Abstract
Pituitary corticotroph SOCS-3 is a novel intracellular regulator of leukemia inhibitory factor (LIF)-mediated proopiomelanocortin gene expression and adrenocorticotropic hormone (ACTH) secretion, inhibiting LIF-activated Janus kinase-signal transducers and activators of transcription (STAT) signaling in a negative autoregulatory loop. We now demonstrate in corticotroph AtT-20 cells that LIF-stimulated endogenous SOCS-3 mRNA expression is blocked in stable transfectants of SOCS-3 wild type or in dominant negative STAT-3 mutants, respectively. We characterized ≈3.8-kb genomic 5′ sequence of murine SOCS-3, including ≈2.9-kb sequence upstream of the transcription start site (+1), which was determined by 5′ rapid amplification of cDNA ends and RNase protection assay. Different 5′ constructs were cloned into the pGL3Basic vector, and luciferase activity was assayed in transiently transfected ACTH-secreting corticotroph AtT-20 cells. A STAT-1/STAT-3 binding element, located at nucleotides −72 to −64, was essential for LIF stimulation of SOCS-3 promoter activity. LIF induced 10-fold increased luciferase activity in a wild-type construct spanning −2757 to +929 bases. However, deletion or point mutation of the STAT-1/STAT-3 binding element abrogated LIF action (2- to 3-fold). Electrophoretic mobility-shift assay analysis confirmed specific binding of STAT-1 and STAT-3 to this region. These results characterize the genomic 5′ region of murine SOCS-3 and identify an important STAT-1/STAT-3 binding element therein. Thus, LIF-stimulated SOCS-3 gene expression is at least in part mediated by STAT-3 and STAT-1. The cytokine inhibitor SOCS-3 acts in a negative loop to autoregulate its own gene expression, thus limiting its accumulation in the corticotroph cell. These results demonstrate a mechanism for corticotroph plasticity with rapid “on” and “off” ACTH induction in response to neuro-immuno-endocrine stimuli, such as LIF.
Keywords: leukemia inhibitory factor, STAT-3, AtT-20 cells
Ligand binding to several type I and type II cytokine receptors causes activation of cytoplasmic Janus kinases (Jaks), tyrosine phosphorylation of signal transducers and activators of transcription (STATs), and further downstream events (1–3). Recently, several groups have described a new family of cytokine-inducible proteins, inhibiting the Jak-STAT signaling cascade. These proteins have been termed suppressors of cytokine signaling (SOCS) (4, 5), STAT-induced STAT inhibitors (SSI) (6, 7), cytokine-inducible SH2 containing protein (CIS) (8–10), and Jak binding protein (JAB) (11, 12). The SOCS-protein family currently consists of CIS and SOCS-1 to SOCS-7 (5, 13, 14). SOCS-protein expression is stimulated by various cytokines in a tissue-specific manner (4, 13, 14). The gene expression of SOCS-1/SSI-1/JAB (4, 6, 11) and SOCS-3/SSI-3/CIS-3 (4, 7, 9), which in this manuscript are referred to as SOCS-1 and SOCS-3, is induced by IL-6 and leukemia inhibitory factor (LIF) in various tissues (4–7, 9, 11, 15). Both SOCS-1 and SOCS-3 bind to the JH1 domain of Jak-2 and inhibit Jak-2 activity (9, 11), as well as IL-6- or LIF-induced tyrosine phosphoylation of gp130 and STAT-3 (4, 7, 9, 11, 15).
The IL-6 cytokine family, consisting of IL-6, IL-11, LIF, oncostatin M, ciliary neurotrophic factor, and cardiotrophin-1, is characterized by their receptors sharing the common receptor subunit gp130 and subsequent activation of the Jak-STAT signaling cascade (1, 3, 16). In addition to IL-1, several cytokines of the IL-6 cytokine family are important neuro-immuno-endocrine modulators of pituitary function (17–19). We previously demonstrated LIF to be a potent auto/paracrine stimulus of pituitary proopiomelanocortin (POMC) gene expression and adrenocorticotropic hormone (ACTH) secretion, modulating the hypothalamus-pituitary-adrenal (HPA) axis response to various inflammatory and stress stimuli (20–22). In vitro experiments using human fetal pituitary cells (23) and the corticotroph cell line AtT-20 (24, 25) showed a profound synergistic action of LIF and corticotropin-releasing hormone on POMC gene expression and ACTH secretion. The LIF-induced signaling cascade in the corticotroph cell involves phosphorylation of gp130, STAT-3, and STAT-1 (15, 23, 26, 27), and LIF-induced POMC expression and ACTH secretion is STAT-3 dependent (28). SOCS-3 gene expression is rapidly induced by LIF in the pituitary in vivo and in corticotroph AtT-20 cells in vitro (15). SOCS-3 inhibits LIF-induced POMC gene expression and ACTH secretion (15), thus providing an intracellular negative feedback on cytokine-induced activation of the HPA axis. We now demonstrate the regulation of pituitary SOCS-3 gene expression by characterizing ≈3.8 kb of the genomic 5′ region of murine SOCS-3, including its promoter region. We show that murine SOCS-3 gene expression is STAT-3 dependent and is negatively autoregulated by SOCS-3 protein itself. We also show that a STAT-1/STAT-3 binding element is functionally critical for murine SOCS-3 promoter activity. Thus, negative autoregulatory feedback of SOCS-3 appears to provide a mechanism for limiting corticotroph SOCS-3 accumulation.
