Abstract
The Notch family of proteins consists of transmembrane receptors that play a critical role in the determination of cell fate. Genetic studies in Caenorhabditis elegans suggest that the presenilin proteins, which are associated with familial Alzheimer’s disease, regulate Notch signaling. Here we show that proteolytic release of the Notch-1 intracellular domain (NICD), an essential step in the activation of Notch signaling, is markedly reduced in presenilin-1 (PS1)-deficient cells and is restored by PS1 expression. Nuclear translocation of the NICD is also markedly reduced in PS1-deficient cells, resulting in reduced transcriptional activation. Mutations in PS1 that are associated with familial Alzheimer’s disease impair the ability of PS1 to induce proteolytic release of the NICD and nuclear translocation of the cleaved protein. These results suggest that PS1 plays a central role in the proteolytic activation of the Notch-1-signaling pathway and that this function is impaired by pathogenic PS1 mutations. Thus, dysregulation of proteolytic function may underlie the mechanism by which presenilin mutations cause Alzheimer’s disease.
Notch-1 is a member of a conserved family of transmembrane receptors that regulate cell fate decisions during development (1, 2). Members of the Notch family are activated by ligands of the Delta-Serrate-LAG2 (DSL) family. Notch-1 undergoes a complex set of proteolytic cleavages during its maturation and upon activation by ligand binding. Ligand binding induces proteolytic cleavage within or proximal to the Notch-1 transmembrane domain, resulting in the release of the Notch-1 intracellular domain (NICD) (3, 4). The NICD can then translocate to the nucleus and activate the transcription of downstream target genes (5).
Mutations in the presenilin genes are a major cause of early-onset familial Alzheimer’s disease (FAD) (6–8). Presenilins are ubiquitously expressed eight-transmembrane domain proteins (9, 10) that are localized predominantly in the endoplasmic reticulum, nuclear membrane, and Golgi complex (11–14). A role for presenilins in the regulation of Notch signaling has been suggested by studies of the Caenorhabditis elegans presenilin homologue sel-12 (15) and by the demonstration that mammalian presenilins can substitute functionally for sel-12 (16, 17). Furthermore, presenilin-1 (PS1)-deficient mice exhibit decreased expression of Notch and the Notch ligand Dll1 in the presomitic mesoderm, as well as defective somite formation and skeletal abnormalities reminiscent of mice with targeted disruption of the Notch-1 gene (18, 19). These findings suggest that PS1 may regulate the Notch-signaling pathway during development, although the cellular mechanism is unknown.
The generation of amyloid-β protein, the major component of senile plaques in Alzheimer’s disease, is markedly reduced in cells derived from PS1-deficient mice because of reduced γ-secretase cleavage in the transmembrane domain of amyloid precursor protein (APP) (20). Because Notch-1 undergoes a similar proteolytic cleavage in response to ligand binding (3, 4), we explored the possibility that PS1 may also regulate this proteolytic event. We now report that PS1 induces the proteolytic release and nuclear translocation of the NICD and that PS1 mutations associated with FAD impair this function. Thus, PS1 plays a central role in the activation of Notch-1 signaling, which is impaired by pathogenic PS1 mutations.
MATERIALS AND METHODS
Cell Culture and Transfection.
Embryonic fibroblast cultures were derived from PS1 wild-type and knockout (PS1-wt and PS1-KO) littermates at embryonic days 15–16 (18), immortalized with a replication-defective v-myc retrovirus, and cultured in DMEM supplemented with 10% FBS. Plasmids were introduced into PS1-wt and PS1-KO cells by the Lipofectamine Plus transfection method (Life Technologies, Gaithersburg, MD), and cells were harvested 48 hr after transfection and lysed in RIPA-DOC buffer (1% Triton X-100/1% sodium deoxycholate/0.1% SDS/0.15 M NaCl/0.05M Tris⋅HCl, pH 7.2) supplemented with protease inhibitors (Complete; Boehringer Mannheim). The pCH110 β-galactosidase expression plasmid was cotransfected to normalize for transfection efficiency.
