Abstract
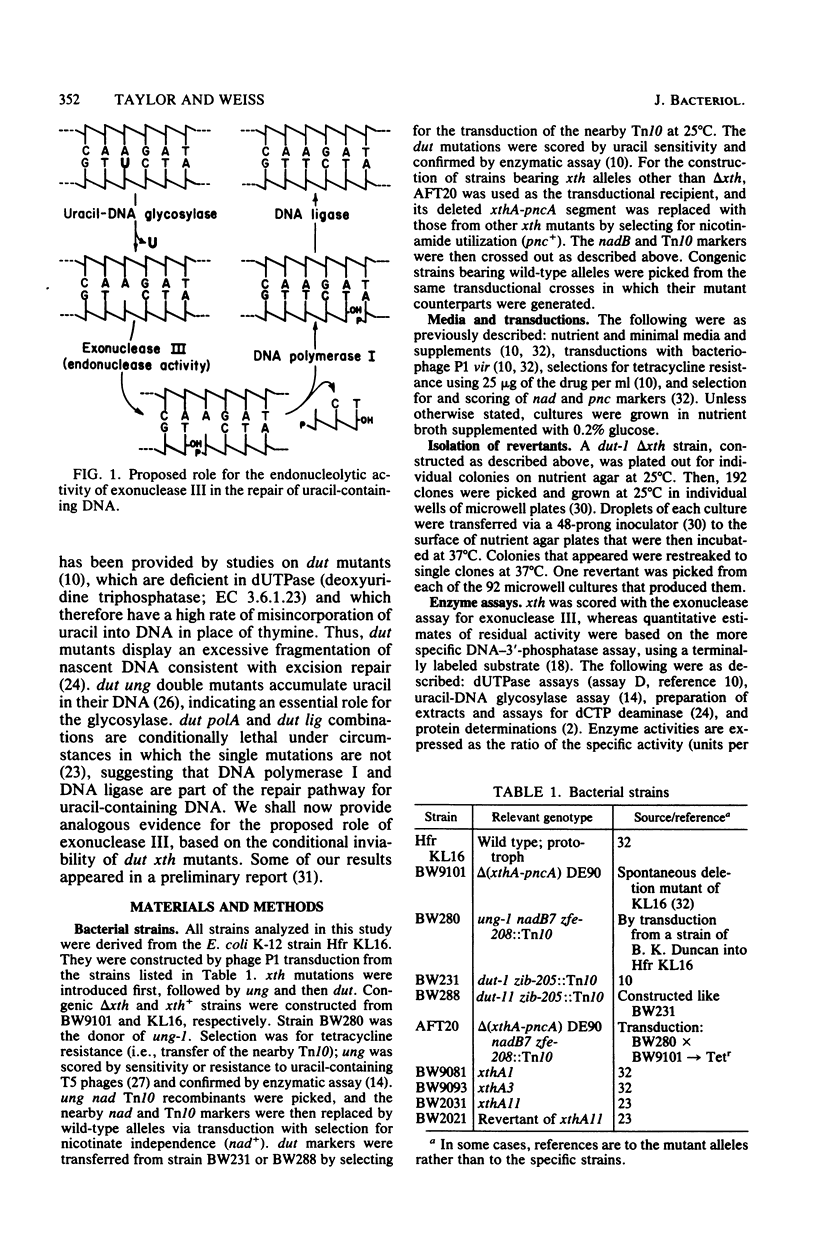
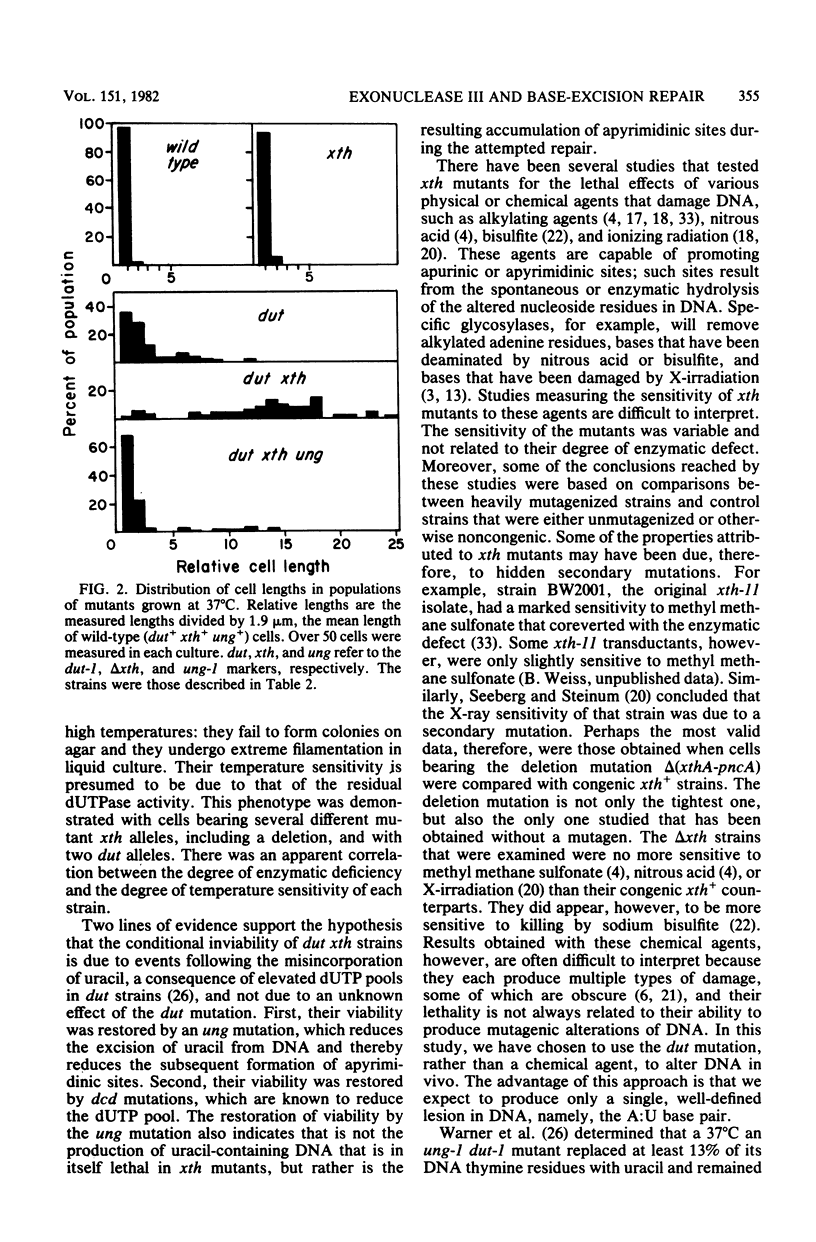
Mutants of Escherichia coli K-12 deficient in both exonuclease III (the product of the xth gene) and deoxyuridine triphosphatase (the dut gene product) are inviable at high temperatures and undergo filamentation when grown at such temperatures. In dut mutants, the dUTP pool is known to be greatly enhanced, resulting in an increased substitution of uracil for thymine in DNA during replication. The subsequent removal of uracil from the DNA by uracil-DNA glycosylase produces apyrimidinic sites, at which exonuclease III is known to have an endonucleolytic activity. The lethality of dut xth mutants, therefore, indicates that exonuclease III is important for this base-excision pathway and suggests that unrepaired apyrimidinic sites are lethal. Two confirmatory findings were as follows. (i) dut xth mutants were viable if they also had a mutation in the uracil-DNA glycosylase (ung) gene; such mutants should not remove uracil from DNA and should not, therefore, generate apyrimidinic sites. (ii) In the majority of the temperature-resistant revertants isolated, viability had been restored by a mutation in the dCTP deaminase (dcd) gene; such mutations should decrease dUTP production and hence uracil misincorporation. The results indicate that, in dut mutants, exonuclease III is essential for the repair of uracil-containing DNA and of apyrimidinic sites.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chetsanga C. J., Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979 Aug 10;6(11):3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Roza R., Friedberg E. C., Duncan B. K., Warner H. R. Repair of nitrous acid damage to DNA in Escherichia coli. Biochemistry. 1977 Nov 1;16(22):4934–4939. doi: 10.1021/bi00641a030. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Rockstroh P. A., Warner H. R. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J Bacteriol. 1978 Jun;134(3):1039–1045. doi: 10.1128/jb.134.3.1039-1045.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A. D., Duncan B. K., Hartman P. E. Nitrous acid damage to duplex deoxyribonucleic acid: distinction between deamination of cytosine residues and a novel mutational lesion. J Bacteriol. 1980 Apr;142(1):335–338. doi: 10.1128/jb.142.1.335-338.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates F. T., 3rd, Linn S. Endonuclease V of Escherichia coli. J Biol Chem. 1977 Mar 10;252(5):1647–1653. [PubMed] [Google Scholar]
