Abstract
Bulk tank milk from 1,429 herds were collected in 3 rounds from 19 different geographic areas. The milk samples were tested by use of indirect LPS-ELISA procedure to detect Salmonella dublin antibodies. From the obtained OD-values herd seroprevalence in the given area was determined and GR-scores calculated for each herd by addition of the number of positive sampling rounds by the 5 geographically closest neighbour herds. In the 19 different areas the calculated prevalence ranged from 0.01 to 0.41. Totally 3,697 GR-scores were given. The mean GR-scores in the areas ranged from 0.0 to 6.5. Higher GR-scores were found in herds changing to seropositive status compared with herds seronegative throughout the study period. The results indicate that the risk for a dairy herd to receive S. dublin infection increases with the disease status among the nearest neighbours and with the prevalence of seropositive herds in the geographic area.
Keywords: Salmonella dublin, cattle, dairy herds, milk samples, LPS-ELISA, epidemiology, seroprevalence, risk
Introduction
Salmonella dublin is a host-adapted serotype causing infection in cattle of all ages [5]. It has been isolated from cattle in all parts of the world [10]. In Denmark this serotype has been isolated in 60%–75% of all diagnosed Salmonella outbreaks in cattle since 1980 [1]. S. dublin may persist in the cattle herds, as animals of all ages may be infected from S. dublin contaminated environments and some animals may become carriers excreting the organism in faeces for years, eventually for life [13]. An ELISA test for IgG antibodies directed against S. dublin LPS has been shown effective in detecting both mammary and faecal carriers [7]. The prevalence of Salmonella infection in dairy herds has been determined by means of serologic testing and percentage of culture-positive calves and environment in California [8]. It is possible to screen milk samples for S. dublin infection in dairy herds [2-4]. The LPS-ELISA has the capability to detect S. dublin infection in dairy herds and to point out newly infected herds by repeated testing [12]. In order to control and eventually eliminate an infection figures for risk and prevalence are beneficial. Spatial analysis of seroconversion, and spatial and temporal analysis of antibiotic resistance in cattle have been performed [11,6]. The purpose of the present study was to estimate the prevalence of S. dublin infection for different geographic areas and to evaluate the risk of receiving the infection if the neighbouring herds have S. dublin infection.
Materials and methods
Test samples
A total of 4,287 bulk tank milk samples were collected from March 1994 to March 1996 in 3 test rounds from a total of 1,429 dairy herds [12]. Nineteen geographic areas were represented. The radius of the areas ranged from 6 to 21 kilometres, and the number of dairy herds ranged from 16 to 258. Two geographic areas (Bornholm and Samsø) were islands. Inside the geographic areas the herds were equally distributed, and almost all dairy herds within the designated areas participated. Consequently approximately 10 percent of all Danish dairy herds participated in the study.
The milk samples were taken in connection with the Danish control program for milk quality. The samples were transported without preservatives at 5°C to the laboratory, where they were centrifuged at 200 × g for 5 min and the fat layer removed. The samples were stored in polypropylene tubes at -18°C until use.
Serology
The ELISA described by [3], was slightly modified for this study. Briefly: microwell plates (Cat. # 475094, Nunc, Denmark) were coated with S. dublin LPS antigen, and an undiluted positive, a weakly positive and a negative control samples were added in duplicate to each plate, each test milk was added undiluted (100 μL) in duplicate, and the plates were incubated overnight at 5°C. The bound antibodies were detected by using a purified immunoglobulin fraction of goat antiserum to bovine IgG (γ), labelled with horseradish peroxidase (Cat. # 14-12-02, Kirkegaard & Perry Lab. Maryland). H2O2 was used as a substrate and OPD (1,2-Ortho-Phenyl-Diamine) as an indicator. A test setup was considered valid if the negative milk control had an OD below 0.2 and the weak positive milk control had OD between 0.6 and 1.0. A test-sample was considered positive with OD values ≥0.30, and negative with OD-values <0.30 [12].
Statistical analyses
For the calculations, statistical and geographic analyses SAS Software (version 6.11, OS/2) was used, while @Risk (Palisade corporation, Newfield) was used for the risk analysis and for calculation of the true prevalences.
Each herd was given a geographical risk score (GR-) from 0 to 15, according to the reactions of the 5 nearest neighbouring dairy herds within its designated area: If all of the 5 neighbours tested negative in all 3 test rounds the herd was given the score 0 – if only 1 of the 5 neighbours tested positive in one test-round the herd was given the score 1 – if 2 herds of the 5 neighbours tested positive each in one test-round or one tested positive in 2 test-rounds the herd was given the score 2 and so on, so that the maximum score 15 was given when all the neighbours tested positive in all 3 test-rounds. As the closest neighbours not necessary were located within the designated geographical area, a certain edge effect will confound the calculations. The size of the effect will be negatively correlated with the number of herds included.