Plasmids.
The NδEF plasmid encodes the transmembrane and entire intracellular domain of murine Notch-1, extending from I1701 to K2530, with a C-terminal Myc tag in the pcDNA3.1 mammalian expression vector (Invitrogen). The constructs NδE, NδE(V1744K), and ICv1744 were provided by Raphael Kopan and have been described (3). The NδEF PEST deletion mutant was generated by cloning the cDNA sequence corresponding to codons 1701–2480 into pcDNA3.1. The constructs were confirmed by sequencing. Wild-type and mutant forms of PS1 were cloned into the pRK7 expression vector and confirmed by sequencing.
Immunoblotting and Reporter Assays.
Cell lysates were resolved by 10% SDS/PAGE, and immunoblotting was performed as described (21). Myc-tagged Notch-1 derivatives were resolved with monoclonal anti-Myc (9E10 from ATCC). PS1 was resolved with rabbit polyclonal antibody 231 to the PS1 N terminus (13). Cleaved and uncleaved Notch-1 proteins were quantitated by using Molecular Dynamics software and analysis of immunoblots in which bands were demonstrated to be present in a linear concentration range, as determined by standard curves. The HES-1 promoter reporter assay was performed as described (5).
Immunofluorescence Microscopy and Subcellular Fractionation.
PS1-wt and PS1-KO fibroblasts were cultured on glass coverslips in a 24-well plate at a density of 40,000 cells per well and transfected with 200 ng of NδEF plasmid DNA per well. Cells were fixed after 48 hr, permeabilized with 0.1% Triton X-100, and blocked with 5% BSA, followed by incubation with anti-Myc (9E10) and Cy3-conjugated anti-mouse IgG (1:400; Jackson ImmunoResearch). Nuclei were visualized by staining with Hoechst dye 3342 (1:2,000; Molecular Probes). Myc-positive cells were scored with a fluorescence microscope by counting eight different coverslips (>200 cells) per condition. Significantly reduced nuclear localization of NδEF in PS1-KO cells was observed in three independent experiments. Subcellular fractionation of PS1-wt and PS1-KO cells was performed as described (22).
RESULTS
PS1-Induced Proteolytic Release of the NICD and Activation of Notch-1 Signaling.
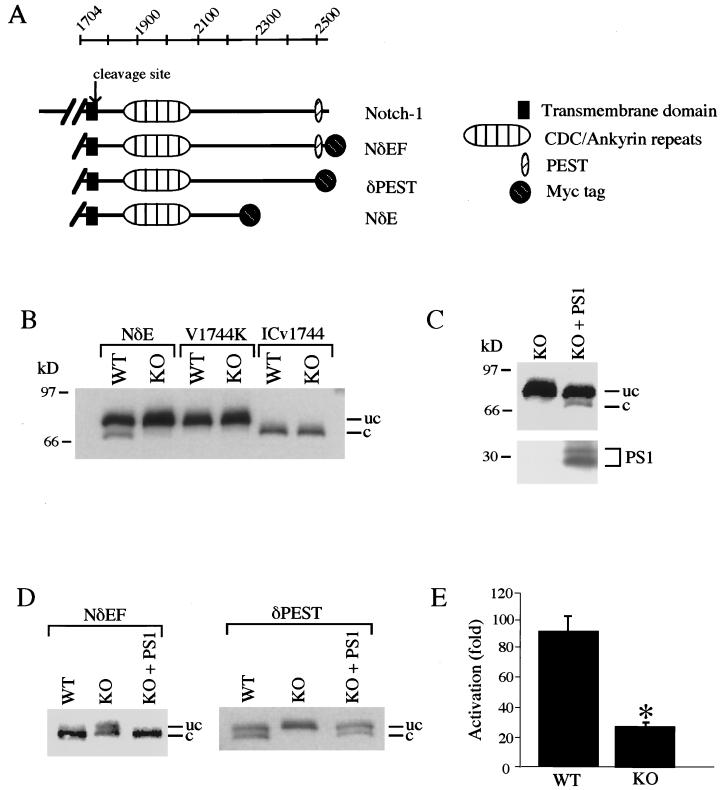
To explore the role of PS1 in Notch-1 signaling, we analyzed the proteolytic processing of a constitutively active, membrane-bound derivative of Notch-1 (NδE) in cultured embryonic fibroblasts derived from PS1-KO and littermate PS1-wt mice (Fig. 1A) (18). Cleavage of both full-length Notch-1 and NδE at residue V1744 results in the release of the NICD (3, 4). Myc-tagged NδE was expressed in PS1-wt and PS1-KO cells by