Abstract
The p53 protein is involved in several central cellular processes, including gene transcription, DNA repair, cell cycling, genomic stability, chromosomal segregation, senescence, and apoptosis. p53 mutations frequently result in an immunocytochemically detectable accumulation of the p53 protein in tumor cells. To evaluate whether p53 gene mutations are required for the onset of hematogeneous tumor cell dissemination, we compared the p53 status of primary and micrometastatic tumor cells. Disseminated carcinoma cells could be detected in bone marrow aspirates obtained from 46 (40%) of 114 patients with various types of epithelial tumors without overt skeleton metastases. There was no correlation between the detection of p53 protein in primary lung carcinomas and the presence of tumor cells in bone marrow. Further analyses revealed that the disseminated carcinoma cells rarely accumulate mutated p53 protein and that 10 cell lines derived thereof did not harbor p53 mutations even in the presence of such mutations in the autologous primary tumors. These observations indicate that tumor cells can leave the primary tumor before mutations of the p53 gene occur and that these mutations are not essential for such early hematogeneous dissemination of cancer cells. Thus, the value of mutated p53 as a target for diagnosis and treatment of micrometastatic disease in cancer patients is questionable.
Blood-borne dissemination of tumor cells is a major risk factor for metastatic relapse and cancer-related mortality in patients with operable solid tumors. With the recent development of sensitive immunocytochemical and molecular methods, individual disseminated tumor cells, undetectable by conventional tumor staging procedures, now can be directly assessed at frequencies of 10−5–10−6 normal cells (for review, see refs. 1–3). Various clinical studies have provided substantial evidence that immunocytochemically identifiable tumor cells in bone marrow (BM) are independent prognosticators of metastatic relapse (4–11). Further, these immunostained cells have the capacity of clonal growth (12, 13), harbor cancer-specific cytogenetic aberrations and mutations (14, 15), and show various tumor-associated characteristics, such as neoexpression of cancer/testis antigens, down-regulation of MHC class I antigens, and overexpression of the oncogene erbB2 and protease receptors (9, 16, 17). Thus, the stained cells are derived from the primary neoplasm, and they appear to be the potential precursors of subsequent overt metastases. Interestingly, most of these cells appeared in a dormant (i.e. nonproliferating) state, as assessed by immunostaining for proliferation-associated proteins such as Ki-67 or p120 (17). This finding may explain the extended latency period between the detection of micrometastatic cells and the occurrence of overt metastasis. The genetic and epigenetic events regulating tumor cell dormancy are not well understood (18). It is unclear whether dormant tumor cells present in secondary organs such as BM lack mutations in genes that control cell proliferation, such as p53.
Of the approximately 6.5 million new cancer cases worldwide each year, 2.4 million are estimated to involve p53 mutations (19–21). The high frequency of p53 mutations in human cancer attests to its importance as a critical component of tumor progression and a target of rational cancer therapy. The p53 protein is involved in several central cellular processes, including gene transcription, DNA repair, cell cycling, genomic stability, chromosomal segregation, senescence, and apoptosis (19, 22–24). At present, it remains unclear whether p53 mutations are required for the early onset of tumor cell dissemination. p53 mutations frequently lead to an accumulation of p53 protein because of an increased stability of the protein, whereas the number of p53 protein molecules in normal, nonmalignant cells usually is limited to about 103–104 per cell (25). Thus immunostaining of p53 protein has been claimed as a marker of malignant disease in diagnostic cytopathology (26). Here, we evaluated whether p53 immunostaining is frequent in micrometastatic tumor cells present in BM of patients with different types of epithelial tumors. Counter to our expectations, immunostaining of p53 in these cells was observed only in a small minority of cancer patients. This finding was in line with the sequence analysis of p53 cDNA from BM-derived micrometastatic cell lines that lacked mutations in the coding region of the p53 gene despite the presence of p53 mutations in the autologous primary tumor.
MATERIALS AND METHODS
Patients and Tissue Preparation.
After obtaining informed consent, we investigated BM aspirates from 114 patients with primary carcinomas of the colorectum (n = 46), stomach (n = 27), lung (n = 25), urogenital tract (n = 5), breast (n = 4), and pancreas (n = 4) as well as three patients with cancer of unknown primary. BM aspirates were obtained from both sides of the upper iliac crest through an aspiration needle. The volumes of all aspirates ranged from 2 to 10 ml (mean: 4 ml), yielding between 1 and 6 × 107 (mean: 2 × 107) mononuclear cells. After centrifugation at 900 g for 30 min through a Ficoll-Hypaque density gradient (Amersham Pharmacia; density 1.077 g/ml) at room temperature, interface cells were cytocentrifuged at 150 g for 5 min on glass slides at room temperature (8 × 104 cells per slide). After overnight air drying, slides were either stained immediately or stored at −80°C before use with preservation of epithelial antigens for at least 2 years (unpublished data). From each aspirate five slides comprising 4 × 105 mononuclear cells were examined (8 × 105 cells per patient), while one additional slide served as Ig isotype control.
