp120 at similar levels, and the loss or gain of an N-linked glycan was suggested by slight differences in mobility in SDS-PAGE gels compared to wild-type Envs (Fig. 3B). Consistent with our previous results (Thomas et al., 2007), UK1 Envs had less efficient processing of gp160 to gp120 compared to other Envs. MDM (from donor A in Fig. 2) and Cf2th cells expressing different levels of CD4 and CCR5 were infected with luciferase reporter viruses pseudotyped with wild-type and mutant Envs. In JRFL Env, a N386D change increased virus entry to a similar extent in MDM and Cf2th cells (50% and 58%, respectively). A N386D mutation in UK1fl2-15 Env decreased the ability to enter MDM by 37%, but had no effect on entry in Cf2th cells (Fig. 3C). Thus, D386 does not preferentially enhance macrophage tropism in these highly macrophage-tropic Envs (O'Brien et al., 1990; Thomas et al., 2007). In contrast to these results, in YU2 and UK1bg4-13 Envs, a N386D change increased virus entry in MDM by 200% and 450%, respectively, compared to wild-type (p=0.008 and 0.049, respectively, Student’s t test), but did not increase entry in Cf2th cells (p=0.14 and 0.27, respectively) (Fig. 3C). A reciprocal D386N change in MACS2br13 Env resulted in a 65% decrease in the ability to infect MDM compared to the wild-type Env (p=0.027), but did not have a significant effect on entry in Cf2th cells (p=0.79) (Fig. 3D). Collectively, Envs with D386 had an increased capacity to enter macrophages and Cf2th cells compared to Envs with N386 (p=0.014, Student’s t test, n=10 (5 Envs, 2 cell types)). These results suggest that loss of an N-linked glycan at position 386 enhances HIV entry in macrophages and transfected Cf2th cells in 5/6 Envs from 4 HAD patients, and that D386 preferentially enhances macrophage tropism in 4/6 Envs from 3 HAD patients (YU2, UK1br, UK1bg4-13, and MACS2br13).
Fig. 3. D386 in Envs with low or intermediate macrophage-tropism increases virus entry into macrophages.
(A) Amino acid sequences from the V4 region of gp120 of brain-derived Envs used in this study were obtained from Genbank (JRFL and YU2, accession numbers U63632 and M93258, respectively) or published studies (Gorry et al., 2002; Thomas et al., 2007) and aligned using Clustal X. Potential N-linked glycosylation sites are highlighted in gray. Sequences are numbered relative to the HXB2 reference sequence. Dots represent residues identical to the Clade B consensus sequence, and dashes represent gaps. (B) 293T cells expressing wild-type or mutant Envs were lysed and analyzed by Western blotting with anti-gp120. The positions of gp160 and gp120 are indicated on the right. (C, D) Cf2th cells expressing different levels of CD4 and CCR5 or MDM were infected with luciferase-expressing reporter viruses pseudotyped with wild-type, N386D mutant (C), or D386N mutant (D) Envs. Cells were lysed 48 to 72 h postinfection and analyzed for luciferase activity. Data were normalized to the amount of luciferase activity of the wild-type Env on cells expressing high CD4/CCR5 or on MDM. Results shown are from duplicate (Cf2th) or triplicate (MDM) samples. Error bars represent standard deviations. *, p<0.05, **, p<0.01, Student’s t test.
Increased glycosylation of UK1br Envs produced in MDM compared to microglia or PBMC
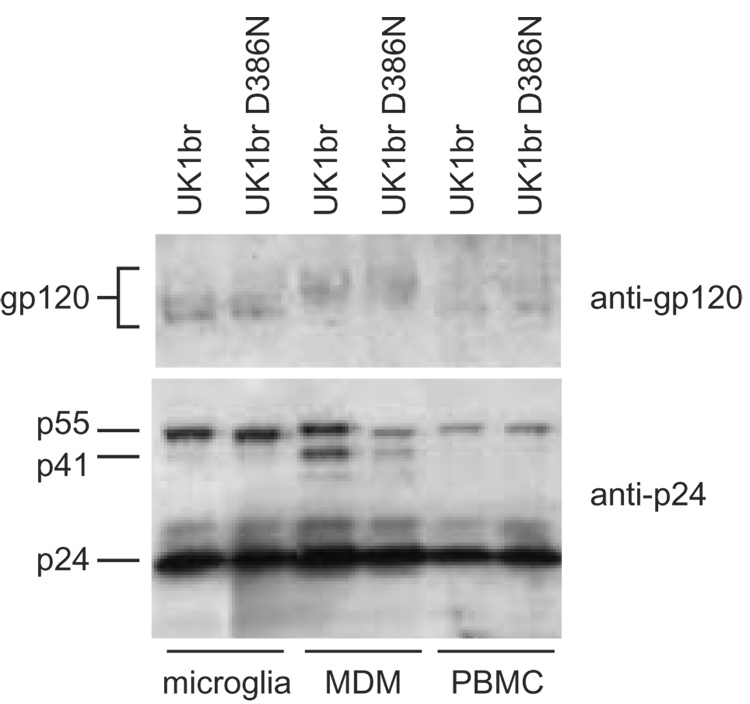
HIV Envs expressed in infected macrophage and PBMC cultures are differentially glycosylated (Lin et al., 2003; Willey et al., 1996). Thus, one possible explanation for the different effects of the UK1br D386N mutation on replication in PBMC, MDM, and microglia is differential N-linked glycosylation of Envs produced in these cell types. To investigate glycosylation of UK1br Env produced in primary cells, we purified virions from supernatants of infected PBMC, MDM, and primary brain cultures and analyzed virion-associated gp120 by Western blotting. Consistent with previous studies (Lin et al., 2003; Willey et al., 1996), UK1br Envs produced in MDM had decreased mobility and a more diffuse migration pattern in SDS-PAGE gels compared to Envs produced in PBMC (Fig. 4). Unexpectedly, Envs produced in MDM migrated more slowly and had a more diffuse pattern in SDS-PAGE gels than Envs produced in microglia (Fig. 4). Together with previous studies demonstrating that differences in mobility of MDM- and PBMC-derived Envs were due to glycosylation (Lin et al., 2003; Willey et al., 1996), these results suggest that UK1br Envs are more highly glycosylated when expressed in MDM compared to PBMC or microglia. These findings are consistent with the possibility that differences in replication of the UK1br wild-type and D386N mutant viruses in these primary cell types are due to differential N-linked glycosylation.
Fig. 4. Western blot of virions from infected microglia, MDM, and PBMC.
Virions from infected microglia, MDM, or PBMC cultures were purified by high-speed centrifugation on a sucrose cushion as described in Materials and Methods, lysed, and analyzed by Western blotting with anti-gp120 or anti-p24. The positions of gp120, p24, and the Gag precursor proteins p55 and p41 are indicated on the left.