transfection and analyzed by immunoblotting for the Myc epitope tag. In PS1-wt cells expressing NδE, an approximately 85-kDa protein representing uncleaved NδE and a lower-molecular-mass fragment (70–75 kDa) corresponding to the NICD were detected (Fig. 1B). To confirm the identities of these bands, we transfected an NδE-derived construct that is truncated at the cleavage site (ICv1744), thus generating a fragment corresponding to the NICD. ICv1744 comigrated with the lower-molecular-mass band generated from NδE (Fig. 1B). We also expressed an NδE construct with a V1744K mutation at the cleavage site, which prevents cleavage (3). NδE(V1744K) comigrated with the higher-molecular-mass NδE band and did not generate the NICD (Fig. 1B).
Figure 1.
PS1 induces proteolytic release of the Notch-1 intracellular domain. (A) Notch-1 constructs utilized in assays of proteolytic cleavage. (B) Release of the Notch-1 intracellular domain is markedly reduced in PS1-KO cells. PS1-wt (WT) and PS1-KO (KO) cells were transfected with Myc epitope-tagged NδE, NδE(V1744K), or ICv1744, and immunoblot analysis was performed with monoclonal anti-Myc. Note that the cleaved form of NδE is generated in PS1-wt cells but not in PS1-KO cells. The NδE mutation V1744K prevents cleavage, and ICv1744 is truncated at the cleavage site (3). The uncleaved (uc) and cleaved (c) NδE proteins are indicated. Immunoreactivity was absent in immunoblots of untransfected cell lysates. (C) PS1 restores Notch-1 proteolytic release in PS1-KO cells. (Upper) NδE immunoblot. (Lower) PS1 immunoblot showing PS1 N-terminal fragments. (D) The NδEF construct containing the full-length Notch-1-intracellular domain is cleaved completely, and cleavage depends on PS1 and the presence of a PEST motif. Note that PS1-wt cells expressing NδEF show only the cleaved protein, whereas PS1-wt cells expressing the PEST deletion mutant show similar amounts of cleaved and uncleaved proteins. Shown are NδE immunoblots of PS1-wt and PS1-KO cells transfected with Myc-tagged NδEF or δPEST constructs. (E) Transcriptional activation of the HES-1 promoter is potentiated by PS1. NδEF-induced transcriptional activation of the HES-1 promoter was markedly reduced in PS1-KO cells. PS1-wt and PS1-KO cells were cotransfected with NδEF and HES-1-luciferase together with a β-galactosidase expression plasmid (5). Values represent the ratio of normalized luciferase activity to normalized activity determined with the luciferase plasmid alone and represent the mean ± SEM, n = 9. ∗, P < 0.01 relative to PS1-wt by ANOVA with post-hoc Student–Newman–Keuls test.