Immunocytochemistry.
For double staining, we used a combined immunogold/enzymatic technique that has been successfully applied to the detection of histogenetic and proliferation-associated markers on disseminated tumor cells (17). Briefly, cells were first incubated for 45 min with 5 μg/ml of mAb Pab 1801 directed to wild-type and mutated p53 protein (Dianova, Hamburg, Germany). After a thorough wash with PBS, gold-conjugated goat anti-mouse Igs (Amersham Pharmacia) were incubated for 45 min. Alternatively, the rabbit antiserum CM1 (Medac, Hamburg, Germany), directed against full-length p53, was used at a 1:200 dilution as primary antibody, and the antibody binding was developed with gold-conjugated goat anti-rabbit Igs (Amersham Pharmacia). Subsequently, the slides were washed and exposed to 2% glutaraldehyde diluted in PBS for 5 min. The next immunoenzymatic step was similar to the single labeling method described above. For detection of tumor cells in BM, cytospin preparations were incubated with one of the following biotinylated mAbs: CK2 (anti-CK18; 2.5 μg/ml, Boehringer Mannheim), Ks19.1 (anti-CK19; 5 μg/ml; Progen, Heidelberg), and A45-B/B3 (5 μg/ml; Micromet, Munich) directed against cytokeratins (CK), such as CK18, CK19, and a common epitope of various CK components, including CK8/18 and CK8/19 heterodimers (27), as well as a mixture of mAbs 17–1A and M79 (20 μg/ml; Centocor) directed against epithelial glycoprotein-40 and mAb SDZ ABL 364 (20 μg/ml; Sandoz Pharmaceutical) against Lewis Y antigens. Appropriate dilutions of the unrelated mouse myeloma proteins served as isotype controls. The biotinylated mAbs were developed with preformed complexes of streptavidin and alkaline phosphatase (Dako).
All slides were examined by two independent observers in a double-blinded fashion. By using sensitive epipolarization microscopy, we were able to detect even weak signals; we did not use a cut-off for staining intensity to call a cell p53-positive. A BM sample was called p53-positive if any of the CK-positive cells detected in this sample were costained with the p53 antibodies. Cells with an inhomogeneous background-like distribution of a few colloidal gold particles were not called p53-positive.
Tissue samples from the primary tumor and five regional lymph node levels were sampled from 47 patients with nonsmall cell lung cancer (19 adenocarcinomas and 23 squamous cell carcinomas) during primary surgery. One part of each lymph node was embedded in paraffin for routine histopathological staining; the other part and a sample of the primary tumor were snap-frozen in liquid nitrogen and stored at −80°C. From each of the primary carcinomas, three representative sections were stained with mAb PAb 1801. The antibody reaction was developed with the alkaline phosphatase antialkaline phosphatase (APAAP) technique combined with the Neufuchsin method for visualizing antibody binding (7). For immunohistochemical detection of tumor cells in lymph nodes, the antiepithelial cell mAb Ber-EP4 was used as described. Ber-EP4 detects two glycopeptides of 34 and 49 kDa on the surface and in the cytoplasm of all epithelial cells. The antibody does not react with mesenchymal tissue, including lymphoid tissue (28).
Sequence Analysis of Primary Tumor Specimens and Disseminated Tumor Cell Lines.
Total RNA was isolated from 10 cell lines recently established from micrometastatic BM tumor cells by immortalization with simian virus 40 (SV40) large T antigen DNA (13). The identities of the cell lines were confirmed by HLA-DRB1* typing using either genomic DNA of cell lines and peripheral blood lymphocytes or DNA isolated from cytospins preparations of the patients, following the oligonucleotide typing system protocol as described by Nevinny-Stickel et al. (29). Total RNA was purified to poly(A)-RNA by using the Oligotex mRNA Mini Kit (Quiagen). The reverse transcription of poly(A)-RNA was performed by using Superscript II reverse transcriptase (GIBCO/BRL) and random hexameric primers according to manufacturer’s instructions. The primers 5′-TTTCCACGACGGTGACAGC-3′ (nucleotides 154–173) and 5′-CTGTCATCCAAGTACTCCACACGCG-3′ (nucleotides 840–818) as well as 5′-ATGAGCGCTGCTCAGATAGC-3′ (nucleotide 720) and 5′-AAGACCCAAAACCCAAAATGG-3′ (nucleotides 1475–1455) were used together with 2 μl of the cDNA in a 52.5-μl reaction mix amplifying the DNA in 30 cycles (delay, 1 min 94°C, 94°C for 30 s, 50°C for 30 s, 72°C for 30 s) during the PCR. Dependent on the primers, the entire 1,316-bp p53 coding region or two overlapping fragments were gained, which spanned the ORF of the p53 cDNA.