D386 influences sensitivity of UK1br to neutralization by the CD4 binding site monoclonal antibody b12
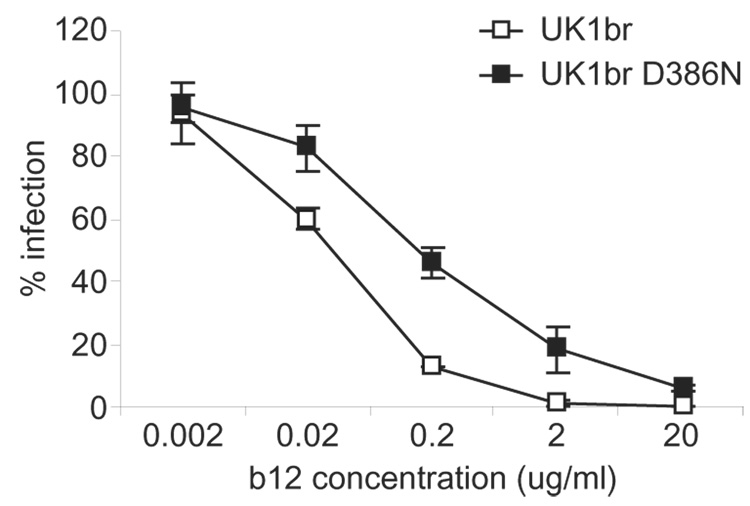
Env glycosylation is important for maintaining structure and shielding conserved structural epitopes from neutralizing antibodies (Burton et al., 2004). To investigate whether the loss of a glycan at position 386 influences sensitivity to neutralizing antibodies, we analyzed neutralization of viruses containing wild-type and D386N mutant UK1br Envs by a panel of monoclonal antibodies (mAbs) (summarized in Table 1). We previously demonstrated that the UK1-br primary isolate is sensitive to neutralizing antibody b12, which maps to an epitope that overlaps the CD4 binding site, and CD4-IgG2 (Gorry et al., 2002). A D386N change resulted in an 8-fold increase in resistance to neutralization by b12 and a 2-fold increase in resistance to CD4-IgG2 (Table 1 and Fig. 5). In contrast, there was no difference in sensitivity of the wild-type or D386N mutant UK1br viruses to neutralization by mAbs directed against other epitopes in gp120 and gp41 (Table 1). Together, these results suggest that D386 influences sensitivity of UK1br Env to neutralization by antibody b12, and raise the possibility that changes in amino acid sequence at this position may affect the conformation and/or exposure of the CD4 and mAb b12 binding sites.
Table 1.
Neutralization of UK1br wild-type and D386N mutant viruses by mAbs and CD4-IgG2
| mAb | Epitope | UK1br | UK1br D386N |
|---|---|---|---|
| b12 | CD4 binding site | 0.04 | 0.31 |
| F105 | CD4 binding site | > 20 | > 20 |
| 17b | CD4-induced | > 20 | > 20 |
| 2G12 | Outer domain glycans | > 20 | > 20 |
| 2F5 | gp41 | 2.87 | 3.11 |
| CD4-IgG2 | CD4 binding site | 0.92 | 1.78 |
mAb or CD4-IgG2 concentration (µg/ml) at which luciferase expression was reduced by 50% compared to infection in the absence of mAb (IC50).
Fig. 5. Neutralization of UK1br wild-type and D386N mutant viruses by mAb b12.
Recombinant viruses expressing UK1br wild-type or D386N mutant Envs were incubated with mAb b12 for 1 h and then used to infect Cf2-Luc cells expressing high levels of CD4 and CCR5. Cells were lysed 36 h postinfection and analyzed for luciferase activity. Data are normalized to the amount of luciferase activity in cells infected with virus in the absence of b12 and expressed as the mean of duplicate samples. Error bars represent standard deviations. Results are representative of 4 independent experiments.
Loss of a glycan at 386 increases exposure of the CD4 and mAb b12 binding sites
To investigate the mechanism by which changes at position 386 might affect the CD4 binding site, we used Swiss PDB Viewer (http://ca.expasy.org/spdbv) to model the amino acid at this position on the YU2 gp120-CD4-17b crystal structure (1G9N) (Kwong et al., 1998). The HIV Clade B consensus amino acid at this position is Asn (Fig. 6A). N386 is located on the highly glycosylated outer domain of gp120 (Kwong et al., 1998; Wyatt et al., 1998), proximal to the base of the V3 loop and the CCR5 binding site, and ~12 Å from the CD4 binding site. Introducing an N386D change in the structural model did not alter predicted interactions with amino acids that contact CD4 (data not shown). Previous studies demonstrated that N386 in monomeric gp120 produced in CHO cells is glycosylated with a high mannose group (Leonard et al., 1990). To better understand how a glycan at this position might affect the CD4 binding site, we aligned the N-acetylglucosamine (NAG) attached to N386 in the YU2 crystal structure with the first NAG of a high mannose group from the crystal structure of HIV mAb 2G12 bound to its Man9GlcNAc2 epitope (1OP5) (Calarese et al., 2003). The addition of the mannose group resulted in a modified N386 in which the carbohydrate side chains were only ~7 Å from CD4, partially occluding the CD4 binding site (Fig. 6B). Together with previous studies demonstrating that removal of the glycan at position 386 affects the conformation of surrounding carbohydrate chains (Scanlan et al., 2002), the structural modeling data suggest that removal of this N-linked glycosylation site may directly or indirectly increase exposure of the CD4 binding site.
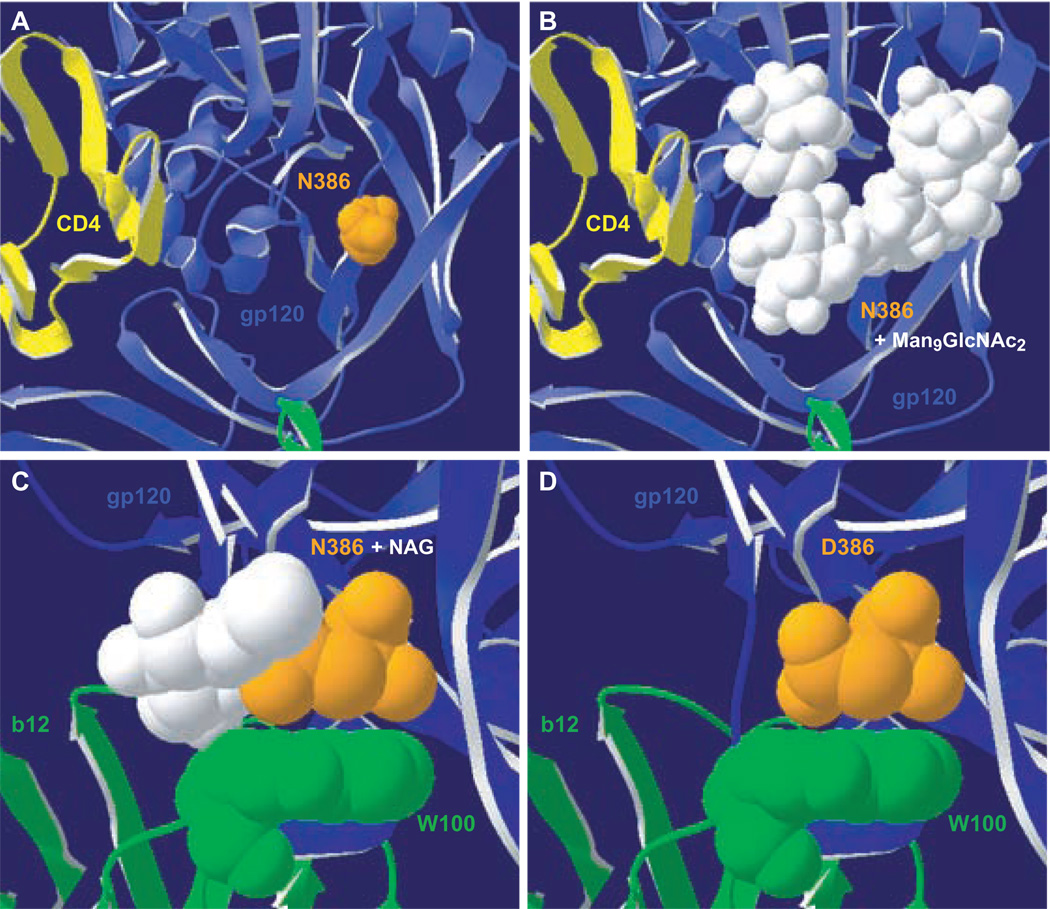
Fig. 6. Structural modeling of position 386 in Envs bound to CD4 or mAb b12.