The proteolytic release of the NICD was markedly reduced in PS1-KO cells, despite robust expression of NδE (Fig. 1B). Introduction of wild-type PS1 cDNA in PS1-KO cells by transfection completely restored the generation of the NICD (Figs. 1C and 3B), suggesting that PS1 plays a role in this Notch-1 cleavage. Because the NδE construct lacks a portion of the C-terminal domain of Notch-1, we determined whether PS1-induced cleavage would also occur in the presence of an intact Notch-1 intracellular domain. To address this question, we examined the cleavage of the Notch-1 construct NδEF, which contains the entire intracellular domain (Fig. 1A). In contrast to NδE, which was only partially cleaved, NδEF was completely cleaved in PS1-wt cells, resulting in the appearance of only the lower-molecular-mass NICD (Fig. 1D Left). Cleavage of NδEF was inhibited in PS1-KO cells, resulting in the appearance of both the cleaved and uncleaved proteins (Fig. 1D). Complete cleavage of NδEF was restored by transfection of PS1-KO cells with PS1 (Fig. 1D). These results suggest that the Notch-1 C-terminal domain modulates PS1-dependent proteolytic cleavage. The Notch-1 C-terminal domain contains a PEST sequence, a motif that mediates proteolytic cleavage by proteasome. To assess the role of the PEST motif in Notch-1 cleavage, we examined the cleavage of an NδEF derivative lacking a PEST domain (Fig. 1A). In contrast to NδEF, δPEST was cleaved only partially, resulting in similar levels of cleaved and uncleaved proteins in PS1-wt cells (Fig. 1D Right). Cleavage of δPEST was inhibited completely in PS1-KO cells and was restored by transfection of PS1 (Fig. 1D). Thus, PS1-induced release of the NICD is enhanced by the Notch-1 PEST motif. These results suggest that PS1 plays a critical role in the proteolytic release of the NICD.
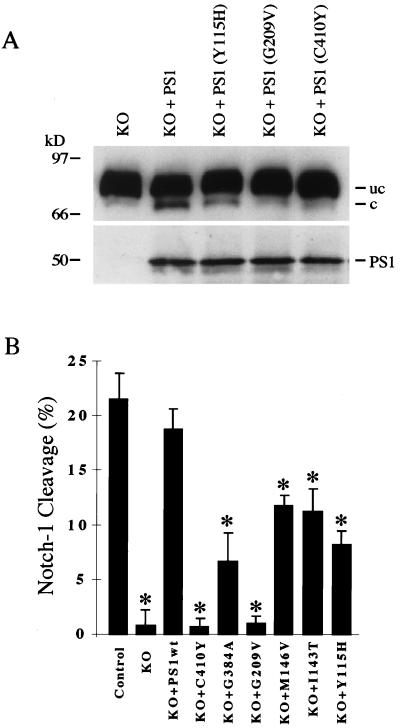
Figure 3.
PS1 mutations associated with familial Alzheimer’s disease impair proteolytic release of the NICD. (A) PS1 mutations associated with familial Alzheimer’s disease impair PS1-induced cleavage. PS1-KO cells transfected with the indicated PS1 mutants exhibit reduced Notch-1 cleavage relative to PS1-KO cells transfected with wild-type PS1. (Upper) NδE immunoblot. (Lower) PS1 immunoblot showing transfected holo-PS1. (B) Quantitative analysis of the effects of six PS1 mutants on Notch-1 cleavage. The ratio of cleaved to uncleaved forms of NδE was quantitated after transfection of NδE in PS1-wt (Control) and PS1-KO (KO) cells and in PS1-KO cells cotransfected with wild-type PS1 (PS1wt) or the indicated PS1 mutants. Note that inhibition of Notch-1 cleavage ranges from 40–60% for PS1 N-terminal mutations to 70–100% for PS1 C-terminal mutations. Notch-1 cleavage was not significantly different between PS1-wt cells and PS1-KO cells transfected with wild-type PS1. Shown is the mean ± SEM, n = 4. ∗, P < 0.05 relative to KO+PS1wt by ANOVA with post-hoc Student–Newman–Keuls test.
To determine whether PS1 affects the biological activity of Notch-1, we assayed for transcription mediated by the HES-1 promoter, a downstream target of the Notch-signaling pathway that can be activated by the NICD (5). The effect of PS1 was assessed by expression of NδEF and a HES-1-luciferase reporter in PS1-KO and PS1-wt cells. HES-1 promoter activity was markedly induced in PS1-wt cells expressing NδEF (Fig. 1E). Activation of HES-1 promoter activity was reduced by 70% in PS1-KO cells relative to PS1-wt cells (P < 0.01) (Fig. 1E). Thus, PS1 potentiates Notch-1-signaling activity.