To isolate tumor-specific chromosomal DNA from the paraffin-embedded primary tumors, histomorphologically recognizable tumor areas were separated from surrounding normal tissue by microdissection. Microdissection was performed from 8-μm sections of the tumor, which had been mounted on lysin-treated slides, deparaffinized by standard procedures, and stained with methyl-green. The microdissected tumor areas were incubated with proteinase K at 55°C for 3–5 days. The DNA was extracted by phenol/chloroform, ethanol-precipitated, and used for PCR (50 ng). Primers for the amplification of the exons 4–9 of the p53 gene were as follows: exon 4 (334 bp), 5′-GCTCTTTTCACCCATCTACAG-3′, 5′-GTCTCATGGAAGCCAG-3′; exon 5 (252 bp), 5′-TCACTTGTGCCCTGACTTTCA-3′, 5′-TCTCCAGCCCCAGCTGCT-3′; exon 6 (215 bp), 5′-TTCCTCACTGATTGCTCTTA-3′, 5′-GACCCCAGTTGCAAACCAG-3′; exon 7 (208 bp), 5′-GCCTGTGTTATCTCCTAG-3′, 5′-GGTGGCAAGTGGCTCCTGA-3′; exon 8/9 (374 bp), 5′-AGGACCTGATTTCCTTACTGC-3′, 5′-GAGGTCCCAAGACTTAGTAC-3′. The PCR primers were separated by centrifugation of the reaction mix through a Centricon-100 spin column (Amicon), and the recovered PCR products were used for direct sequencing.
Purified PCR products (100 ng) were sequenced by the nonradioactive cycle sequencing method by using 10 pmol of the p53 primers mentioned above in the Taq-DNA polymerase sequencing kit and dye-labeled dideoxynucleotides on the 373A-Stretch system (Applied Biosystems). PCRs were performed on a model 9600 thermocycler (Perkin–Elmer). If necessary, the primers 5′-ACGTGCAAGTCACAGACTTGG-3′ and 5′-GGGACAGCATCAAATCATCC-3′ were used to get double-strand information at the 5′ end of the ORF. Six p53 cDNA PCR products and the p53 exons 4–9 were sequenced by a commercial laboratory (Sequiserve, Vaterstetten, Germany).
RESULTS AND DISCUSSION
We have used mAb CK2 against CK component no. 18 (CK18) as probe for the detection of tumor cells in BM of 63 patients with various types of epithelial cancer, including tumors of the colon/rectum (n = 33), lung (n = 13), stomach (n = 12), breast (n = 3), pancreas (n = 1), and one cancer of unknown primary. The prognostic relevance of tumor cells identified with this antibody has been sustained in several clinical studies (7–10, 30). The basic principle used in the identification of occult epithelial tumor cells in the bone marrow is the ability to distinguish between cells of different histogenesis, in this case cells of hematopoietic vs. epithelial origin. Many studies have used cytokeratins as epithelial marker antigens; these proteins are intermediate filaments that are stably expressed and abundant in normal epithelial cells and epithelial tumors. The specificity of CK2 for the detection of carcinoma cells in BM samples is high but not absolute; in a recent study we observed positive findings in six of 215 noncarcinoma control patients (7). However, the majority of CK2-positive cells in BM of cancer patients exhibit tumor-associated characteristics, such as overexpression of the erbB2 oncogene or down-regulation of MHC class I antigens (16, 17), which indicate their malignant nature.
