Abstract
The INK4a-ARF locus encodes two distinct tumor suppressors, p16INK4a and p19ARF. Whereas p16INK4a restrains cell growth through preventing phosphorylation of the retinoblastoma protein, p19ARF acts by attenuating Mdm2-mediated degradation of p53, thereby stabilizing p53. Recent data indicate that Mdm2 shuttles between the nucleus and the cytoplasm and that nucleo-cytoplasmic shuttling of Mdm2 is essential for Mdm2’s ability to promote p53 degradation. Therefore, Mdm2 must export p53 from the nucleus to the cytoplasm where it targets p53 for degradation. We show here that coexpression of p19ARF blocks the nucleo-cytoplasmic shuttling of Mdm2. Moreover, subnuclear localization of Mdm2 changes from the nucleoplasm to the nucleolus in a shuttling time-dependent manner, whereas p19ARF is exclusively located in the nucleolus. In heterokaryons containing Mdm2 and p19ARF, the longer the Mdm2 shuttling is allowed, the more Mdm2 protein colocalizes with p19ARF in the nucleolus, implying that Mdm2 moves from the nucleoplasm to the nucleolus and then associates with p19ARF there. Furthermore, whether or not Mdm2 colocalizes with p19ARF in the nucleolus, p19ARF prevents Mdm2 shuttling. This observation suggests that Mdm2 might be exported through the nucleolus and p19ARF could inhibit the nuclear export of Mdm2 by tethering Mdm2 in the nucleolus. Taken together, p19ARF could stabilize p53 by inhibiting the nuclear export of Mdm2.
Mammalian cell division is controlled in the G1 phase of the cell cycle by two tumor suppressor proteins, the retinoblastoma protein (Rb) and p53 (1, 2). While Rb regulates the exit from the G1 phase of the cell cycle, p53 serves to maintain genomic integrity by arresting cell growth or triggering apoptosis in response to various types of cellular stress (1, 2). The p53 protein has a short half-life and is maintained at low levels in normal cells. Upon stimulation by cellular stress, such as DNA damage, p53 is transiently stabilized in the nucleus (3) where it becomes active as a transcription factor and performs its antiproliferative function (2, 4). Thus, the regulation of p53’s biological activity is largely accomplished by regulating its protein levels as well as its transactivation activity. The Mdm2 oncoprotein, encoded by a p53-inducible gene (5), is a critical cellular antagonist of p53, which targets p53 for degradation and blocks the transactivation activity of p53 (6–9).
The INK4a-ARF locus encodes two proteins, p16INK4a and p19ARF, that affect the functions of Rb and p53, respectively (10, 11). P16INK4a acts as an inhibitor of cyclin D1-dependent kinases 4 or 6 and prevents the phosphorylation of Rb, thereby maintaining an active Rb and blocking the exit from the G1 phase. P19ARF inhibits cell growth by interacting with Mdm2, thereby blocking Mdm2-mediated degradation of the p53 protein and neutralizing Mdm2’s inhibition of p53 activity (12, 13). However, the question remains; how does p19ARF attenuate Mdm2-regulated p53 turnover or activity?
It has been shown that Mdm2-mediated degradation of p53 occurs through a proteasome-dependent pathway in the cytoplasm (6, 7, 14). Recent data have shown that Mdm2 continuously shuttles between the nucleus and the cytoplasm and that nucleo-cytoplasmic shuttling of Mdm2 is essential for its ability to promote p53 degradation (14–16), indicating that Mdm2 must export p53 from the nucleus to the cytoplasm, and then it targets p53 to the cytoplasmic proteasome. These observations raise the possibility that p19ARF could stabilize p53 via blocking nucleo-cytoplasmic shuttling of Mdm2.
By using a heterokaryon assay, we demonstrate in this report that coexpression of p19ARF prevents nucleo-cytoplasmic shuttling of the Mdm2 oncoprotein. In addition, we show that while p19ARF is exclusively located in the nucleolus, subcellular localization of the Mdm2 protein changes temporally from the nucleoplasm to the nucleolus in the heterokaryons containing Mdm2 and p19ARF. The longer the Mdm2 shuttling is allowed, the more Mdm2 protein colocalizes with p19ARF in the nucleolus. Irrespective of whether Mdm2 colocalizes with p19ARF, the p19ARF tumor suppressor inhibits the nuclear export of Mdm2. From these data, we suggest that Mdm2 might be exported from the nucleus to the cytosol through the nucleolus and p19ARF could prevent this process by tethering Mdm2 in the nucleolus.