Nuclear Translocation of the NICD Is PS1-Dependent.
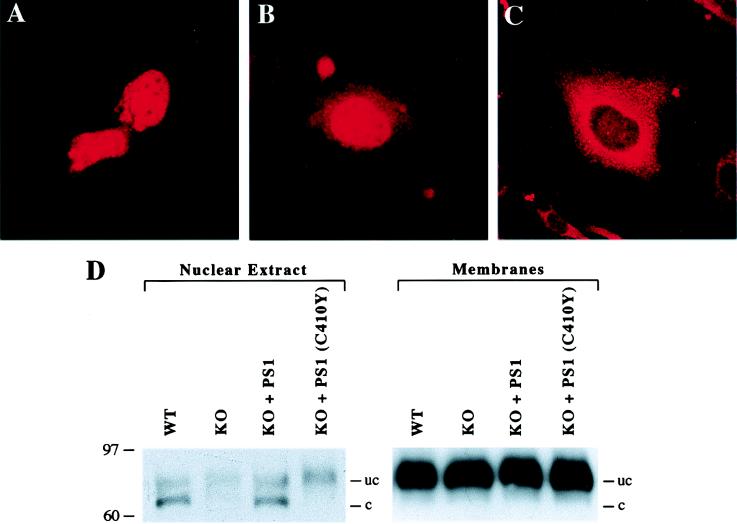
Proteolytic release of the NICD results in its translocation to the nucleus (3–5). To determine whether nuclear translocation is affected by PS1, immunofluorescence microscopy was performed on PS1-wt and PS1-KO cells after transfection of Myc-tagged NδEF by using the anti-Myc mAb. Transfected PS1-wt cells showed predominantly nuclear staining, consistent with the almost complete cleavage of the NδEF construct (Fig. 1D), and lower levels of cytoplasmic staining (Fig. 2 A and B). In contrast, NδEF-transfected PS1-KO cells showed predominantly cytoplasmic staining in a vesicular distribution, and most cells did not show nuclear staining (Fig. 2C). Quantitative analysis showed that 72 ± 5% of transfected PS1-wt cells showed nuclear staining, whereas only 27 ± 5% of transfected PS1-KO cells showed nuclear staining (mean ± SD, P < 0.001 by ANOVA).
Figure 2.
PS1 potentiates nuclear translocation of the Notch-1 intracellular domain. PS1-wt and PS1-KO cells were transfected with Myc-tagged NδEF. The cells were fixed 48 hr after transfection, and immunofluorescence microscopy was performed with monoclonal anti-Myc. (A) PS1-wt cells show predominantly nuclear immunoreactivity. (B) PS1-wt cell that shows prominent nuclear immunoreactivity and lower levels of cytoplasmic staining. (C) PS1-KO cells show cytoplasmic immunoreactivity in a vesicular distribution; most cells do not show nuclear immunoreactivity. (D) The cleaved Notch-1 fragment is detected in the nucleus of PS1-wt but not PS1-KO cells. Nuclear and membrane fractions were prepared from NδE-transfected PS1-wt and PS1-KO cells, and the distribution of cleaved and uncleaved Notch-1 proteins was determined by immunoblot analysis. Note that in PS1-wt cells, the cleaved Notch-1 fragment is detected in the nuclear extract but not in the membrane fraction, whereas uncleaved NδE is present in the membrane fraction. In PS1-KO cells, the cleaved fragment is not detected at significant levels in the nuclear extract, but appears after transfection of PS1. The cleaved fragment is not restored in the nuclear extract after transfection of the PS1 mutant C410Y. The difference in cytoplasmic membrane immunoreactivity for NδEF in A and B vs. NδE in D is due to the increased cleavage of the NδEF construct (Fig. 1D).