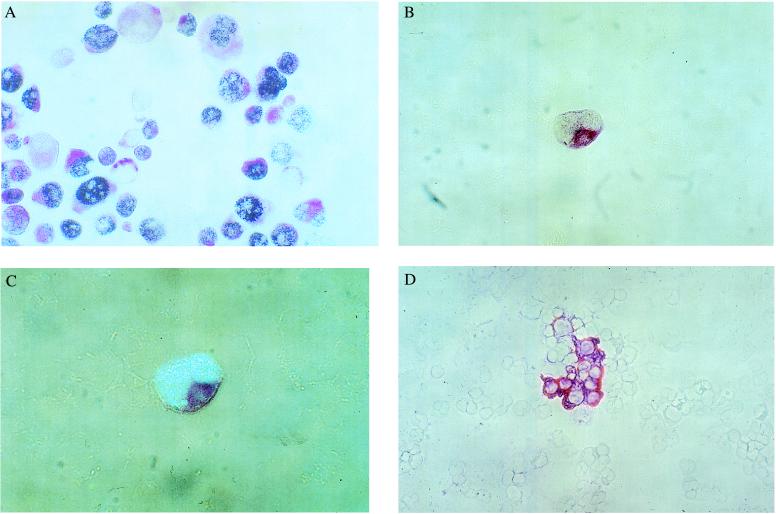
In 29 (46%) of 63 carcinoma patients, CK18-positive cells were found (Table 1); the frequency of these cells in each specimen varied from 1 to 333 per 4 × 105 BM cells. p53-positive/CK18-positive cells were found only in four of the 63 (6.3%) patients, whereas any of the CK18-positive cells in the remaining 25 specimens consistently lacked a detectable p53 signal (Table 1). By using additional mAbs against different marker proteins, we confirmed the finding that p53 immunostaining of disseminated carcinoma cells in BM is a rare event, which was not observed in any of the additional 51 patients analyzed (Table 1). As control for double-positive staining, we stained the pancreatic cancer cell line PancTu-I (Fig. 1A), known to express cytokeratins and to accumulate a mutated p53 protein (31). Two of the patients with double-positive cells in their BM presented with a colorectal carcinoma (TNM-stages: T1N0M0, T3N0M0) and two with gastric cancer (T3N2M1, T4N2M1); in three of those specimens only some CK18-positive cells (1 of 3, 2 of 3, and 3 of 22 cells) costained for p53, indicating tumor cell heterogeneity. The few double-positive cells showed a nuclear staining pattern (Fig. 1 B and C). Our analysis was limited by the fact that most patients showed fewer than 10 CK18-positive cells per sample; however, in six samples we found between 22 and 333 CK18-positive cells and none of those cells reacted with the p53 antibodies (Fig. 1D).
Table 1.
Immunocytochemical detection of p53 protein in epithelial tumor cells identified with various epithelial marker antigens in BM of cancer patients
| Epithelial marker | No. of BM samples, %
|
|||
|---|---|---|---|---|
| Total | Marker+ | p53+/marker+ | p53−/marker+ | |
| Total | 114 | 46 (40.3) | 4 (3.5) | 42 (36.8) |
| CK18 | 63 | 29 (46.0) | 4 (6.3) | 25 (39.7) |
| CK19 | 14 | 5 (35.7) | 0 | 5 (35.7) |
| Pan-CK | 17 | 8 (47.1) | 0 | 8 (47.1) |
| EGP-40/Lewis-Y | 20 | 4 (20.0) | 0 | 4 (20.0) |
Patients with carcinomas of the colon/rectum (n = 46), stomach (n = 27), stomach (n = 27), lung (n = 25), urogenital tract (n = 5), breast (n = 4), pancreas (n = 4), and cancer of unknown primary (n = 3).
Figure 1.
Double staining of (A) p53-positive/CK18-positive PancTu pancreas cancer cells (magnification: ×85), (B) a dividing p53-positive/CK18-positive gastric cancer cell (magnification: ×136), (C) same cell as shown in B, epi-polarization microscopy (magnification: ×170), and (D) p53-negative/CK18-positive lung cancer cells (magnification: ×85). All specimens were stained with mAb CK2 against CK18 (red cytoplasmatic stain) and mAb Pab 1801 against p53 protein (blue nuclear stain).
Because p53 mutations leading to an increased half-life of p53 protein are common in carcinomas, it is unlikely that all of our patients with CK18-positive/p53-negative cells in their BM have primary tumors without such mutations. From 47 patients with nonsmall cell lung cancer (adenocarcinomas, squamous cell carcinoma), frozen tissue sections of the primary tumors were available for staining with mAb PAb 1801 against p53. In contrast to the analysis of BM cytospins, we used a simpler single-labeling protocol because the tumor cell areas in the lung tissue sections could be identified by morphological examination. Staining of consecutive sections with CK2 showed homogenous expression of CK18 in all tumor cells. Expression of p53 was found in 22 (46.8%) of primary tumors (Table 2); however, the fraction of p53-positive tumor cells per specimen varied considerably from 1% to 100% with the majority of tumors displaying more than 30% positive cells (Table 2). There was no correlation to micrometastatic BM involvement (Table 2), providing further indirect evidence that p53 mutations associated with an accumulation of p53 protein are not required for early hematogeneous tumor cell dissemination. Micrometastases with wild-type p53 were found in patients who showed a significant but still heterogeneous expression of p53 protein in their autologous tumors; the percentage of positive cells in these tumors was greater than 30% (Table 2). Thus, the p53-negative tumor cells in BM of these patients may be derived from the p53-negative subclone of the tumor. In contrast, the immunohistochemical detection of micrometastases in lymph nodes, classified as “tumor-free” by histopathological examination, correlated with p53 staining of the respective primary tumors (Table 2). Our finding suggests that p53 mutations may favor lymphatic dissemination of lung carcinoma cells. This assumption also is supported by the recent detection of p53-positive cells in lymph nodes of patients with nonsmall cell lung cancer (32).