Swiss PDB Viewer was used to model changes at position 386 on the V4 region of the YU2 gp120-CD4-17b CD4i Ab crystal structure (1G9N) (Kwong et al., 1998) (A, B) or the HXB2 gp120-mAb b12 crystal structure (2NY7) (Zhou et al., 2007) (C, D). Blue, gp120. Yellow, CD4. Green, 17b CD4i Ab (A, B) or mAb b12 (C, D). Orange, position 386. White, carbohydrate moieties. (A) Location of N386 in the YU2 crystal structure. Position 386 is ~12Å from the CD4 binding site. (B) The N-acetylglucosamine (NAG) attached to N386 was aligned with NAG1 from a high mannose group (Man9GlcNAc2) derived from the crystal structure of mAb 2G12 bound to its carbohydrate epitope (1OP5) (Calarese et al., 2003) to produce a model of gp120 with intact glycosylation at position 386. (C) N386 and the attached NAG make contacts with W100 of mAb b12. (D) Swiss PDB Viewer was used to model an N386D change in HXB2, which results in elimination of the NAG but retains the conformation of the b12 W100 and Env amino acid 386 side chains.
To investigate the mechanism by which D386 might enhance sensitivity to mAb b12, we modeled the N386D change on the recently described HXB2 gp120-b12 crystal structure (Zhou et al., 2007). N386 and the attached NAG contact Trp 100 (W100) of the heavy chain of b12, with the amino acid side chains stacking together (Fig. 6C) and a hydrogen bond between the backbone of W100 and the NAG (data not shown) (Zhou et al., 2007). Modeling a D386 change in the structure eliminated the hydrogen bond between W100 and the NAG of N386, but D386 and W100 of b12 side chains had a similar stacking conformation as the wild-type N386 (Fig. 6D). Thus, a D386N change would be predicted to enhance b12 binding and neutralization by restoring the hydrogen bond between the NAG on N386 and the mAb heavy chain. However, a D386N mutation in UK1br Env had the opposite effect and increased resistance to neutralization by b12. These data suggest that a mechanism other than direct binding of b12 to D386, such as increased exposure of the b12 epitope, underlies the increase in b12 neutralization sensitivity mediated by D386.
Relationship between neutralization sensitivity to mAb b12 and enhanced macrophage tropism
To investigate whether increased neutralization sensitivity to mAb b12 is associated with enhanced macrophage tropism in a larger data set, we tested b12 neutralization of viruses expressing UK7br, ADA, or MACS2br13 Envs, and performed a meta-analysis of these results together with b12 neutralization data from 4 published studies (Binley et al., 2004; Gray et al., 2005; Thomas et al., 2007; Wei et al., 2003) (Table 2). For this analysis, we compiled results for HIV Envs or isolates that have been well-characterized for their macrophage tropism (Collman et al., 1989; Dunfee et al., 2006; Gray et al., 2005; Li et al., 1991; O'Brien et al., 1990; Thomas et al., 2007). Highly macrophage-tropic viruses had increased sensitivity to b12 neutralization (median 50% neutralization (IC50) 0.16 µg/ml, n=10) compared to Envs with low or intermediate macrophage tropism (median IC50 4.21 µg/ml, n=8) (p=0.0044, Mann-Whitney test). Furthermore, 9 of 11 (82%) of Envs with b12 IC50 ≤ 2 µg/ml were highly macrophage-tropic, compared with only 1 of 7 (14%) of Envs with b12 IC50 > 2 µg/ml (p=0.013, Fisher’s exact test). Thus, there is a significant association between increased sensitivity to b12 neutralization and enhanced macrophage tropism. In contrast, there was no correlation between sensitivity to other gp120 mAbs and enhanced macrophage tropism in this data set (Binley et al., 2004; Gray et al., 2005; Thomas et al., 2007; Wei et al., 2003) (Table 1). Viruses with Env variant D386 were sensitive to b12 neutralization (n=4, IC50 0.03 to 2.8 µg/ml), whereas viruses with N386 had variable sensitivity to b12 neutralization (n=14, IC50 0.03 to 25 µg/ml) (Table 2). Together, these findings suggest that increased exposure of the b12 epitope is associated with enhanced tropism of HIV for macrophages. The influence of the N-linked glycosylation site at position 386 on b12 neutralization sensitivity, however, is strain-dependent.
Table 2.
Neutralization sensitivity of HIV-1 Envs and viral isolates with variable macrophage tropism to mAb b12
| Env clone or isolatea | M-tropism | Amino acid at 386 | b12 IC50b | Reference |
|---|---|---|---|---|
| UK1br | high | D | 0.03 | |
| SF162 | high | N | 0.03 | Binley et al., 2004 |
| JRFL | high | N | 0.09 | Binley et al., 2004 |
| NB6 | high | Dc | <0.1 | Gray et al., 2005 |
| UK7br | high | N | 0.16 | |
| NB2 | high | Nc | 0.16 | Gray et al., 2005 |
| BaL | high | N | 0.19 | Binley et al., 2004 |
| ADA | high | N | 0.91 | |
| NB7 | high | Nc | 2 | Gray et al., 2005 |
| NB8 | high | Dc | 2.8 | Gray et al., 2005 |
| MACS2br13 | intermediate | D | 0.48 | |
| YU2 | intermediate | N | 2.3 | Wei et al., 2003 |
| MACS2fl8-12 | intermediate | N | 5.22 | Thomas et al., 2007 |
| JRCSF | low | N | 0.16 | Binley et al., 2004 |
| NB25 | low | Nc | 3.2 | Gray et al., 2005 |
| NB27 | low | Nc | 7 | Gray et al., 2005 |
| NB24 | low | Nc | 12 | Gray et al., 2005 |
| NB23 | low | Nc | 25 | Gray et al., 2005 |
Data represents results of neutralization assays performed using cloned HIV Envs except for BaL (viral quasispecies) and the data from Gray et al. (primary isolates).