MATERIALS AND METHODS
Plasmids.
The Mdm2 expression plasmid, pCMV-mdm2, has been described (9). The p19ARF cDNA was obtained by PCR using a retrovirus vector encoding the hemagglutinin-tagged p19ARF (a gift from C. J. Sherr, St. Jude Children’s Research Hospital, Memphis) as template and the following primers: 5′-GATCGGATCCATGGGTCGCAGGTTCTTGGTCACTGTGAG-3′ and 5′-GATCCTCGAGCTATGCCCGTCGGTCTGGGCGACGTTCC-3′. The p19ARF expression plasmid was constructed by subcloning the p19ARF cDNA into the plasmid pcDNA3 (Invitrogen) between BamHI and XhoI restriction sites.
Cells and Transfection.
2KO cell line was derived from mouse embryo fibroblasts that lacked the p53 and mdm2 genes (provided by G. Lozano, M. D. Anderson Cancer Center, Houston). Both 2KO and HeLa cell lines were maintained in DMEM containing 10% FBS and transfected by using Superfect (Qiagen, Chatsworth, CA) according to the manufacturer’s protocol.
The Heterokaryon Assay.
The assay was performed as described (14) with the following modifications. HeLa cells (3 × 105) were transfected with 2 μg expression plasmid for Mdm2 or cotransfected with 2 μg Mdm2 plasmid and 3 μg p19ARF plasmid. Twenty-four hours after transfection, these cells were cocultured with 1 × 106 murine 2KO cells on a 1.8 × 1.8-cm glass coverslip in a 35-mm dish. Twelve hours later, the cells were treated with 50 μg/ml of cycloheximide for 20 min, and then cell fusion was induced by 50% (wt/vol) polyethylene glycol 8000 (Sigma) in DMEM. The cells subsequently were incubated at 37°C for 1 hr or 5 hr in the presence of cycloheximide. Then, they were fixed with 4% paraformaldehyde in PBS for 15 min and followed by permeabilization with 0.2% Triton X-100 in PBS. During and after fixation, all incubations were done at room temperature, and the cells were washed three times with PBS containing 0.5% BSA and 0.05% Tween-20 between incubations. After treatment with the blocking solution (PBS containing 0.5% BSA, 10% horse serum, and 0.05% Tween-20) for 30 min, the cells were incubated with an anti-Mdm2 monoclonal hybridoma supernatant (2A10) for 1 hr, followed by the incubation with an anti-p19ARF polyclonal antibody raised in rabbit (1:500) for 1 hr, which is specifically reactive to mouse-p19ARF (a gift from C. J. Sherr). Subsequently, the cells were incubated with the following reagents: biotinylated goat anti-mouse IgG (Amersham Pharmacia) (1:500) for 30 min, streptavidin-Alexa-568 (Molecular Probes) (1:1,000) for 20 min, and Alexa 488-conjugated goat anti-rabbit IgG (Molecular Probes) (1:1,000) for 20 min. Finally, the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (5 μg/ml) for 10 min and mounted with 90% glycerol in PBS containing N-propylgallate (1 mg/ml). Cell staining was observed by confocal fluorescence microscopy.
RESULTS
A heterokaryon assay was developed to determine whether p19ARF could regulate nucleo-cytoplasmic shuttling of Mdm2. In this assay, HeLa cells (in which ectopic expression of p19ARF inhibited Mdm2-triggered degradation of p53) (12, 13) were transfected with an expression plasmid for Mdm2 or cotransfected with plasmids encoding Mdm2 and p19ARF. Thirty-six hours after transfection, these cells were fused to murine 2KO cells, which were null for both the mdm2 and p53 genes, while blocking de novo protein synthesis with cycloheximide (20 min before cell fusion and throughout the experiment). The fused cells or heterokaryons subsequently were incubated at 37°C for 1 hr in the presence of cycloheximide, and then they were fixed and stained for Mdm2 and p19ARF. To distinguish between human (HeLa) and murine (2KO) nuclei, the cells were simultaneously stained with DAPI, where murine nuclei could be readily identified by the punctate pattern of fluorescence (Fig. 1 Ac and Bc) (17). Subcellular distribution of Mdm2 and p19ARF was determined by confocal immunofluorescent analysis. In agreement with previous observations (15, 16), Mdm2 is predominantly localized to the nucleus (Fig. 1Aa and Fig. 2Aa). The 2KO cells do not express Mdm2, so the appearance of Mdm2 in the 2KO nucleus that is part of a heterokaryon containing the transfected HeLa nucleus indicates that Mdm2 migrated from the HeLa nucleus to the cytoplasm and subsequently entered the 2KO nucleus. Thus, nucleo-cytoplasmic shuttling of Mdm2 can be determined by this assay.