Table 2.
Immunohistochemical detection of p53 protein in primary nonsmall cell lung cancer: Correlation to lymphatic and hematogeneous micrometastasis
| Parameter | No. of patients | p53-positive primary tumors (% of all patients)
|
p53-negative primary tumors (%) | |||
|---|---|---|---|---|---|---|
| Total | 1–30% p53+ cells | 31–60% p53+ cells | 61–100% p53+ cells | |||
| All patients | 47 | 22 (46.8) | 4 (8.5) | 9 (19.1) | 9 (19.1) | 25 (53.2) |
| Lymph node | ||||||
| micrometastasis | ||||||
| BerEp4-positive | 11 | 8 (72.7) | 1 (9.1) | 3 (27.3) | 4 (36.4) | 3 (27.3) |
| BerEp4-negative | 36 | 14 (38.9)* | 3 (8.3) | 6 (16.7) | 5 (13.9) | 22 (61.1) |
| BM micrometastasis | ||||||
| CK18-positive | 18 | 7 (38.9) | 0 (0) | 4 (22.2) | 3 (16.7) | 11 (61.1) |
| CK18-negative | 29 | 15 (51.7) | 4 (13.8) | 5 (17.2) | 6 (20.7) | 14 (48.3) |
P < 0.05, total number of patients with p53-positive tumors compared to total number of patients with p53-negative tumors.
It cannot be excluded that micrometastatic tumor cells may harbor selectively p53 mutations that do not result in a stabilized form of p53 protein detectable by immunocytochemistry. Greenblatt et al. (21) recently analyzed 84 studies that compared immunohistochemistry and sequencing in the same tumor sets; positive staining was found in 44% of tumors by immunohistochemistry, whereas 36% contained p53 mutations. Thus the status of the p53 protein by immunohistochemistry should not be equated with wild-type or mutant genotype but requires confirmatory DNA analysis. Because the number of carcinoma cells detected per aspirate was too small (about one tumor cell per 105 BM cells) to directly analyze the p53 cDNA sequence, we sequenced the p53 cDNA of 10 micrometastatic cancer cell lines that had been established from BM micrometastases by immortalization with a cDNA coding for the SV40 large T antigen (13, 33). The cell lines (Table 3) were derived from five patients with prostate cancer (PC-R1, PC-S1, PC-E1, PC-H1, and PC-L1), two with nonsmall cell lung cancer (LC-D1 and LC-M1), one with colon cancer (CC-B1), and two with breast cancer (BC-H1 and BC-K1); none of these patients had clinical signs of overt metastases. The identities of the cell lines were confirmed by HLA typing (29); the tumor-derived origin of these cell lines recently was demonstrated by expression of prostate cancer-associated proteins (in the case of prostate cancer) and the expression of MAGE antigens (33). MAGE antigens belong to the family of cancer/testis antigens that are not expressed in any normal human tissue except testis (34, 35). Moreover, by using comparative genomic hybridization and multiplex fluorescence in situ hybridization (36), we found several tumor-specific cytogenetic aberrations such as the deletion of the short arm of chromosome 8 in PC-R1 cells (unpublished observation). The cDNAs of all of the micrometastatic cell lines showed the wild-type p53 cDNA sequence with the exception of the AccII/BstU1 polymorphism, characterized by a G in position 429 instead of a C, resulting in an arginine in codon 72 instead of a proline, which was detected in nine cell lines (Table 3). As positive control, the p53 cDNA of the colon cancer cell line HT-29 was sequenced and the published mutation in exon 8 (37), a G > A mutation in position 1032, resulting in a arginine to histidine exchange in codon 273, could be detected.
Table 3.