The distribution of GR-scores in the 4 groups: repeatedly positive herds, herds changing form positive to negative, herds changing from negative to positive, and herds fluctuating from negative to positive and reverse were compared to the repeatedly negative herds using chi-square from the Proc catmod procedure in SAS.
The true prevalence of S. dublin infection was calculated for each area using the Beta-distribution, Beta (α1, α2), where α1 = (r + 1) and α2 = (n - r + 1) and r = the true number of infected herds in all test rounds, and n = the total number of herds [9]. Where the true number of infected herds was obtained by adding the false negative and subtracting the false positive while correcting for the specificity and sensitivity of the test previously estimated to 0.89 and 0.88 respectively [12].
For estimation of the number of false negative samples the outcome of a negative binomial distribution with parameters (s,p), where s = number of test-positive herds + 1 and p = sensitivity was used [9].
Spearman rank correlation was used to determine the correlation between the calculated true prevalence, the proportion of herds changing from test-negative to test-positive, the mean GR-scores given to the group of herds, which tested constantly negative, the mean GR-scores given to the group of herds changing from test-negative to test-positive.
Results
A total of 3,697 GR-scores were given reflecting the test-results from the 5 nearest neighbours. A total of 1.058 herds (74.1%) were constantly negative in all 3 test-rounds, 90% of these herds were given less than 6 GR-scores. A total of 117 herds (8.1%) turned positive during the study, 90% of these herds were given 9 GR-scores and less.
Within the geographic areas the mean GR-scores ranged from 0.0 to 6.5 with mean GR-score = 2.6. The mean score was significantly higher for herds changing from test-negative to test-positive (GR = 3.4) during the study period than for herds that remained test-negative (GR = 2.1) throughout the study period (chi-square: 16.26, p < 0.0001). The highest mean scores were, however, obtained for herds, that tested positive in all test rounds (GR = 4.8) (Table 1). For each area and for each round the true prevalence was calculated with the mean true prevalence ranging from 0.01 to 0.41. The percentage of herds changing from test-negative to test-positive ranged from 0.0 to 13.5. The mean number of GR-scores for the group of herds, which were constantly negative ranged from 0.0 to 6.8, and for the group of herds, which turned positive the number of GR-score ranged from 0.0 to 7.3.
Table 1.
Distribution of scores according to serological status.
| Number of Herds (%)* | Number of GR-scores (%)* | Mean GR-scores* | P-value | |
| All Herds | 1429 (100) | 3697 (100) | 2.6 | |
| Herds test-negative in all rounds | 1058 (74.0) | 2251 (60.9) | 2.1 | |
| Herds test-positive in all rounds | 143 (10.0) | 689 (18.6) | 4.8 | 0.0000 |
| Herds changing from test-positive to test-negative | 55 (3.8) | 197 (5.3) | 3.6 | 0.0003 |
| Herds changing form test-negative to test-positive | 117 (8.2) | 394 (10.7) | 3.4 | 0.0001 |
| Herds fluctuating from test-negative to test-positive to test-negative and reverse | 56 (3.9) | 166 (4.5) | 3.0 | 0.0330 |
*The number of herds, GR-scores, and mean GR-scores are stated according to whether the herds tested negative in all test-rounds, tested positive in all test-rounds or changed test-status during the study period. The GR-scores were divided into 4 groups: repeatedly positive herds, herds changing from positive to negative, herds changing from negative to positive, and herds fluctuating from negative to positive and reverse. The groups were compared to the repeatedly negative herds using chi-square.
For each of the 19 geographic areas the calculated true prevalence was compared to the proportion of herds changing from test-negative to test-positive, and to the mean GR-scores given to herds testing negative in all test-rounds and herds changing from test-negative to test-positive respectively. Spearman rank correlation between the calculated true prevalence and the proportion of herds changing from test-negative to test-positive was 0.63 (p = 0.004). Spearman rank correlation between the calculated true prevalence and the mean GR-scores given to herds testing negative all round and to herds changing form test-negative to test-positive was 0.94 (p = 0.000) and 0.66 (p = 0.002), respectively (Fig. 1).
Figure 1.
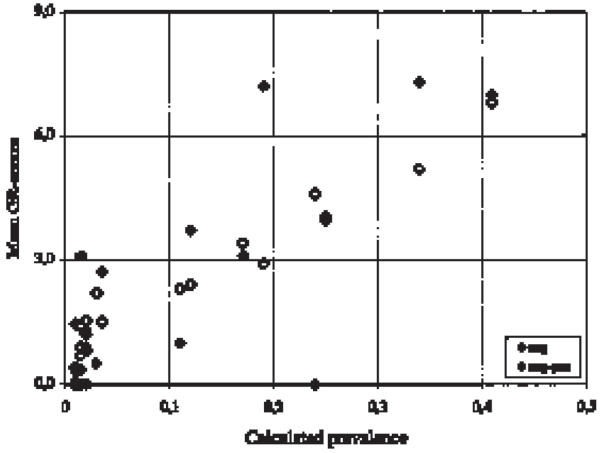
Comparison of prevalence and mean geographical risk-scores. For each area the calculated prevalence are compared to the mean GR-risk scores given to herds testing negative in all test-rounds, and the herds changing from test-negative to test-positive respectively.