µg/ml
J. Sterjovski and P. R. Gorry, unpublished data
Loss of a glycan at position 386 is associated with dementia in vivo
To determine if the loss of an N-linked glycan at position 386 is associated with brain infection or dementia in vivo, we examined 284 matched brain- and blood- or lymphoid-derived Envs from 22 patients in 10 published studies (Dunfee et al., 2006; Gartner et al., 1997; Gorry et al., 2002; Korber et al., 1994; Liu et al., 2000; Martin-Garcia et al., 2006; Ohagen et al., 2003; Pang et al., 1991; Shapshak et al., 1995; Thomas et al., 2007). D386 or another amino acid variant that results in loss of the NXS/T motif at position 386 appeared in 18% of brain-derived sequences (n=152) and 20% of blood or lymphoid-derived sequences (n=132) (Table 3). Thus, loss of an N-linked glycan at position 386 is not associated with brain compartmentalization. To determine if loss of a glycan at this site is associated with HAD in this data set, we compared sequences from HAD and non-HAD patients. Amino acid variants that resulted in the loss of the glycosylation motif appeared in 26% of sequences from 13 HAD patients (n=185), compared to only 7% of sequences from 9 non-HAD patients (n=99) (Table 3). In contrast, variants that eliminated any of the 4 other NXS/T motifs in the Clade B consensus V4 region appeared at similar or lower frequencies in HAD compared to non-HAD patients (data not shown). Thus, the loss of an N-linked glycan at position 386 was significantly associated with dementia (p<0.001, Fisher’s exact test, with Bonferroni correction for multiple comparisons). Although amino acid variants resulting in elimination of the NXS/T motif appeared in 22% of brain-derived sequences from HAD patients (n=103) compared to 10% from non-HAD patients (n=49), the most significant association of these variants with HAD was in the blood or lymphoid compartment. Amino acid variants resulting in the loss of a glycosylation site at position 386 appeared in blood or lymphoid-derived Envs in 30% of sequences from HAD patients (n=82) compared to only 4% of sequences from non-HAD patients (n=50; p<0.001) (Table 3). These results suggest that D386 and other variants that result in elimination of an N-linked glycosylation site at position 386 are associated with dementia in vivo.
Table 3.
Loss of an N-linked glycosylation site at position 386 is associated with HADa
| Amino acid at 386 | Glycosylation site at 386 | |||||
|---|---|---|---|---|---|---|
| Comparison | Nb | D | K | S | Present | Absent |
| By tissue (22 patients) | ||||||
| Brain (n=152) | 0.83c | 0.14 | 0.03 | 0 | 0.82 | 0.18 |
| Lymphoid (n=132) | 0.82 | 0.10 | 0.04 | 0.04 | 0.80 | 0.20 |
| By disease | ||||||
| HAD (13 patients) | ||||||
| Total (n=185) | 0.76 | 0.16* | 0.05 | 0.03 | 0.74 | 0.26** |
| Brain (n=103) | 0.79 | 0.16 | 0.04 | 0.01 | 0.78 | 0.22 |
| Lymphoid (n=82) | 0.73 | 0.15* | 0.06 | 0.06 | 0.70 | 0.30** |
| non-HAD (9 patients) | ||||||
| Total (n=99) | 0.95 | 0.05* | 0 | 0 | 0.93 | 0.07** |
| Brain (n=49) | 0.92 | 0.08 | 0 | 0 | 0.90 | 0.10 |
| Lymphoid (n=50) | 0.98 | 0.02* | 0 | 0 | 0.96 | 0.04** |
Sequences were obtained from published studies (Dunfee et al., 2006; Gartner et al., 1997; Gorry et al., 2002; Korber et al., 1994; Liu et al., 2000; Martin-Garcia et al., 2006; Ohagen et al., 2003; Pang et al., 1991; Shapshak et al., 1995; Thomas et al., 2007) and Genbank.
N is the Clade B consensus residue.
The frequency is calculated by dividing the number of sequences with each amino acid variant or with or without the NXS/T glycosylation motif (where X is any amino acid except proline) at position 386 by the total number of sequences. Sequences were limited to 2 to 10 per patient.
p < 0.05.
p < 0.001. p values calculated using Fisher’s exact test.
Discussion
In this study, we demonstrated that D386 and other amino acids that result in the loss of an N-linked glycosylation site in the V4 region of HIV gp120 are more frequent in brain and lymphoid Envs from HAD patients (26%, n=185) compared to non-HAD patients (7%, n=99; p<0.001). D386 preferentially enhances HIV tropism for macrophages but not microglia or PBMC, possibly as a result of differential modification of N-linked glycosylation motifs in these primary cell types. A D386N mutation in UK1br Env increased resistance to b12 neutralization by 8-fold compared to wild-type. These results, together with molecular modeling studies, suggest that loss of a glycan at position 386 increases exposure of the CD4 binding site. These findings also suggest an association between exposure of the b12 epitope, which overlaps the CD4 binding site, and enhanced HIV tropism for macrophages. Consistent with this idea, increased b12 neutralization sensitivity is associated with enhanced macrophage tropism of other Clade B Envs as well (p=0.0044, Mann-Whitney test).
Molecular modeling with the D386 variant in the YU2 crystal structure (1G9N) (Kwong et al., 1998) suggests that removal of the high mannose group attached to position 386, which is located on the heavily glycosylated outer domain of gp120, increases exposure of the CD4 binding site. Restoring the N-glycan at 386 increased UK1br resistance to neutralization by sCD4-Ig by only 2-fold, consistent with our finding that a D386N mutation had only a minor effect on the capacity of UK1br Envs to use low CD4. In contrast, the N283T mutation, which influences gp120 affinity for CD4 but has no significant effect on b12 neutralization (R. Dunfee and D. Gabuzda, unpublished data), has a significant effect on virus entry into cells expressing low CD4 (Dunfee et al., 2006). The observation that UK1br D386N mutant virus was 8-fold more resistant to neutralization by b12 than the wild-type virus was unexpected, since molecular modeling with the HXB2 gp120-b12 crystal predicted that a D386N mutation would enhance b12 binding by restoring interactions between the glycan at N386 and the mAb heavy chain. The influence of this glycan on b12 sensitivity is strain-dependent, however (Table 2). D386 may work cooperatively with changes in the V1/V2, C2, V3, and/or C3 regions, since these regions also influence sensitivity to b12 neutralization (Koch et al., 2003; Mo et al., 1997; Pugach et al., 2004). Alternatively, changes in glycan structure at position 386 could affect the conformation and packing of other carbohydrates on the outer domain of gp120 to modulate exposure of the CD4 and mAb b12 binding sites. The finding that loss of an N-linked glycan at position 386 in an HXB2 Env adapted to use only CXCR4 for entry enhanced sensitivity to neutralization by HIV-infected patient sera (Edwards et al., 2001) provides further evidence that changes at position 386 influence exposure of the CD4 binding site and conserved neutralization epitopes on gp120. Position 386 is located at the base of the V3 loop and proximal to the coreceptor binding site. Thus, changes at this position may influence Env interactions with CCR5 as well.