Sequence analysis of the p53 gene (exons 4–9) of the primary tumor and of the p53 cDNA of the respective micrometastatic cell lines
| Patient identifier | Tumor type | Tumor stage | Primary tumor p53 gene
|
Micrometaststic cell lines p53 cDNA
|
||
|---|---|---|---|---|---|---|
| Sequenced exons | Mutation, codon | Sequenced region, bp | Mutation, codon | |||
| PC-E1 | Prostate Ca | pT3N0M0 | 4,5,6,7,8,9 | No* | 215–1396 | No* |
| PC-R1 | Prostate Ca | pT4N0M0 | 4,5,6,7,8,9 | No* | 179–1369 | No* |
| PC-S1 | Prostate Ca | pT3N0M0 | n.d. | n.d. | 178–1396 | No* |
| PC-L1 | Prostate Ca | pT3N0M0 | n.d. | n.d. | 174–1396 | No* |
| PC-H1 | Prostate Ca | pT3N0M0 | 4,5,6,7,8,9 | 273, CGT → TGT* | 215–1396 | No* |
| 284, ACA → ATA | ||||||
| LC-D1 | Lung Ca | pT3N2M0 | 4,5,6,7,8,9 | No** | 215–1396 | No* |
| LC-M1 | Lung Ca | pT4N0M0 | 4,5,6,7,8,9 | No* | 173–1396 | No* |
| CC-B1 | Colon Ca | pT3N0M0 | n.d. | n.d. | 215–1396 | No |
| BC-H1 | Breast Ca | pT2N0M0 | 4,5,6,7,8,9 | No* | 215–1396 | No* |
| BC-K1 | Breast Ca | pT1N0M0 | 4,5,6,7,8,9 | 169, Δ15 bp* | 215–1396 | No* |
The p53 cDNA has a length of 1,760 bp. The start codon ATG begins at bp 215 and the stop codon ends at bp 1386 of the p53 cDNA. n.d., not determined.
Wild-type p53 sequences with the homozygous BstUI polymorphism CGC at codon 72; the other p53 sequences have CCC at codon 72.
Heterozygous for the BstUI polymorphism.
To prove that disseminated cancer cells with a wild-type p53 gene can be derived from an autologous primary tumor that carries a p53 mutation, we obtained genomic DNA from paraffin-embedded archival tumor sections of seven of these patients, and the hot-spot regions of p53 (exons 4–9) were analyzed. As shown in Table 3, the primary tumor of patient PC-H1 harbored two distinct point mutations at codon 273 (Arg > Cys) and codon 284 (Thr > Ile), and the tumor of patient BC-K1 harbored a deletion mutation of 15 bp upstream of the codon 169. In contrast, the autologous micrometastatic cells from both patients carried the wild-type p53 gene.
Interestingly, the primary tumor cells of LC-D1 were heterozygous for the BstUI polymorphism at codon 72, whereas the sequence of the p53 cDNA showed only one species; the expected mixture of two p53 cDNAs corresponding to the two alleles was not observed. The biological relevance of the AccII/BstUI polymorphism, which recently was implicated in the development of human papilloma-virus-associated cancer (38) and detected in all but one of our micrometastatic cell lines remains unclear. The arginine form of p53 binds more efficiently the HPV E6 protein and is more susceptible to degradation than is the proline form. Thus, it cannot be excluded that this polymorphism also might support the inactivation of the wild-type p53 by SV40 T Ag; however, codon 72 encodes an amino acid that lies outside the SV40 T Ag-binding domain of p53 (39).
Although our findings do not exclude the possibility that p53 mutations may occur in disseminated tumor cells in the BM, they do not appear to be a common genetic characteristic of these cells. This observation also may explain why transfection of disseminated carcinoma cells in BM with SV40 T antigen cDNA was required to establish these cell lines. SV40 T antigen can bind and inactivate wild-type p53, which results in the loss of p53 function (39). If mutations, inactivating p53, would be frequent in BM tumor cells, the transfection with SV40 T antigen cDNA would be redundant, at least with regard to its interaction with p53. Further support for the hypothesis that p53 might control the dormant state of micrometastatic cells was derived from the failure to immortalize these cells with a mutant SV40 T antigen that lacks the p53 binding domain (data not shown). Thus, malignant cells may leave the primary carcinoma at an early stage of its development when p53 is not mutated yet, and targeting mutated p53 (19) is unlikely to be a successful strategy to detect and to eliminate these potential precursors of subsequent overt metastases.