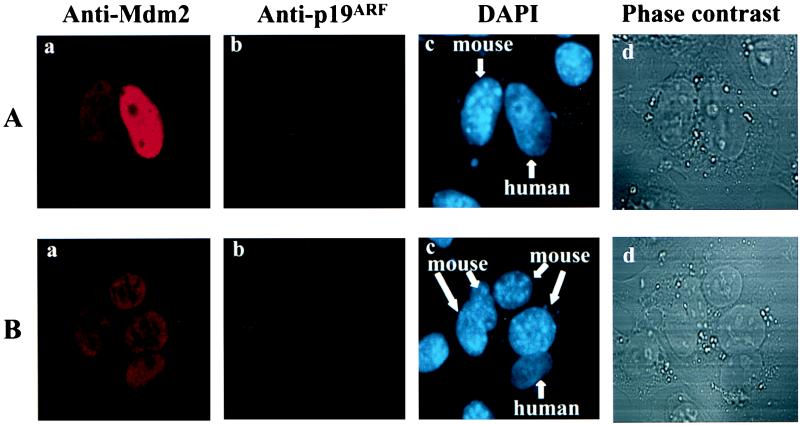
Figure 1.
Mdm2 shuttles between the nucleus and the cytoplasm. HeLa cells were transiently transfected with an expression plasmid for Mdm2. Thirty-six hours after transfection, the heterokaryon assay was performed, in which cells were incubated for 1 hr at 37°C to allow protein shuttling after cell fusion. Cell staining was observed by confocal fluorescence microscopy. The red signal represents the Mdm2 protein (a in A and B), and p19ARF is represented by the green signal (b). The nuclear staining by DAPI is represented by the blue color (c). As shown in c, murine nuclei (2KO) are readily distinguished from human nuclei (HeLa) by their molted appearance. Phase-contrast optics were used to confirm the fusion of cells (d). (A) Mdm2 shuttling in a heterokaryon containing a transfected HeLa nucleus and a 2KO nucleus. (B) Mdm2 shuttles in a heterokaryon that contains a transfected HeLa nucleus and four murine nuclei.
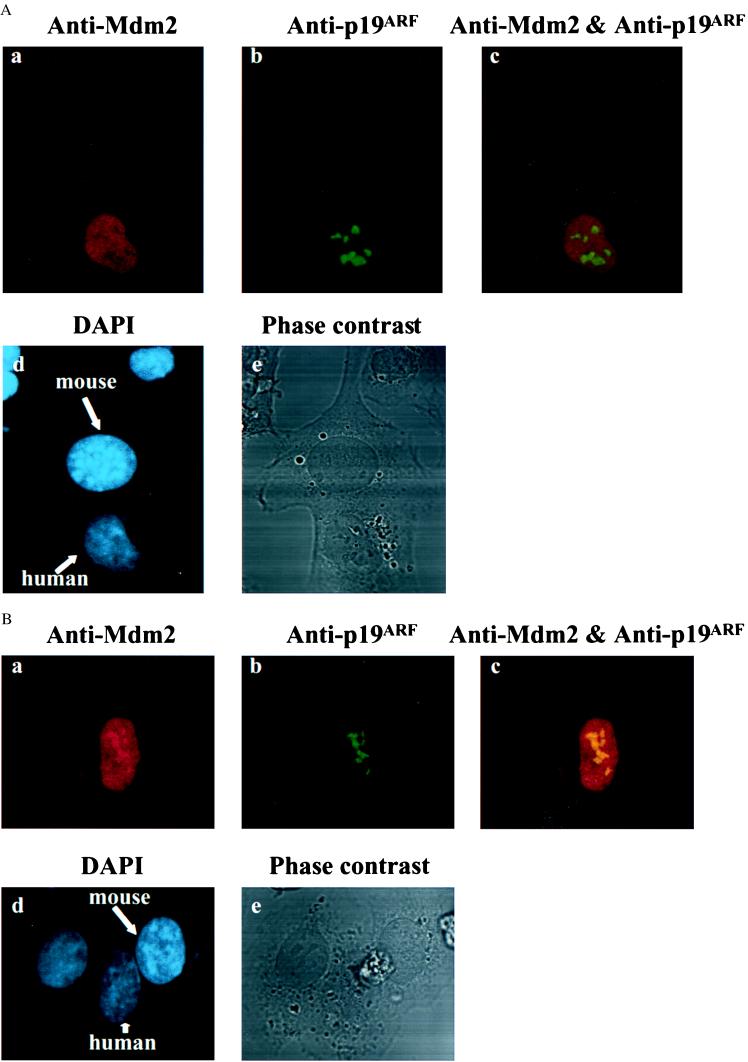
Figure 2.