We then analyzed the distribution of cleaved and uncleaved Notch-1 proteins in membrane and nuclear fractions from NδE-transfected PS1-wt and PS1-KO cells. In contrast to NδEF, the NδE construct is only partially cleaved (Fig. 1 B and C), enabling the resolution of both nuclear and nonnuclear Notch-1 proteins. In PS1-wt cells, the cleaved NICD fragment was detected only in the nuclear fraction, whereas uncleaved NδE was detected predominantly in the membrane fraction (Fig. 2D). The cytosolic S100 fraction did not contain detectable Notch-1 proteins (data not shown). In PS1-KO cells, the cleaved NICD fragment was not detected at significant levels in the nuclear fraction, despite the presence of high levels of uncleaved NδE in the membrane fraction (Fig. 2D). Transfection of wild-type PS1 in PS1-KO cells restored the appearance of the cleaved NICD fragment in the nuclear fraction (Fig. 2D). Cytoplasmic Notch-1 immunoreactivity was much greater in PS1-wt cells transfected with NδE (Fig. 2D) than in cells transfected with NδEF (Fig. 2 A and B), correlating with partial cleavage of NδE vs. almost complete cleavage of NδEF (Fig. 1 B and D). These results suggest that PS1 facilitates nuclear translocation of the NICD.
Proteolytic Release of the NICD Is Impaired by PS1 Mutations.
We then determined whether PS1 mutations associated with FAD affect the proteolytic release of the NICD. As described above, PS1-KO cells exhibit markedly reduced proteolytic release of NICD, which is restored by transfection of PS1. Transfection of PS1 mutants in PS1-KO cells resulted in reduced cleavage of NδE compared with wild-type PS1, despite similar levels of PS1 expression (Fig. 3A). Quantitative analysis of proteolytic cleavage was performed by determining the ratio of cleaved to uncleaved Notch-1 proteins. This analysis showed that six different FAD mutations significantly reduced Notch-1 cleavage (Fig. 3B). Mutations in the PS1 C-terminal domain inhibited Notch-1 cleavage by 70–100%, whereas PS1 N-terminal mutations inhibited Notch-1 cleavage by approximately 40–60% (Fig. 3B). The PS1 mutation C410Y also markedly reduced nuclear translocation of the cleaved NICD fragment (Fig. 2D). These results suggest that PS1-induced proteolytic release of the NICD is reduced by FAD mutations.
DISCUSSION
These experiments indicate that PS1 plays a central role in the proteolytic release of the Notch-1 intracellular domain, leading to nuclear translocation of the cleaved Notch-1 fragment and activation of transcription. PS1 is not absolutely required for Notch-1 cleavage, because residual cleavage of the NδEF construct and activation of the HES-1 promoter are detected in PS1-KO cells. Future studies will determine whether the residual activation of Notch-1 cleavage is mediated by PS2. These results also provide a biochemical demonstration that the cleaved Notch-1 fragment is translocated to the nucleus, complementing previous genetic and transcriptional evidence for this step in Notch signaling (3–5). Although it is likely that PS1 induces nuclear translocation by facilitating proteolytic cleavage, we cannot exclude the possibility that PS1 may also affect a postcleavage step in the translocation process. These findings could account for the potentiation of Notch/lin-12 signaling by the C. elegans presenilin sel-12 (15) and for abnormalities in somite formation and skeletal development in PS1-deficient mice (18, 19).
PS1 could facilitate Notch-1 cleavage by a direct effect on proteolysis or by the regulation of protein trafficking. One possibility is that PS1 could be the protease that cleaves Notch-1. However, Notch-1 is cleaved at the cell surface or in endocytic vesicles after ligand binding (3–5), whereas PS1 is localized predominantly in the nuclear membrane, endoplasmic reticulum, and Golgi complex (11–14). Alternatively, PS1 may facilitate Notch-1 cleavage by activating the protease or by promoting the trafficking of the protease or Notch-1 to the cell surface. The latter possibility would be consistent with reduced plasma membrane localization of the Notch homologue lin-12 in C. elegans with loss of function of the presenilin sel-12 (23). Interestingly, the γ-secretase cleavage of APP, which generates amyloid-β protein (20), and the Notch-1 cleavage, which generates the NICD, are both markedly reduced in PS1-deficient cells. Furthermore, we have found that several peptide aldehydes inhibit γ-secretase and Notch-1 cleavage in parallel (data not shown). Thus, the same or similar proteases may mediate APP and Notch-1 cleavage.