Acknowledgments
We thank A. Volgger, T. Siart, D. Kiefer, and S. Baier for excellent technical assistance and Dr. C. Kaltz for preparing the tumor-DNA samples. The SV40 vector was a generous gift from E. Fanning (Department of Molecular Biology, Vanderbilt University, Nashville, TN). We gratefully acknowledge the following persons for providing bone marrow samples from carcinoma patients: Dr. D. Weckermann, Urologische Klinik, Augsburg; Dr. M.W. Köllermann, Urologische Klinik, Horst-Schmitt-Kliniken, Wiesbaden; Dr. M. Storck, Klinik für Herz- und Thoraxchirurgie, Universität Ulm; Dr. S. Braun, Frauenklinik, Ludwig-Maximilians-Universität, Munich, and Dr. B. Weber, Tumorklinik Bad Trissl. We thank Dr. M. Osborne, Max-Planck- Institut Göttingen and Dr. H. Bodenmüller, Boehringer Mannheim, Tutzing, Germany for mAb CK2 and the Micromet GmbH, Martinsried, Germany, for the mAb A45-B/B3. This work was supported by the Deutsche Forschungsgemeinschaft and Deutsche Krebshilfe, Bonn, Germany.
ABBREVIATIONS
- BM
bone marrow
- CK
cytokeratin
- SV40
simian virus 40
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Sidransky D. Science. 1997;278:1054–1059. doi: 10.1126/science.278.5340.1054. [DOI] [PubMed] [Google Scholar]
- 2.Calaluce R, Miedema B W, Yesus Y W. J Surg Oncol. 1998;67:194–202. doi: 10.1002/(sici)1096-9098(199803)67:3<194::aid-jso11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Braun S, Pantel K. Breast Cancer Res Treatment. 1998;52:201–216. doi: 10.1023/a:1006164914610. [DOI] [PubMed] [Google Scholar]
- 4.Cote R J, Rosen P P, Lesser M L, Old L J, Osborne M P. J Clin Oncol. 1991;9:1749–1756. doi: 10.1200/JCO.1991.9.10.1749. [DOI] [PubMed] [Google Scholar]
- 5.Diel I J, Kaufmann M, Costa S D, Holle R, von Minckwitz G, Solomayer E F, Kaul S, Bastert G. J Natl Cancer Inst. 1996;88:1652–1664. doi: 10.1093/jnci/88.22.1652. [DOI] [PubMed] [Google Scholar]
- 6.Cote R J, Beattie E J, Chaiwun B, Shi S R, Harvey J, Chen S-C, Sherrod A E, Groshen S, Taylor C R. Ann Surg. 1995;222:415–425. doi: 10.1097/00000658-199522240-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pantel K, Izbicki J, Passlick B, Angstwurm M, Häussinger K, Thetter O, Riethmüller G. Lancet. 1996;347:649–653. doi: 10.1016/s0140-6736(96)91203-9. [DOI] [PubMed] [Google Scholar]
- 8.Lindemann F, Schlimok G, Dirschedl P, Witte J, Riethmüller G. Lancet. 1992;340:685–689. doi: 10.1016/0140-6736(92)92230-d. [DOI] [PubMed] [Google Scholar]
- 9.Heiss M M, Allgayer H, Gruetzner K U, Funke I, Babic R, Jauch K-W, Schildberg F W. Nat Med. 1995;1:1035–1039. doi: 10.1038/nm1095-1035. [DOI] [PubMed] [Google Scholar]
- 10.Jauch K W, Heiss M M, Gruetzner U, Funke I, Babic R, Eissner H J, Riethmueller G, Schildberg F W. J Clin Oncol. 1996;14:1810–1817. doi: 10.1200/JCO.1996.14.6.1810. [DOI] [PubMed] [Google Scholar]
- 11.Thorban S, Roder J, Nekarda H, Funk A, Siewert J, Pantel K. J Natl Cancer Inst. 1996;88:1222–1227. doi: 10.1093/jnci/88.17.1222. [DOI] [PubMed] [Google Scholar]
- 12.Ross A A, Cooper B W, Lazarus H M, Mackay W, Moss T J, Ciobanu N, Tallmann M S, Kennedy M J, Davidson N E, Sweet D, et al. Blood. 1993;82:2605–2610. [PubMed] [Google Scholar]
- 13.Pantel K, Dickmanns A, Zippelius A, Klein C, Shi J, Hoechtlen-Vollmar W, Schlimok G, Weckermann D, Oberneder R, Fanning E, Riethmüller G. J Natl Cancer Inst. 1995;87:1162–1168. doi: 10.1093/jnci/87.15.1162. [DOI] [PubMed] [Google Scholar]
- 14.Müller P, Weckermann D, Riethmüller G, Schlimok G. Cancer Genet Cytogenet. 1996;88:8–16. doi: 10.1016/0165-4608(95)00189-1. [DOI] [PubMed] [Google Scholar]