Coexpression of p19ARF blocks the nucleo-cytoplasmic shuttling of Mdm2. HeLa cells were cotransfected with plasmids encoding Mdm2 and p19ARF. Thirty-six hours after cotransfection, the heterokaryon assay was performed, in which cells were incubated for 1 hr at 37°C to allow protein shuttling after cell fusion. The red signal represents the Mdm2 protein (a in both A and B), and p19ARF is represented by the green signal (b). The nuclear staining by DAPI is represented by the blue color (d). Phase-contrast optics were used to confirm the fusion of cells (e). Although A shows that Mdm2 did not have an identical subnuclear localization with that of p19ARF (no yellow signal in Ac), B exhibited a level of colocalization of two proteins (yellow signal in Bc). In both cases, the nucleo-cytoplasmic shuttling of Mdm2 was blocked by p19ARF (a in both A and B).
In the heterokaryon that contained a HeLa nucleus transfected with the Mdm2 expression plasmid alone (no p19ARF signal in the HeLa nucleus) (Fig. 1 A a and b) and a 2KO nucleus (Fig. 1 A c and d), Mdm2 shuttled from the human to the murine nucleus (Fig. 1Aa). Similarly, when a HeLa cell expressing Mdm2, but not p19ARF, was fused with several 2KO cells, Mdm2 moved into every murine nucleus in the heterokaryon (Fig. 1B). This finding is consistent with the previous observation that Mdm2 undergoes nucleo-cytoplasmic shuttling (14). To further confirm this phenomenon, the nucleo-cytoplasmic shuttling of Mdm2 was assessed in many randomly chosen heterokaryons that contained at least one HeLa nucleus transfected with mdm2 alone and one 2KO nucleus. As shown in Table 1, of 32 such heterokaryons, nucleo-cytoplasmic shuttling of Mdm2 was detected in 27 (84.4%), indicating that the nucleo-cytoplasmic shuttling of Mdm2 is a common physiological process in these cells.
Table 1.
P19ARF blocks nucleo-cytoplasmic shuttling of Mdm2
| Transfected genes | Total number of heterokaryons examined | Number of heterokaryons in which Mdm2 shuttles | Percentage of heterokaryons in which Mdm2 shuttles |
|---|---|---|---|
| Mdm2 | 32 | 27 | 84.4 |
| Mdm2 and p19ARF | 26 | 0 | 0 |
HeLa cells were transfected with an expression plasmid for Mdm2 or cotransfected with plasmids encoding Mdm2 and p19ARF. Thirty-six hours after transfection, the nucleo-cytoplasmic shuttling of Mdm2 was determined by the heterokaryon assay as described. The heterokaryons that contained at least one transfected HeLa nucleus and one 2KO nucleus were randomly chosen and examined for Mdm2 shuttling.
By contrast, in the heterokaryons that contained a HeLa nucleus cotransfected with both Mdm2 and p19ARF plasmids and a 2KO nucleus (or nuclei), the Mdm2 protein was not detected in the 2KO nucleus (Fig. 2 A and B). Moreover, the nucleo-cytoplasmic shuttling of Mdm2 was observed in none of 26 randomly chosen heterokaryons that contained at least one HeLa nucleus cotransfected with the Mdm2 and p19ARF plasmids and one 2KO nucleus (Table 1). These results indicate that coexpression of p19ARF with Mdm2 blocked nucleo-cytoplasmic shuttling of Mdm2.
As reported previously (18), p19ARF is located in the nucleolus with a speckled pattern. (Fig. 2 Ab and Bb). However, the subnuclear localization of Mdm2 is not consistently identical to that of p19ARF in the HeLa nuclei if heterokaryons were incubated for 1 hr at 37°C (to allow protein shuttling) after induction of cell fusion (Fig. 2A a-c). Under this circumstance, Mdm2 displayed a partial colocalization with p19ARF in the nucleolus in about 40–50% of the cells cotransfected with Mdm2 and p19ARF (Fig. 2B a-c and data not shown), whereas in others Mdm2 exhibited an even distribution in the nucleoplasm as opposed to the nucleolus localization of p19ARF (Fig. 2A a-c). Nevertheless, whether or not Mdm2 was colocalized with p19ARF, p19ARF appeared to abrogate the nucleo-cytoplasmic shuttling of Mdm2 (Fig. 2 Aa and Ba and data not shown).