As a mean true prevalence of 0.10 divided the geographic area into two sections fairly uniform according to the numbers of geographic areas, the material was stratified according to this mean true prevalence: One group of areas (n = 11) with a mean true prevalence lower than 0.10 (range: 0.01 to 0.03) and one group of areas (n = 8) with a mean true prevalence higher than 0.10 (range: 0.11 to 0.41). The distribution of test-negative herds compared to the herds which changed form test-negative to test-positive in the two prevalence areas is shown in Fig. 2. Statistical analysis showed no significant difference in the mean scores for the herds in the low prevalence areas (639 herds, chi-square = 0.82, p = 0.364). In the high prevalence areas (790 herds) the mean score was significantly higher for the herds changing from negative to positive during the study period (mean score = 4.4), compared to the herds that remained negative (mean score = 3.5), chi-square = 4.31 (p = 0.038).
Figure 2.
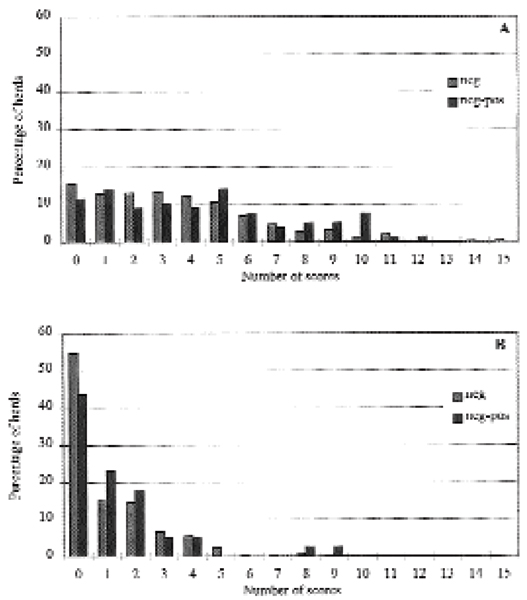
Comparison of test-negative herds and herds changing from test-negative to test-positive in areas with high prevalenence (>0.10) and areas with low prevalence (<0.10). A: The distribution of GR-scores in the high prevalence areas where 84 herds of totally 790 herds changed from negative to positive. B: The distribution of GR-scores in the low prevalence areas where 33 herds of totally 639 herds changed form negative to positive.
In the low prevalence areas (n = 11) the correlation between the calculated true prevalence and the mean GR-scores given to the herds, which tested constantly negative was 0.67 (p = 0.024). In the high prevalence areas (n = 8) the correlation between the calculated true prevalence and the mean GR-scores given to the herds, which tested constantly negative was 0.95 (p = 0.0003). All the calculations were corrected for ties (Fig. 1).
For the estimation of edge effect 2 areas (Bornholm and Samsø) were left out of the calculation, as these areas were islands. For the rest of the areas no correlation between the number of herds in the designated areas and the number of GR-scores was found.
Discussion
For cattle diseases spatial analysis have been used to estimate the effect of clustering in space. [11] detected clustering of bluetongue virus serotype 1 in Queensland. [6] tried to determine whether ampicillin- and tetracycline-resistant strains of Pasteurella multocida and P. haemolytica were spatially and temporally clustered, and found spatial clustering of resistant isolates in California. Both studies used Cuzick and Edwards' test to detect spatial clustering by comparing coordinates of cases and controls. In the present study the coordinates were used to point out the neighbouring dairy herds within a geographic area. Queensland covers around 1,727,500 km2, California covers 405,940 km2, while Denmark covers 43,032 km2. Thus the present study claims to yield a closer look into the areas under examination. Differences between 19 geographic areas were evaluated, and we investigated the use of a new herd parameter, the GR-score, that estimates the frequency of Salmonella infection in the neighbouring herds. In each area the results for the 5 nearest dairy herds were compared and each herd were given GR-scores after the results in the neighbouring dairies. Though the herds at the edge of an area are only receiving GR-scores from herds at one to 3 surrounding sides, this edge effect was left out of consideration as no correlation between number of herds, and number of mean GR-scores was found in the geographic areas.