PS1 mutations associated with FAD impair the ability of PS1 to induce proteolytic release and nuclear translocation of the NICD. This finding is consistent with the observation that pathogenic PS1 mutants show loss of function in the rescue of a sel-12-deficient phenotype in C. elegans (16, 17). Although these observations appear to be inconsistent with reports that a PS1 mutant can rescue developmental defects in PS1-deficient mice (24, 25), our results indicate that most PS1 mutants exhibit only partial loss of function. Thus, high-level expression of mutant PS1 transgenes may rescue a PS1-deficient phenotype. This would be consistent with the observation that some A246E PS1 transgenic lines with lower transgene expression levels only partially rescue the PS1-deficient phenotype (25). It will be of interest to determine whether the PS1 mutations that show the greatest impairment of Notch-1 cleavage are able to rescue the PS1-deficient phenotype in mice.
It is intriguing that PS1 mutations alter the proteolytic cleavage of APP (26–29), Notch-1, and β-catenin (21) and impair nuclear translocation of Notch-1 and β-catenin (30). The altered proteolytic cleavage of APP, which gives rise to increased levels of a pathogenic, 42-aa form of amyloid-β protein, has been implicated as a potential cause of the neurodegenerative process in AD (31). However, it remains to be determined whether altered proteolytic cleavage of other substrates, such as Notch-1 and β-catenin (21), contribute to the neurodegenerative process. Taken together, these observations suggest that dysregulation of proteolytic function may underlie the mechanism by which presenilin mutations cause Alzheimer’s disease.
Acknowledgments
We thank Raphael Kopan for providing Notch constructs and Alain Israel for providing the HES-1-luciferase reporter. We also thank Zhuohua Zhang for helpful discussion. This work was supported by National Institutes of Health Grants NS33325 and NS30352, a grant from Novartis Ltd. (to B.A.Y.), and a National Institutes of Health Mental Retardation Research Center Core Grant (P30-HD18655). P.N. was supported by a fellowship from the Edward R. and Anne G. Lefler Foundation.
ABBREVIATIONS
- PS1
presenilin-1
- FAD
familial Alzheimer’s disease
- KO
knockout
- wt
wild type
- NICD
Notch-1 intracellular domain
- APP
amyloid precursor protein
Note Added in Proof
Several recent reports also provide evidence of a role for PS1 in the proteolytic cleavage of Notch and APP (32–35).
References
- 1.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 2.Greenwald I. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 3.Schroeter E H, Kisslinger J A, Kopan R. Nature (London) 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 4.Struhl G, Adachi A. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 5.Jarriault S, Brou C, Logeat F, Schroeter E, Kopan R, Israel A. Nature (London) 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 6.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature (London) 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 7.Levy-Lahad E, Wasco W, Poorkaj P, Romano D M, Oshima J, Pettingell W H, Yu C, Jondro P D, Schmidt SD, Wang K, et al. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Ma J, Potter H. Proc Natl Acad Sci USA. 1995;92:12180–12184. doi: 10.1073/pnas.92.26.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doan A, Thinakaran G, Borchelt D, Slunt H, Ratovitsky T, Podisny M, Selkoe D, Seeger M, Gandy S E, Price D L, Sisodia S S. Neuron. 1996;17:1023–1030. doi: 10.1016/s0896-6273(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Greenwald I. Neuron. 1996;17:1015–1021. doi: 10.1016/s0896-6273(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 11.Kovacs D M, Fausett H J, Page K J, Kim T W, Moir R D, Merriam D E, Hollister R D, Hallmark O G, Mancini R, Felsenstein K M, et al. Nat Med. 1996;2:224–229. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- 12.Walter J, Capell A, Grunberg J, Pesold B, Schindzielorz A, Prior R, Podisny M B, Fraser P, St. George Hyslop P, Selkoe E, et al. Mol Med. 1996;2:673–691. [PMC free article] [PubMed] [Google Scholar]