MATERIALS AND METHODS
Materials.
Recombinant murine LIF, IL-6, and IL-11 were purchased from R & D Systems. Mouse liver Marathon-Ready cDNA, Advantage-GC cDNA polymerase, mouse GenomeWalker Kit, and Advantage-GC genomic polymerase were from CLONTECH. Maxiscript T7 polymerase kit and ribonuclease protection kit RPA-II were from Ambion (Austin, TX), and polyclonal STAT-1 p84/p91 (M-22) and STAT-3 (H-190) antibodies were from Santa Cruz Biotechnology. Mouse genomic DNA, Erase-a Base system, pGL3 Basic, and pSV-β-galactosidase vector were from Promega, and TOPO-TA PCR2.1 was from Invitrogen.
Cell Culture.
Cell culture of AtT-20/D16v-F2 cells was performed as described (15, 21). Individual clones of AtT-20 cells, overexpressing SOCS-3 (AtT-20S), mock-transfected (AtT-20M), wild-type (wt) STAT-3 (AtT-20W), or dominant negative STAT-3 mutants (AtT-20F and AtT-20D) were isolated after stable transfection (15, 28). Three separate individual clones with high stable overexpression of the respective construct were selected with G418 (1 mg/ml) for the experiments.
Northern Blot Analysis.
Northern blot analysis was performed as described (15, 21). To detect endogenous SOCS-3 mRNA in AtT-20S cells, without hybridization to exogenous SOCS-3 mRNA derived from stable overexpression of SOCS-3, a probe spanning exon 1 and the untranslated 5′ region of exon 2 was used. Otherwise, the previously described (15, 19) murine SOCS-3 probe spanning most of the coding region was used.
5′ Rapid Amplification of cDNA Ends (RACE) and RNase Protection Assay.
5′ RACE was performed with a premade, adaptor-ligated Marathon-Ready double-stranded (ds) cDNA derived from pooled BALB/c mouse liver (29) and Advantage-GC cDNA polymerase by using gene-specific primary and nested antisense primers 5′-CAGTAGAATCCGCTCTCCTGCAGCTTG-3′ and 5′-CTCGCTTTTGGAGCTGAAGGTCTTGAG-3′. Products were cloned into PCR2.1 vector, and multiple single clones were sequenced.
RNase protection assay was performed with a RPA-II kit, following the manufacturer’s recommendations. A fragment spanning nucleotides +160 to −273 was cloned into PCR2.1 and linearized, and a [32P]UTP-labeled antisense probe was generated.
PCR-Based Characterization of the 5′ Genomic Region.
The 5′ genomic region of SOCS-3 was cloned by using a PCR-based technique (30) with premade adaptor-ligated genomic DNA fragments, derived from ICR Swiss mice (Genomewalk kit). PCR and subsequent nested PCR were performed by automatic hot-start as touchdown-PCR (Advantage-GC genomic Polymerase) and gene-specific antisense primers 5′-CAGTAGAATCCGCTCTCCTGCAGCTTG-3′ and 5′-CTCGCTTTTGGAGCTGAAGGTCTTGAG-3′. Further genomic walks in 5′ direction were performed with gene-specific antisense primers 5′-CTTCCTACCTAGTCCCGAAGCGAAATC-3′, 5′-CAGATGTTGGCAGCCGTGAAGTCTAC-3′, 5′-GCGGGCGAGTGTAGAGTCAGAGTTAGAG-3′, and 5′-CGATTCCTGGAACTGCCCGGCCGGTCTTC-3′, as well as 5′-CTCAGTGGGCTTTCTGACCTGCCCTCTTG-3′ and 5′-GACTACACAGAGTAGCTTGGGCTAGGAG-3′. Products were cloned into PCR2.1, and single clones were sequenced.
Different Constructs of the 5′ Genomic Region of SOCS-3.
3′ Truncated forms of the full-length 3.7-kb construct in pGL3Basic vector (clone 6) were generated by PCR from genomic DNA.