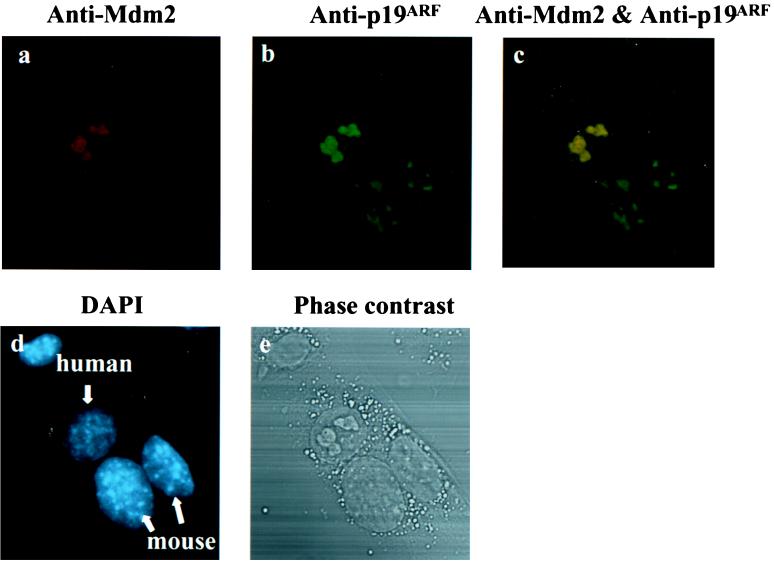
These data imply that at least some Mdm2 protein migrated from the nucleoplasm to the nucleolus and then it was tethered by p19ARF in the nucleolus. It previously has been shown that p19ARF can bind to Mdm2 and form protein complexes in vivo (12, 13, 19). If this is the case, in the heterokaryons expressing Mdm2 and p19ARF, the longer the shuttling time is allowed, the more Mdm2 protein will colocalize with p19ARF in the nucleolus (tethered by p19ARF). To test this possibility, the heterokaryon assay was performed, in which the heterokaryons were incubated at 37°C for 5 hr to allow Mdm2 shuttling after cell fusion. Under this experimental condition, Mdm2 exhibited a much higher level of colocalization with p19ARF in the nucleolus (compare Fig. 3 to Fig. 2B), and it was not detected in the mouse nucleus. In addition, in 90% of heterokaryons containing Mdm2 and p19ARF, both proteins colocalized in the nucleolus. In contrast, in the heterokaryons expressing Mdm2 alone, much less Mdm2 protein could be detected (because Mdm2 was a short-lived protein and most Mdm2 had been degraded), and the residual Mdm2 protein was located in the nucleoplasm of both HeLa and 2KO cells (data not shown).
Figure 3.
More Mdm2 protein colocalizes with p19ARF in the nucleolus when Mdm2 shuttling is allowed for a longer period of time. HeLa cells were cotransfected with plasmids encoding Mdm2 and p19ARF. Thirty-six hours after cotransfection, the heterokaryon assay was performed, in which cells were incubated for 5 hr at 37°C to allow protein shuttling after cell fusion. The red signal represents the Mdm2 protein (a), and p19ARF is represented by the green signal (b). The green signal observed in the mouse nucleus (b) represents the endogenous p19ARF in mouse 2KO cells. The nuclear staining by DAPI is represented by the blue color (d). Phase-contrast optics were used to confirm the fusion of cells (e). Under this experimental condition, Mdm2 shows a much higher level of colocalization with p19ARF in the nucleolus (compare a-c to Fig. 2B). In addition, nucleo-cytoplasmic shuttling of Mdm2 was blocked by p19ARF (a).