Totally 668 herds (46.7%) were given none or one GR-score. If the GR-scores had been equally distributed between the herds studied, the repeatedly negative herds would have received 2.736 GR-scores; instead these herds were given 485 GR-scores less (17.7%). The test-positive herds should be given only 370 GR-scores at equal distribution, instead these herds were given 319 GR-scores more (86.2%), indicating an additional risk among the test-positive herds of having a test-positive neighbour (Table 1).
That herd seroconversion is primarily determined by seroprevalence in the given geographic area is supported both by the calculated Spearman rank correlations, which was significant at 5% condidence-level, and by the calculations based on stratification into high prevalence areas and low prevalence areas (Fig. 2), where the mean number of GR-scores is 1.1 in repeatedly test-negative herds and 1.4 for the herds changing from test-negative to test-positive in the low prevalence areas, while the corresponding figures are 3.5 and 4.4 in the high prevalence areas. This difference was only statistically significant in the latter population of herds. Thus in the low prevalence areas (<0.10) acquisition of S. dublin infection do not appear to be directly dependent of close contact with infected neighbour herds, but may be contracted otherwise e.g. by animal trade. Indications were found for an increased risk associated with infected neighbour herds. Unfortunately we are presently not able to evaluate this effect compared to factors operating on each area in general, such as herd size, climate and management factors, all though we expect most cases of infection to occur during the grazing season, as a result of the close contact between animals at pasture. In addition the size of the areas and the number of participating herds within these were not the same in all the geographic areas examined, however the distance between neighbour herds may influence the risk for receiving the infection.
In conclusion, a higher risk for changing serostatus from negative to positive for S. dublin infection was found if a dairy herd was located in a high prevalence area, and if the neighbouring herds were infected.
References
- Danish Veterinary Laboratory Annual Reports Copenhagen, Denmark. 1980.
- Hoorfar J, Feld NC, Schirmer AL, Bitsch V, Lind P. Serodiagnosis of Salmonella dublin Infection in Danish Dairy Herds Using O-Antigen Based Enzyme-linked Immunosorbent Assay. Can J Vet Res. 1994;58:268–274. [PMC free article] [PubMed] [Google Scholar]
- Hoorfar J, Lind P, Bitsch V. Evaluation of an O antigen Enzyme-linked Immunosorbent Assay for Screening of Milk Samples for Salmonella dublin Infection in Dairy Herds. Can J Vet Res. 1995;59:142–148. [PMC free article] [PubMed] [Google Scholar]
- Hoorfar J, Wedderkopp A, Lind P. Comparison between persisting anti-lipopolysaccharide antibodies and culture at postmortem in salmonella-infected cattle herds. Vet Microbiol. 1996;50:81–94. doi: 10.1016/0378-1135(95)00199-9. [DOI] [PubMed] [Google Scholar]
- Richardson A. Salmonellosis in Cattle. Vet Rec. 1975;96:329–331. doi: 10.1136/vr.96.15.329. [DOI] [PubMed] [Google Scholar]
- Singer RS, Case JT, Carpenter TE, Walker RL, Hirsh DC. Assessment of spatial and temporal clustering of ampicillin- and tetracycline-resistant strains of Pasturella multocida and P. Haemolytica isolated form cattle in California. JAVMA. 1998;212:1001–1005. [PubMed] [Google Scholar]
- Smith BP, House JK, Dilling GW, Roden LD, Spier SJ. Identification of Salmonella dublin carrier cattle. Proceedings of Salmonella and Salmonellosis. 1992. pp. 225–230.
- Smith BP, Roden LD, Thurmond MC, Dilling GW, Konrad H, Pelton JA, Picanso JP. Prevalence of salmonellae in cattle and in the environment on California dairies. JAVMA. 1994;205:467–471. [PubMed] [Google Scholar]
- Vose D. Quantitiative risk anlysis: a gudide to Monte Carlo simulation modeling. Wielly, Chichester, England; 1996. p. 57. [Google Scholar]
- Walker LR. Proceedings 27th annual convention. American Association of Bovine Practitioners; 1995. Salmonella dublin infection in cattle in California; pp. 8–9. [Google Scholar]
- Ward MP, Carpenter TE, Johnson SJ. Spatial analysis of seroconversion og sentinel cattle to bluetongue viruses in Queensland. A Vet Joul. 1996;74:128–131. doi: 10.1111/j.1751-0813.1996.tb14812.x. [DOI] [PubMed] [Google Scholar]
- Wedderkopp A, Strøger U, Bitsch V, Lind P. Testing of bulk tank milk for Salmonella dublin infection in dairy herds. Can J Vet Res. 2001;65 [PMC free article] [PubMed] [Google Scholar]
- Wray C, Wadsworth QC, Richards DW, Morgan JH. A three-year study of Salmonella dublin infection in a closed dairy herd. Vet Rec. 1989;124:532–535. doi: 10.1136/vr.124.20.532. [DOI] [PubMed] [Google Scholar]