- Gates F. T., Linn S. Endonuclease from Escherichia coli that acts specifically upon duplex DNA damaged by ultraviolet light, osmium tetroxide, acid, or x-rays. J Biol Chem. 1977 May 10;252(9):2802–2807. [PubMed] [Google Scholar]
- Gossard F., Verly W. G. Properties of the main endonuclease specific for apurinic sites of Escherichia coli (endonuclease VI). Mechanism of apurinic site excision from DNA. Eur J Biochem. 1978 Jan 16;82(2):321–332. doi: 10.1111/j.1432-1033.1978.tb12026.x. [DOI] [PubMed] [Google Scholar]
- Hochhauser S. J., Weiss B. Escherichia coli mutants deficient in deoxyuridine triphosphatase. J Bacteriol. 1978 Apr;134(1):157–166. doi: 10.1128/jb.134.1.157-166.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., D'Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981 Apr 30;290(5809):797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974 Jul 30;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Ljungquist S. A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J Biol Chem. 1977 May 10;252(9):2808–2814. [PubMed] [Google Scholar]
- Ljungquist S., Lindahl T., Howard-Flanders P. Methyl methane sulfonate-sensitive mutant of Escherichia coli deficient in an endonuclease specific for apurinic sites in deoxyribonucleic acid. J Bacteriol. 1976 May;126(2):646–653. doi: 10.1128/jb.126.2.646-653.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milcarek C., Weiss B. Mutants of Escherichia coli with altered deoxyribonucleases. I. Isolation and characterization of mutants for exonuclease 3. J Mol Biol. 1972 Jul 21;68(2):303–318. doi: 10.1016/0022-2836(72)90215-x. [DOI] [PubMed] [Google Scholar]
- O'Donovan G. A., Neuhard J. Pyrimidine metabolism in microorganisms. Bacteriol Rev. 1970 Sep;34(3):278–343. doi: 10.1128/br.34.3.278-343.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeberg E., Steinum A. L. Repair of x-ray-induced deoxyribonucleic acid single-strand breaks in xth mutants of Escherichia coli. J Bacteriol. 1980 Mar;141(3):1424–1427. doi: 10.1128/jb.141.3.1424-1427.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R. Genetic effects of bisulfite (sulfur dioxide). Mutat Res. 1977;39(2):149–175. doi: 10.1016/0165-1110(77)90020-3. [DOI] [PubMed] [Google Scholar]