DISCUSSION
The results presented here demonstrate that overexpression of p19ARF in a cell nucleus blocks the nucleo-cytoplasmic shuttling of the Mdm2 protein (Table 1, Fig. 2 A and B, and Fig. 3). Because Mdm2 shuttling to the cytoplasm regulates p53 protein levels (half-life) in the cell (14–16), these data provide a mechanistic explanation of how p19ARF regulates p53 levels. When Mdm2 and p19ARF each were expressed separately in cells, they showed distinct subnuclear localizations; i.e., Mdm2 was located in the nucleoplasm whereas p19ARF resided in the nucleolus (Fig. 2Aa) (18). In heterokaryons containing both p19ARF and Mdm2, p19ARF was located exclusively in the nucleolus, but the localization of the Mdm2 protein changed in a shuttling time-dependent manner. The longer Mdm2 was allowed to shuttle, the more Mdm2 protein was colocalized with p19ARF in the nucleolus. However, irrespective of whether Mdm2 colocalized with p19ARF, p19ARF prevented nuclear export of Mdm2.
The nucleolus is the region for rDNA localization, rRNA synthesis, and ribosome assembly (20, 21). It is likely to be involved in the nuclear export of rRNAs and ribosomal proteins. It has been shown that the nuclear export of 5S rRNA was mediated by the TFIIIA protein that contains a nuclear export signal similar to that observed in Mdm2 (22, 23). Thus, the results presented here suggest that Mdm2 might be exported through the nucleolus and p19ARF could block Mdm2 shuttling by tethering Mdm2 in the nucleolus. Indeed, previous results have demonstrated a Mdm2-p19ARF protein complex exists in cells (12, 13, 19). Interestingly, the Mdm2 protein binds to the L5 ribosomal protein and 5S rRNA, and the ring finger domain of Mdm2 binds to RNA sequences found in 28S RNA in the large ribosomal subunit (14, 24, 25). To further test this hypothesis, the p19ARF mutants that are defective for binding to Mdm2 or localizing to the nucleolus and the Mdm2 mutants that lack the ability to interact with p19ARF should be examined. Because Mdm2 must associate with CRM-1 (exportin–1) and Ran-GTP (15) to successfully shuttle, p19ARF might prevent this process by altering Mdm2 modification or sequestering Mdm2, thereby inhibiting the interactions among these proteins.
Because HeLa cells must be used in this assay because of their fusion efficiency, p53 levels in the cells were kept low (the human papilloma virus E6 protein is present and degrades p53) (26) and thus the possible impact of these experimental conditions on Mdm2 and p19ARF localization is an uncontrolled variable. However, it is clear that p19ARF inhibits Mdm2-directed p53 degradation in HeLa cells (12, 13) and that it also blocks Mdm2 shuttling in these cells. Thus, these results can explain how p19ARF regulates p53 levels, and they reveal a way by which p53 levels can be controlled in a cell.
Acknowledgments
We thank Dr. C. J. Sherr for anti-p19ARF polyclonal antibody and stimulating discussions and Dr. G. Lozano for 2KO cells. We are grateful to J. Goodhouse for expert assistance with confocal microscopy and Drs. J. Roth and M. Dobbelstein for help in the heterokaryon assay. This work was supported by a grant from the National Cancer Institute (Pol-CA41086) (to A.J.L.). W.T. was supported by a National Institutes of Health Cancer Training Grant.
ABBREVIATIONS
- Rb
retinoblastoma protein
- DAPI
4′,6-diamidino-2-phenylindole
References
- 1.Weinsberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 2.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Maltzman W, Czyzyk L. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 5.Barak Y, Juven T, Haffner R, Oren M. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 7.Kubbutat M H G, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 8.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 9.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Nature (London) 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 10.Sherr C J, Roberts J M. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 11.Sherr C J. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 12.Pomerantz J, Schreiber-Agus N, Liégeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, et al. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 14.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freedman D A, Levine A J. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao W, Levine A J. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moser F G, Dorman B P, Ruddle F H. J Cell Biol. 1975;66:676–680. doi: 10.1083/jcb.66.3.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 19.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M, Sherr C J. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheer U, Weisenberger D. Curr Opin Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 21.Fomproix N, Gebrane-Younes J, Hernandez-Verdun D J. J Cell Sci. 1998;111:359–372. doi: 10.1242/jcs.111.3.359. [DOI] [PubMed] [Google Scholar]
- 22.Guddat U, Bakken A H, Pieler T. Cell. 1990;60:619–628. doi: 10.1016/0092-8674(90)90665-2. [DOI] [PubMed] [Google Scholar]
- 23.Fridell R A, Fischer U, Luhrmann R, Meyer B E, Meinkoth J L, Malim M H, Cullen B R. Proc Natl Acad Sci USA. 1996;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marechal V, Elenbaas B, Piette J, Nicolas J-C, Levine A J. Mol Cell Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine A J. Mol Med. 1996;2:439–451. [PMC free article] [PubMed] [Google Scholar]
- 26.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]