Brain macrophages and microglia are both derived from the mononuclear phagocyte lineage, yet they differ in some phenotypic and functional characteristics (Gonzalez-Scarano and Martin-Garcia, 2005). Whether HIV tropism for macrophages and microglia is similar (Ghorpade et al., 1998; Gorry et al., 2001), or overlapping but distinct (Strizki et al., 1996), has been debated. The D386N mutation in UK1br decreased viral entry and replication in macrophages but had no effect on viral replication in microglia. In contrast, the N283 variant, which increases gp120 affinity for CD4, enhances viral tropism for both macrophages and microglia (Dunfee et al., 2006). MDM-derived Envs migrated more slowly on SDS-PAGE gels than microglia- or PBMC-derived Envs, consistent with increased gp120 glycosylation. Thus, D386 may preferentially enhance HIV replication in macrophages as a consequence of differential N-linked glycosylation in these primary cell types.
The mechanism by which loss of an N-linked glycan at position 386 contributes to enhanced tropism for macrophages, but not PBMC or microglia, is unknown. Restoring the glycosylation site with a D386N mutation in UK1br Envs had a minor effect on virus entry in Cf2th cells at all levels of CD4, raising the possibility that D386 enhances Env interactions with CD4, and/or CCR5, albeit to a minor extent. One possible explanation for preferential enhancement of macrophage tropism by D386 is differential glycosylation of the NXS/T motif at 386 in PBMC, macrophages, and microglia. An alternative possibility is that changes in glycosylation at position 386 influence Env interactions with molecules important for initial attachment to macrophages, such as DC-SIGN, macrophage mannose receptor, syndecans, or other attachment factors (Binley et al., 2006; Nguyen and Hildreth, 2003; Saphire et al., 2001). Further studies are needed to investigate these possibilities.
The CNS has lower concentrations of neutralizing antibodies compared to peripheral tissues (Goudsmit et al., 1987; Kaul et al., 2001), and may therefore be a favorable environment for replication and persistence of HIV variants with exposed receptor binding sites. Most HIV-infected individuals do not develop neurological dysfunction until after development of immunosuppression and progression to AIDS. Loss of immune control associated with disease progression is one possible reason why HAD typically occurs only in the late stages of HIV infection (Gonzalez-Scarano and Martin-Garcia, 2005). van Marle et al (Van Marle et al., 2002) demonstrated that sera from HAD patients was less efficient at neutralizing HIV than sera from non-HAD patients, suggesting that patients with HAD may have a decreased ability to generate neutralizing antibodies. The significantly higher frequency of D386, a viral variant that enhances b12 neutralization sensitivity, in blood- and lymphoid-derived Envs from HAD patients compared to non-HAD patients, is consistent with the possibility that loss of a glycan at 386 may have evolved more readily in these individuals because they have lower levels of neutralizing antibodies that target the b12 epitope. In a longitudinal study of neutralizing antibody responses, changes at positions 400–406 in the V4 region were indicative of adaptive mutations selected by immune responses (Wei et al., 2003). In our Env sequence data set, position 406 was the only N-linked glycosylation motif in the V4 region that was eliminated at a significantly higher frequency in non-HAD patients (49%, n=99) compared to HAD patients (26%, n=185; p<0.001, Fisher’s exact test, with Bonferroni correction for multiple comparisons) (data not shown). These observations and the association between enhanced macrophage tropism and increased exposure of the b12 epitope demonstrated here and by others (Gray et al., 2005) raise the possibility that macrophage-tropic strains may evolve more readily in individuals with low levels of neutralizing antibodies directed against outer domain regions of Env.
The identification and characterization of the loss of an N-linked glycan at position 386 in the V4 region of gp120 suggests that increased exposure of the b12 epitope overlapping the CD4 binding site enhances HIV macrophage tropism, and provide evidence that determinants of macrophage and microglia tropism are overlapping but distinct. These findings also provide insights that may facilitate development of immunogens that elicit broadly neutralizing antibodies as well as therapeutics to prevent CNS infection and neurologic injury in HIV-infected patients.
Materials and methods
Cells
293T cells and canine thymocyte cell line Cf2th (Choe et al., 1996) were cultured in DMEM medium supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 100 µg/ml penicillin and streptomycin. Cf2-Luc cells, which are derived from Cf2th cells and stably express luciferase under control of HIV-1 LTR (Etemad-Moghadam et al., 2000), were cultured in medium supplemented with 0.7 mg/ml G418 (Mediatech, Herndon, VA). Peripheral blood mononuclear cells (PBMC) were isolated from the blood of healthy HIV-1-negative donors as described (Gorry et al., 2001), stimulated with 2 µg/ml phytohemagglutinin (PHA; Sigma, St. Louis, MO), and cultured in RPMI 1640 medium supplemented with 10% (vol/vol) FBS, 100 µg/ml penicillin and streptomycin, and 20 U/ml interleukin-2 (Boehringer Mannheim, Germany). CD8+ T cells were depleted with anti-CD8-conjugated magnetic beads (Miltenyi Biotech, Auburn, CA). Monocyte-derived macrophages (MDM) were isolated from PBMC by plastic adherence and cultured in RPMI 1640 medium supplemented with 10% FBS, 100 µg/ml penicillin and streptomycin, and 10 ng/ml macrophage colony stimulating factor (M-CSF; R&D Systems, Minneapolis, MN) (Gorry et al., 2001). Primary human fetal brain cultures containing a mixture of astrocytes, neurons, and microglia were prepared and maintained in 48-well tissue culture plates as described (Gorry et al., 2001; Gorry et al., 2002) and cultured in DMEM medium supplemented with 10% bovine calf serum (HyClone, Logan, UT), 100 µg/ml penicillin and streptomycin, 1 mM sodium pyruvate (Invitrogen, Carlsbad, CA), and 10 ng/ml M-CSF.
Virus isolates, HIV Env cloning, and sequence analysis
Primary viruses MACS2-br and UK1-br were previously isolated from autopsy brain tissues from AIDS patients with HAD (Gorry et al., 2001). Env genes amplified from genomic DNA extracted from PBMC on day 7 post-infection with these primary isolates or from autopsy brain tissue and cloned into pCR3.1 were described previously (Dunfee et al., 2006; Gorry et al., 2002; Thomas et al., 2007). Env protein expression was verified by Western blot with goat anti-gp120 (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, Bethesda, MD) (Gorry et al., 2002). Sequences were aligned using Clustal X. D386N and N386D mutant Env plasmids were created by PCR-based mutagenesis and changes were verified by DNA sequencing. Differences in migration of UK1br wild-type and D386N mutant Envs on SDS-PAGE gels, consistent with changes in glycosylation, were verified by Western blot with goat anti-gp120 of 293T lysates incubated with 0.01 µg/ml trypsin (Sigma, St. Louis, MO) for 2 h at 37°C, run on a 4–20% SDS-PAGE gradient gel, and analyzed by Western blot with goat anti-gp120.
Fusion assays
293T cells were cotransfected with 15 µg of Env-expressing plasmid and 2 µg of pLTR-Tat using calcium phosphate. Cf2-Luc cells (Gorry et al., 2002) were cotransfected with different amounts of pcDNA3-CD4 and pcDNA3-CCR5 as indicated using Lipofectamine 2000 (Invitrogen). The total amount of DNA in each transfection was normalized using pcDNA3. 2.5 × 104 293T cells and 2.5 × 105 Cf2-Luc cells were mixed in 48-well plates and incubated at 37°C for 8 to 12 h. Cells were lysed and assayed for luciferase activity. 293T cells cotransfected with a nonfunctional Env (pSVIII-ΔKSenv) and pLTR-Tat were used to determine background levels.