- 13.Busciglio J, Hartmann H, Lorenzo A, Wong C, Baumann K, Sommer B, Staufenbiel M, Yankner B. J Neurosci. 1997;17:5101–5107. doi: 10.1523/JNEUROSCI.17-13-05101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Xu M, Zhou H, Ma J, Potter H. Cell. 1997;90:917–927. doi: 10.1016/s0092-8674(00)80356-6. [DOI] [PubMed] [Google Scholar]
- 15.Levitan D, Greenwald I. Nature (London) 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 16.Levitan D, Doyle T G, Brousseau D, Lee M D, Thinakaran G, Slunt H H, Sisodia S S, Greenwald I. Proc Natl Acad Sci USA. 1996;93:14940–14944. doi: 10.1073/pnas.93.25.14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumeister R, Leimer U, Zweckbronner I, Jakubek C, Grunberg J, Haass C. Genes Funct. 1997;2:149–159. doi: 10.1046/j.1365-4624.1997.00012.x. [DOI] [PubMed] [Google Scholar]
- 18.Shen J, Bronson R T, Chen D F, Xia W, Selkoe D, Tonegawa S. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 19.Wong P, Zheng H, Chen H, Becher M W, Sirinathsinghji J S, Trumbauer M E, Chen H Y, Price D L, Van der Ploeg L H T, Sisodia S S. Nature (London) 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- 20.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, Hartmann H, Do V M, Abramowski D, Sturchler-Pierrat C, Staufenbiel M, Sommer B, van de Wetering M, Clevers H, Saftig P, et al. Nature (London) 1998;395:698–702. doi: 10.1038/27208. [DOI] [PubMed] [Google Scholar]
- 22.Dingham J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levitan D, Greenwald I. Development. 1998;123:3599–3606. doi: 10.1242/dev.125.18.3599. [DOI] [PubMed] [Google Scholar]
- 24.Davis J A, Naruse S, Chen H, Eckman C, Younkin S, Price D L, Borchelt D R, Sisodia S S, Wong P C. Neuron. 1998;20:603–609. doi: 10.1016/s0896-6273(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 25.Qian S, Jiang P, Guan X M, Singh G, Trumbauer M E, Yu H, Chen H Y, Van de Ploeg L H, Zheng H. Neuron. 1998;20:611–617. doi: 10.1016/s0896-6273(00)80999-x. [DOI] [PubMed] [Google Scholar]
- 26.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird T D, Hardy J, Hutton M, Kukull W, et al. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 27.Duff K, Eckman C, Zehr C, Yu X, Prada C M, Perez-tur J, Hutton M, Buee L, Harigaya Y, Yager D, et al. Nature (London) 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 28.Borchelt D R, Thinakaran G, Eckman C B, Lee M K, Davenport F, Ratovitsky T, Prada C M, Kim G, Seekins S, Yager D, et al. Neuron. 1996;17:1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 29.Citron M, Westaway D, Xia W, Carlson G, Diehl T, Levesque G, Johnson-Wood K, Lee M, Seubert P, Davis A, et al. Nat Med. 1997;3:67–72. doi: 10.1038/nm0197-67. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura M, Yu G, Levesque G, Zhang D M, Ruel L, Chen F, Milman P, Holmes E, Liang Y, Kawarai T, et al. Nat Med. 1999;5:164–169. doi: 10.1038/5526. [DOI] [PubMed] [Google Scholar]
- 31.Yankner B A. Neuron. 1996;16:921–932. doi: 10.1016/s0896-6273(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 32.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, et al. Nature (London) 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 33.Struhl G, Greenwald I. Nature (London) 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 34.Ye Y, Lukinova N, Fortini M E. Nature (London) 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 35.Wolfe M S, Xia W, Ostaszewski B L, Taylor Kimberley W, Selkoe D J. Nature (London) 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]