- Simmons R. R., Friedberg E. C. Enzymatic degradation of uracil-containing deoxyribonucleic acid. V. Survival of Escherichia coli and coliphages treated with sodium bisulfite. J Bacteriol. 1979 Mar;137(3):1243–1252. doi: 10.1128/jb.137.3.1243-1252.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B. K., Lehman I. R. Excision repair of uracil incorporated in DNA as a result of a defect in dUTPase. J Mol Biol. 1977 Dec 5;117(2):293–306. doi: 10.1016/0022-2836(77)90128-0. [DOI] [PubMed] [Google Scholar]
- Tye B. K., Nyman P. O., Lehman I. R., Hochhauser S., Weiss B. Transient accumulation of Okazaki fragments as a result of uracil incorporation into nascent DNA. Proc Natl Acad Sci U S A. 1977 Jan;74(1):154–157. doi: 10.1073/pnas.74.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Demple B. F., Deutsch W. A., Kane C. M., Linn S. Apurinic/apyrimidinic endonucleases in repair of pyrimidine dimers and other lesions in DNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4602–4606. doi: 10.1073/pnas.77.8.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Duncan B. K., Garrett C., Neuhard J. Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1981 Feb;145(2):687–695. doi: 10.1128/jb.145.2.687-695.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Thompson R. B., Mozer T. J., Duncan B. K. The properties of a bacteriophage T5 mutant unable to induce deoxyuridine 5'-triphosphate nucleotidohydrolase. Synthesis of uracil-containing T5 deoxyribonucleic acid. J Biol Chem. 1979 Aug 25;254(16):7534–7539. [PubMed] [Google Scholar]
- Weiss B. Endonuclease II of Escherichia coli is exonuclease III. J Biol Chem. 1976 Apr 10;251(7):1896–1901. [PubMed] [Google Scholar]
- Weiss B., Milcarek C. Mass screening for mutants with altered DNases by microassay techniques. Methods Enzymol. 1974;29:180–193. doi: 10.1016/0076-6879(74)29020-7. [DOI] [PubMed] [Google Scholar]
- White B. J., Hochhauser S. J., Cintron N. M., Weiss B. Genetic mapping of xthA, the structural gene for exonuclease III in Escherichia coli K-12. J Bacteriol. 1976 Jun;126(3):1082–1088. doi: 10.1128/jb.126.3.1082-1088.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajko D. M., Weiss B. Mutations simultaneously affecting endonuclease II and exonuclease III in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Feb;72(2):688–692. doi: 10.1073/pnas.72.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieg J., Maples V. F., Kushner S. R. Recombinant levels of Escherichia coli K-12 mutants deficient in various replication, recombination, or repair genes. J Bacteriol. 1978 Jun;134(3):958–966. doi: 10.1128/jb.134.3.958-966.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]