Entry assays
The KpnI to BamHI region of env was PCR amplified and cloned into pNL4-3 to generate replication-competent chimeric viruses (Ohagen et al., 2003). HIV luciferase reporter viruses were generated by cotransfection of 293T cells with pNL4-3env‾luc, an HIV provirus with env deleted and nef replaced by luciferase, and an Env-expressing plasmid as described (Gorry et al., 2002). Cf2th or Cf2-Luc cells were cotransfected with pcDNA3-CD4 and pcDNA3-CCR5 as above. Transfected cells were infected with 104 ³H cpm reverse transcriptase (RT) units of virus stock. Cells were lysed 48 to 60 h post-infection and assayed for luciferase activity. MDM were prepared as above in 48-well plates and infected with 2 × 104 ³H cpm RT units of virus stock. Cells were lysed 6 days post-infection and assayed for luciferase activity.
HIV replication kinetics
3 × 106 PBMC were prepared as described (Gorry et al., 2001) and incubated with 5 × 104 ³H cpm RT units of virus stock for 3 h at 37°C. MDM were prepared by plastic adherence as described above in 24-well tissue culture plates, cultured in the presence of M-CSF for 7 days prior to infection, and incubated with 2 × 104 ³H cpm RT units for 3 h at 37°C. Primary human fetal brain cultures containing a mixture of astrocytes, neurons, and microglia were prepared and maintained in 48-well tissue culture plates as described (Gorry et al., 2001; Gorry et al., 2002) and incubated with equivalent amounts of virus stock (104 ³H cpm RT units) for 16 h at 37°C. 50% media changes were performed every 3 to 7 days for 28 to 35 days. Virus replication was monitored by p24 ELISA (Perkin Elmer, Boston, MA).
Virion preparation and Western blot
Virions were purified from cell culture supernatant of infected PBMC, MDM, or primary human fetal brain cultures by high-speed centrifugation on a 20% sucrose gradient and resuspended in lysis buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 0.05 M Tris hydrochloride buffer [pH 7.5], 0.15 M NaCl, 1 mM EDTA, Complete protease inhibitor cocktail (Roche, Indianapolis, IN)) at a concentration of 1 ng p24 antigen/µl. Equivalent amounts of virions were run on a 4–20% SDS-PAGE gradient gel, and analyzed by Western blotting with goat anti-gp120 as above or with rabbit anti-p24 (Abbott Laboratories, Abbott Park, IL).
Neutralization assays
Replication-competent chimeric virus stocks were incubated with a range of concentrations of human monoclonal Abs (mAbs), or CD4-IgG2 1 h prior to infection of Cf2-Luc cells transiently expressing CD4 and CCR5. Cells were harvested 48 h post infection and assayed for luciferase activity. mAbs b12 (Roben et al., 1994), 2G12 (Buchacher et al., 1994), and 2F5 (Buchacher et al., 1994; Purtscher et al., 1994) were obtained from D. Burton and P. Parren, and H. Katinger (2G12 and 2F5), respectively, through the AIDS Research and Reference Reagents Program. mAb 17b (Thali et al., 1993) was kindly provided by J. Robinson. mAb F105 was kindly provided by M. Posner and L. Cavacini. The plasmid expressing CD4-IgG2 was kindly provided by M. Farzan.
Acknowledgements
We thank J. Cunningham, R. Desrosiers, and J. Sodroski for critical reading of the manuscript and helpful discussions. This work was supported by NIH grant NS37277. Core facilities were supported by Harvard University Center for AIDS Research and DFCI/Harvard Cancer Center grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albright AV, Shieh JT, Itoh T, Lee B, Pleasure D, O'Connor MJ, Doms RW, Gonzalez-Scarano F. Microglia express CCR5, CXCR4, and CCR3, but of these, CCR5 is the principal coreceptor for human immunodeficiency virus type 1 dementia isolates. J Virol. 1999;73(1):205–213. doi: 10.1128/jvi.73.1.205-213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Ngo-Abdalla S, Moore P, Bobardt M, Chatterji U, Gallay P, Burton DR, Wilson IA, Elder JH, de Parseval A. Inhibition of HIV Env binding to cellular receptors by monoclonal antibody 2G12 as probed by Fc-tagged gp120. Retrovirology. 2006;3:39. doi: 10.1186/1742-4690-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of antihuman immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10(4):359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300(5628):2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85(7):1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- Collman R, Hassan NF, Walker R, Godfrey B, Cutilli J, Hastings JC, Friedman H, Douglas SD, Nathanson N. Infection of monocyte-derived macrophages with human immunodeficiency virus type 1 (HIV-1). Monocyte-tropic and lymphocyte-tropic strains of HIV-1 show distinctive patterns of replication in a panel of cell types. J Exp Med. 1989;170(4):1149–1163. doi: 10.1084/jem.170.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Arminio Monforte A, Cinque P, Mocroft A, Goebel FD, Antunes F, Katlama C, Justesen US, Vella S, Kirk O, Lundgren J. Changing incidence of central nervous system diseases in the EuroSIDA cohort. Ann Neurol. 2004;55(3):320–328. doi: 10.1002/ana.10827. [DOI] [PubMed] [Google Scholar]
- Desrosiers RC, Hansen-Moosa A, Mori K, Bouvier DP, King NW, Daniel MD, Ringler DJ. Macrophage-tropic variants of SIV are associated with specific AIDS-related lesions but are not essential for the development of AIDS. Am J Pathol. 1991;139(1):29–35. [PMC free article] [PubMed] [Google Scholar]
- Doms RW. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276(2):229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- Dore GJ, Correll PK, Li Y, Kaldor JM, Cooper DA, Brew BJ. Changes to AIDS dementia complex in the era of highly active antiretroviral therapy. Aids. 1999;13(10):1249–1253. doi: 10.1097/00002030-199907090-00015. [DOI] [PubMed] [Google Scholar]
- Dore GJ, McDonald A, Li Y, Kaldor JM, Brew BJ. Marked improvement in survival following AIDS dementia complex in the era of highly active antiretroviral therapy. Aids. 2003;17(10):1539–1545. doi: 10.1097/00002030-200307040-00015. [DOI] [PubMed] [Google Scholar]
- Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, Kunstman K, Wolinsky SM, Gabuzda D. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci U S A. 2006;103(41):15160–15165. doi: 10.1073/pnas.0605513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TG, Hoffman TL, Baribaud F, Wyss S, LaBranche CC, Romano J, Adkinson J, Sharron M, Hoxie JA, Doms RW. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J Virol. 2001;75(11):5230–5239. doi: 10.1128/JVI.75.11.5230-5239.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Sun Y, Nicholson EK, Fernandes M, Liou K, Gomila R, Lee J, Sodroski J. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J Virol. 2000;74(9):4433–4440. doi: 10.1128/jvi.74.9.4433-4440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner S, McDonald RA, Hunter EA, Bouwman F, Liu Y, Popovic M. Gp120 sequence variation in brain and in T-lymphocyte human immunodeficiency virus type 1 primary isolates. J Hum Virol. 1997;1(1):3–18. [PubMed] [Google Scholar]