- 15.Dietmaier W, Hartmann A, Wallinger S, Heinmöller E, Kerner T, Endl E, Jauch K-W, Hofstädter F, Rüschoff J. Am J Pathol. 1999;154:83–95. doi: 10.1016/S0002-9440(10)65254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pantel K, Schlimok G, Kutter D, Schaller G, Genz T, Wiebecke B, Backmann R, Funke I, Riethmüller G. Cancer Res. 1991;51:4712–4715. [PubMed] [Google Scholar]
- 17.Pantel K, Schlimok G, Braun S, Kutter D, Schaller G, Funke I, Izbicki J, Riethmüller G. J Natl Cancer Inst. 1993;85:1419–1424. doi: 10.1093/jnci/85.17.1419. [DOI] [PubMed] [Google Scholar]
- 18.Uhr J W, Scheuermann R H, Street N E, Vitetta E S. Nat Med. 1997;3:505–509. doi: 10.1038/nm0597-505. [DOI] [PubMed] [Google Scholar]
- 19.Harris C C. J Natl Cancer Inst. 1996;88:1442–1455. doi: 10.1093/jnci/88.20.1442. [DOI] [PubMed] [Google Scholar]
- 20.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 21.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4877. [PubMed] [Google Scholar]
- 22.Prokocimer M, Rotter V. Blood. 1994;84:2381–2411. [PubMed] [Google Scholar]
- 23.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 24.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 25.Iggo R, Gatter K, Bartek J, Lane D, Harris A L. J Cell Biochem. 1990;16G:97–101. [Google Scholar]
- 26.Hall P A, Ray A, Lemoine N R, Midley C A, Krausz T, Lane D P. Lancet. 1991;338:513. doi: 10.1016/0140-6736(91)90586-e. [DOI] [PubMed] [Google Scholar]
- 27.Stigbrand T, Andres C, Bellanger L, Bishr Omary M, Bodenmuller H, Bonfrer H, Brundell J, Einarsson R, Erlandsson A, Johansson A, et al. Tumor Biol. 1998;19:132–152. doi: 10.1159/000029984. [DOI] [PubMed] [Google Scholar]
- 28.Izbicki J R, Hosch S B, Pichlmeier U, Rehders A, Busch C, Niendorf A, Passlick B, Broelsch C E, Pantel K. N Engl J Med. 1997;337:1188–1194. doi: 10.1056/NEJM199710233371702. [DOI] [PubMed] [Google Scholar]
- 29.Nevinny-Stickel C, Hinzpeter M, Andreas A. Eur J Immunogenet. 1991;18:323–332. doi: 10.1111/j.1744-313x.1991.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 30.Schlimok G, Funke I, Bock B, Schweiberer B, Witte J, Riethmüller G. J Clin Oncol. 1990;8:831–837. doi: 10.1200/JCO.1990.8.5.831. [DOI] [PubMed] [Google Scholar]
- 31.Kalthoff H, Schmiegel W, Roeder C, Kasche D, Schmidt A, Lauer G, Thiele H G, Honold G, Pantel K, Riethmüller G, et al. Oncogene. 1993;8:289–298. [PubMed] [Google Scholar]
- 32.Dobashi K, Sugio K, Osaki T. J Thorac Cardiovasc Surg. 1997;114:339–346. doi: 10.1016/S0022-5223(97)70178-8. [DOI] [PubMed] [Google Scholar]
- 33.Putz E, Witter K, Offner S, Stosiek P, Zippelius A, Johnson J, Zahn R, Riethmüller G, Pantel K. Cancer Res. 1999;59:241–248. [PubMed] [Google Scholar]
- 34.Itoh K, Hayashi A, Toh Y, Imai Y, Yamada A, Nishida T, Shichijo S. Int Rev Immunol. 1997;14:153–171. doi: 10.3109/08830189709116850. [DOI] [PubMed] [Google Scholar]
- 35.Van den Eynde B J, Boon T. Int J Clin Lab Res. 1997;27:81–86. doi: 10.1007/BF02912440. [DOI] [PubMed] [Google Scholar]
- 36.Speicher M R, Gwyn Ballard S, Ward D C. Nat Genet. 1996;12:368–375. doi: 10.1038/ng0496-368. [DOI] [PubMed] [Google Scholar]
- 37.Rodrigues N R, Rowan A, Smith M E F, Kerr I B, Bodmer W F, Gannon J V, Lane D P. Proc Natl Acad Sci USA. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh I M, Matlashewski G, Banks L. Nature (London) 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 39.Fanning E. J Virol. 1992;66:1289–1293. doi: 10.1128/jvi.66.3.1289-1293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]