- Ghorpade A, Xia MQ, Hyman BT, Persidsky Y, Nukuna A, Bock P, Che M, Limoges J, Gendelman HE, Mackay CR. Role of the beta-chemokine receptors CCR3 and CCR5 in human immunodeficiency virus type 1 infection of monocytes and microglia. J Virol. 1998;72(4):3351–3361. doi: 10.1128/jvi.72.4.3351-3361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Scarano F, Martin-Garcia J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5(1):69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ, Bell JE, Bannert N, Crawford K, Wang H, Schols D, De Clercq E, Kunstman K, Wolinsky SM, Gabuzda D. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J Virol. 2001;75(21):10073–10089. doi: 10.1128/JVI.75.21.10073-10089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, Moore JP, Wolinsky SM, Gabuzda D. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol. 2002;76(12):6277–6292. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray L, Sterjovski J, Churchill M, Ellery P, Nasr N, Lewin SR, Crowe SM, Wesselingh SL, Cunningham AL, Gorry PR. Uncoupling coreceptor usage of human immunodeficiency virus type 1 (HIV-1) from macrophage tropism reveals biological properties of CCR5-restricted HIV-1 isolates from patients with acquired immunodeficiency syndrome. Virology. 2005;337(2):384–398. doi: 10.1016/j.virol.2005.04.034. [DOI] [PubMed] [Google Scholar]
- He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay CR, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385(6617):645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- Hoffman TL, LaBranche CC, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci U S A. 1999;96(11):6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Imamichi H, Brown CR, Hirsch VM, Martin MA. The emergence and characterization of macrophage-tropic SIV/HIV chimeric viruses (SHIVs) present in CD4+ T cell-depleted rhesus monkeys. J Leukoc Biol. 2003;74(5):772–780. doi: 10.1189/jlb.0503196. [DOI] [PubMed] [Google Scholar]
- Kaul M, Garden GA, Lipton SA. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature. 2001;410(6831):988–994. doi: 10.1038/35073667. [DOI] [PubMed] [Google Scholar]
- Kaul M, Zheng J, Okamoto S, Gendelman HE, Lipton SA. HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death Differ. 2005;12 Suppl 1:878–892. doi: 10.1038/sj.cdd.4401623. [DOI] [PubMed] [Google Scholar]
- Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313(2):387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol. 2001;75(7):3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber BT, Kunstman KJ, Patterson BK, Furtado M, McEvilly MM, Levy R, Wolinsky SM. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68(11):7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265(18):10373–10382. [PubMed] [Google Scholar]
- Lewin SR, Sonza S, Irving LB, McDonald CF, Mills J, Crowe SM. Surface CD4 is critical to in vitro HIV infection of human alveolar macrophages. AIDS Res Hum Retroviruses. 1996;12(10):877–883. doi: 10.1089/aid.1996.12.877. [DOI] [PubMed] [Google Scholar]
- Li S, Juarez J, Alali M, Dwyer D, Collman R, Cunningham A, Naif HM. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J Virol. 1999;73(12):9741–9755. doi: 10.1128/jvi.73.12.9741-9755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol. 1991;65(8):3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Simmons G, Pohlmann S, Baribaud F, Ni H, Leslie GJ, Haggarty BS, Bates P, Weissman D, Hoxie JA, Doms RW. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J Virol. 2003;77(2):1337–1346. doi: 10.1128/JVI.77.2.1337-1346.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tang XP, McArthur JC, Scott J, Gartner S. Analysis of human immunodeficiency virus type 1 gp160 sequences from a patient with HIV dementia: evidence for monocyte trafficking into brain. J Neurovirol. 2000;6 Suppl 1:S70–S81. [PubMed] [Google Scholar]
- Martin-Garcia J, Cao W, Varela-Rohena A, Plassmeyer ML, Gonzalez-Scarano F. HIV-1 tropism for the central nervous system: Brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology. 2006;346(1):169–179. doi: 10.1016/j.virol.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Martin J, LaBranche CC, Gonzalez-Scarano F. Differential CD4/CCR5 utilization, gp120 conformation, and neutralization sensitivity between envelopes from a microglia-adapted human immunodeficiency virus type 1 and its parental isolate. J Virol. 2001;75(8):3568–3580. doi: 10.1128/JVI.75.8.3568-3580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur JC, Haughey N, Gartner S, Conant K, Pardo C, Nath A, Sacktor N. Human immunodeficiency virus-associated dementia: an evolving disease. J Neurovirol. 2003;9(2):205–221. doi: 10.1080/13550280390194109. [DOI] [PubMed] [Google Scholar]
- Mo H, Stamatatos L, Ip JE, Barbas CF, Parren PW, Burton DR, Moore JP, Ho DD. Human immunodeficiency virus type 1 mutants that escape neutralization by human monoclonal antibody IgG1b12. off. J Virol. 1997;71(9):6869–6874. doi: 10.1128/jvi.71.9.6869-6874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Ringler DJ, Kodama T, Desrosiers RC. Complex determinants of macrophage tropism in env of simian immunodeficiency virus. J Virol. 1992;66(4):2067–2075. doi: 10.1128/jvi.66.4.2067-2075.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A, Marsden M, Halcrow K, Hughes ES, Brettle RP, Bell JE, Simmonds P. Mosaic structure of the human immunodeficiency virus type 1 genome infecting lymphoid cells and the brain: evidence for frequent in vivo recombination events in the evolution of regional populations. J Virol. 1999;73(10):8720–8731. doi: 10.1128/jvi.73.10.8720-8731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenburg JK, Brodt HR, Herndier BG, Bickel M, Bacchetti P, Price RW, Grant RM, Schlote W. HIV-related neuropathology, 1985 to 1999: rising prevalence of HIV encephalopathy in the era of highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2002;31(2):171–177. doi: 10.1097/00126334-200210010-00007. [DOI] [PubMed] [Google Scholar]
- Nguyen DG, Hildreth JE. Involvement of macrophage mannose receptor in the binding and transmission of HIV by macrophages. Eur J Immunol. 2003;33(2):483–493. doi: 10.1002/immu.200310024. [DOI] [PubMed] [Google Scholar]
- O'Brien WA, Koyanagi Y, Namazie A, Zhao JQ, Diagne A, Idler K, Zack JA, Chen IS. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348(6296):69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- Ohagen A, Devitt A, Kunstman KJ, Gorry PR, Rose PP, Korber B, Taylor J, Levy R, Murphy RL, Wolinsky SM, Gabuzda D. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J Virol. 2003;77(22):12336–12345. doi: 10.1128/JVI.77.22.12336-12345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S, Vinters HV, Akashi T, O'Brien WA, Chen IS. HIV-1 env sequence variation in brain tissue of patients with AIDS-related neurologic disease. J Acquir Immune Defic Syndr. 1991;4(11):1082–1092. [PubMed] [Google Scholar]
- Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol. 2004;78(13):6915–6926. doi: 10.1128/JVI.78.13.6915-6926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C, McArthur JC, Johnson RT, Griffin DE, Glass JD, Dewey R, Chesebro B. Distinct HIV-1 env sequences are associated with neurotropism and neurovirulence. Curr Top Microbiol Immunol. 1995;202:89–104. doi: 10.1007/978-3-642-79657-9_7. [DOI] [PubMed] [Google Scholar]
- Power C, McArthur JC, Johnson RT, Griffin DE, Glass JD, Perryman S, Chesebro B. Demented and nondemented patients with AIDS differ in brain-derived human immunodeficiency virus type 1 envelope sequences. J Virol. 1994;68(7):4643–4649. doi: 10.1128/jvi.68.7.4643-4649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C, McArthur JC, Nath A, Wehrly K, Mayne M, Nishio J, Langelier T, Johnson RT, Chesebro B. Neuronal death induced by brain-derived human immunodeficiency virus type 1 envelope genes differs between demented and nondemented AIDS patients. J Virol. 1998;72(11):9045–9053. doi: 10.1128/jvi.72.11.9045-9053.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffer BA, Pohlmann S, Edinger AL, Carlin D, Sanchez MD, Reitter J, Watry DD, Fox HS, Desrosiers RC, Doms RW. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J Virol. 2002;76(6):2595–2605. doi: 10.1128/JVI.76.6.2595-2605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach P, Kuhmann SE, Taylor J, Marozsan AJ, Snyder A, Ketas T, Wolinsky SM, Korber BT, Moore JP. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology. 2004;321(1):8–22. doi: 10.1016/j.virol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Purtscher M, Trkola A, Gruber G, Buchacher A, Predl R, Steindl F, Tauer C, Berger R, Barrett N, Jungbauer A, et al. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10(12):1651–1658. doi: 10.1089/aid.1994.10.1651. [DOI] [PubMed] [Google Scholar]
- Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, 3rd, Burton DR. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68(8):4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacktor N, Lyles RH, Skolasky R, Kleeberger C, Selnes OA, Miller EN, Becker JT, Cohen B, McArthur JC. HIV-associated neurologic disease incidence changes:: Multicenter AIDS Cohort Study, 1990–1998. Neurology. 2001;56(2):257–260. doi: 10.1212/wnl.56.2.257. [DOI] [PubMed] [Google Scholar]
- Sacktor N, McDermott MP, Marder K, Schifitto G, Selnes OA, McArthur JC, Stern Y, Albert S, Palumbo D, Kieburtz K, De Marcaida JA, Cohen B, Epstein L. HIV-associated cognitive impairment before and after the advent of combination therapy. J Neurovirol. 2002;8(2):136–142. doi: 10.1080/13550280290049615. [DOI] [PubMed] [Google Scholar]
- Saphire AC, Bobardt MD, Zhang Z, David G, Gallay PA. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J Virol. 2001;75(19):9187–9200. doi: 10.1128/JVI.75.19.9187-9200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1⤍2 mannose residues on the outer face of gp120. J Virol. 2002;76(14):7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Smit TK, Morgello S, Tourtellotte W, Gelman B, Brew BJ, Saksena NK. Env gp120 sequence analysis of HIV type 1 strains from diverse areas of the brain shows preponderance of CCR5 usage. AIDS Res Hum Retroviruses. 2006;22(2):177–181. doi: 10.1089/aid.2006.22.177. [DOI] [PubMed] [Google Scholar]
- Shapshak P, Nagano I, Xin K, Bradley W, McCoy CB, Sun NC, Stewart RV, Yoshioka M, Petito C, Goodkin K, et al. HIV-1 heterogeneity and cytokines. Neuropathogenesis. Adv Exp Med Biol. 1995;373:225–238. doi: 10.1007/978-1-4615-1951-5_31. [DOI] [PubMed] [Google Scholar]
- Shapshak P, Segal DM, Crandall KA, Fujimura RK, Zhang BT, Xin KQ, Okuda K, Petito CK, Eisdorfer C, Goodkin K. Independent evolution of HIV type 1 in different brain regions. AIDS Res Hum Retroviruses. 1999;15(9):811–820. doi: 10.1089/088922299310719. [DOI] [PubMed] [Google Scholar]
- Sharma DP, Zink MC, Anderson M, Adams R, Clements JE, Joag SV, Narayan O. Derivation of neurotropic simian immunodeficiency virus from exclusively lymphocytetropic parental virus: pathogenesis of infection in macaques. J Virol. 1992;66(6):3550–3556. doi: 10.1128/jvi.66.6.3550-3556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit TK, Wang B, Ng T, Osborne R, Brew B, Saksena NK. Varied tropism of HIV-1 isolates derived from different regions of adult brain cortex discriminate between patients with and without AIDS dementia complex (ADC): evidence for neurotropic HIV variants. Virology. 2001;279(2):509–526. doi: 10.1006/viro.2000.0681. [DOI] [PubMed] [Google Scholar]
- Strizki JM, Albright AV, Sheng H, O'Connor M, Perrin L, Gonzalez-Scarano F. Infection of primary human microglia and monocyte-derived macrophages with human immunodeficiency virus type 1 isolates: evidence of differential tropism. J Virol. 1996;70(11):7654–7662. doi: 10.1128/jvi.70.11.7654-7662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67(7):3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K, Bell JE, Wolinsky SM, Gabuzda D. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology. 2007;360(1):105–119. doi: 10.1016/j.virol.2006.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcour V, Shikuma C, Shiramizu B, Watters M, Poff P, Selnes O, Holck P, Grove J, Sacktor N. Higher frequency of dementia in older HIV-1 individuals: the Hawaii Aging with HIV-1 Cohort. Neurology. 2004;63(5):822–827. doi: 10.1212/01.wnl.0000134665.58343.8d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Marle G, Rourke SB, Zhang K, Silva C, Ethier J, Gill MJ, Power C. HIV dementia patients exhibit reduced viral neutralization and increased envelope sequence diversity in blood and brain. Aids. 2002;16(14):1905–1914. doi: 10.1097/00002030-200209270-00007. [DOI] [PubMed] [Google Scholar]
- Wang J, Crawford K, Yuan M, Wang H, Gorry PR, Gabuzda D. Regulation of CC chemokine receptor 5 and CD4 expression and human immunodeficiency virus type 1 replication in human macrophages and microglia by T helper type 2 cytokines. J Infect Dis. 2002;185(7):885–897. doi: 10.1086/339522. [DOI] [PubMed] [Google Scholar]
- Wang TH, Donaldson YK, Brettle RP, Bell JE, Simmonds P. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J Virol. 2001;75(23):11686–11699. doi: 10.1128/JVI.75.23.11686-11699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Willey RL, Shibata R, Freed EO, Cho MW, Martin MA. Differential glycosylation, virion incorporation, and sensitivity to neutralizing antibodies of human immunodeficiency virus type 1 envelope produced from infected primary T-lymphocyte and macrophage cultures. J Virol. 1996;70(9):6431–6436. doi: 10.1128/jvi.70.9.6431-6436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Yi Y, Chen W, Frank I, Cutilli J, Singh A, Starr-Spires L, Sulcove J, Kolson DL, Collman RG. An unusual syncytia-inducing human immunodeficiency virus type 1 primary isolate from the central nervous system that is restricted to CXCR4, replicates efficiently in macrophages, and induces neuronal apoptosis. J Neurovirol. 2003;9(4):432–441. doi: 10.1080/13550280390218706. [DOI] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]