5′ Truncated forms of clone 6 were generated by digestion with SstI and NheI, followed by unidirectional digestion with exonuclease III (Erase-a-Base kit) (31) and subsequent religation.
Mutated forms of clone 6 were generated by overlap extension PCR (32) with Pfu polymerase and 5% DMSO, by using external sense primer 5′-CATCGCGACGCCCCCGCCTCT-3′ and antisense primer 5′-GAAACCCGAGGGCCCCAGTCTG-3′ with exclusive restriction sites for NruI or ApaI, respectively. Internal mutagenizing primers caused deletions of nucleotides −78 to −58 and −99 to −60, respectively. Similarly, the STAT binding element region at −72 to −64 was mutated. Gel-purified PCR products and the original template were digested and purified, and the mutated fragments were religated into the original 3.7-kb construct in pGL3Basic and verified by sequencing.
Luciferase Assay.
For transient transfection experiments, 2 × 105 cells were plated in 6-well plates, incubated for 24 hr, and transfected by using Lipofectamine and 0.5 μg of constructs in pGL3Basic vector and 1.0 μg pSV-β-galactosidase. Transfected cells were first incubated for 24 hr in serum-free DMEM, followed by 6 hr of cytokine treatment and subsequent measurement of luciferase activity. In experiments using different promoter constructs, transfection efficiency was verified by β-galactosidase activity.
Electromobility Shift Assay (EMSA).
Nuclear extracts of AtT-20 cells and EMSA were performed as described (25). AtT-20 cells were grown to 80% confluency and serum-deprived 24 hr before treatment with 10−9 M LIF, followed by cell lysis and preparation of nuclear extracts. For the EMSA, 20-μg nuclear extracts were preincubated for 15 min at room temperature in 20 μl of binding buffer (10 mM Tris⋅HCl/50 mM NaCl/1 mM EDTA/1 mM DTT/0.1% NP-40/5% glycerol/1 mg/ml BSA, pH 7.5) with 1 μg of poly(dI-dC). A 32P-labeled ds oligonucleotide corresponding to sequence −75 to −55 of SOCS-3 promoter (5′-−75CAGTTCCAGGAATCGGGGGGC−55-3′) and used as probe (60,000 cpm, 5 fmol per reaction) then was added for 20 min. In competition experiments, 100-fold molar excess unlabeled competitor oligonucleotides were added to the preincubation reaction with the ds oligonucleotide corresponding to sequence −75 to −55 of SOCS-3 promoter; this same oligonucleotide mutated at positions −72, −69, −67, and −64 (underlined) (5′-−75CAGATCGACGATTCGGGGGGC−55-3′) or the AP-2 recognition site oligonucleotide (5′-GATCGAACTGACCGCCCGCCGCCCGT-3′). For supershift experiments, 2 μg polyclonal STAT-1 p84/p91 or STAT-3 antibody was added to the preincubation reaction and incubated for an additional 60 min at 4°C. Protein-DNA complexes were run on a 6% nondenaturing polyacrylamide gel in 0.5× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA), gels were dried, and autoradiographs were exposed.
Statistical Analysis.
Statistical analysis was performed by unpaired t test. All values are mean ± SEM.
RESULTS
5′ Genomic Sequence of Murine SOCS-3 and Determination of the Transcription Start Site by 5′ RACE and RNase Protection Assay.
Based on the sequence information from the 5′ genome walk, a full-length 5′ product of murine SOCS-3 spanning ≈3.8 kb of genomic sequence was generated by using the following sense and antisense primers: 5′-GACGTTCCTAAAAGCATGCATGTCACCCAG-3′ and 5′-GGATCTGCGCGGCGGTGGCTGCAGCTGCTT-3′. Cloning of the product into PCR2.1 was followed by orientation verification, sequencing, restriction enzyme digestion with SstI and XhoI, and subcloning of a ≈3.7-kb construct into pGL3Basic (clone 6). Sequence information was obtained for the whole ≈3.8 kb (Fig. 1) (GenBank accession no. AF117732).
Figure 1.
Nucleotide sequence of the full-length ≈ 3.8-kb genomic 5′ region of murine SOCS-3. Exons are single underlined. The transcription start site is defined as +1. The translation initiation codon ATG and a putative TATA-box are indicated in bold letters. Two putative STAT binding elements are indicated in bold and underlined. The complete available sequence of the genomic 5′ region of murine SOCS-3 has been deposited in GenBank (accession no. AF117732).
5′ RACE revealed the existence of an untranslated exon 1 (+1 to +299), separated from exon 2 (starting at +856) by an intron (+300 to +855). By using RNase protection assay, the main transcription start site was defined and is referred to as +1 (Fig. 1 and 2). The previously determined translation initiation site for murine SOCS-3 (GenBank accession no. U88328) (4) was in exon 2 at +946 (Fig. 1).
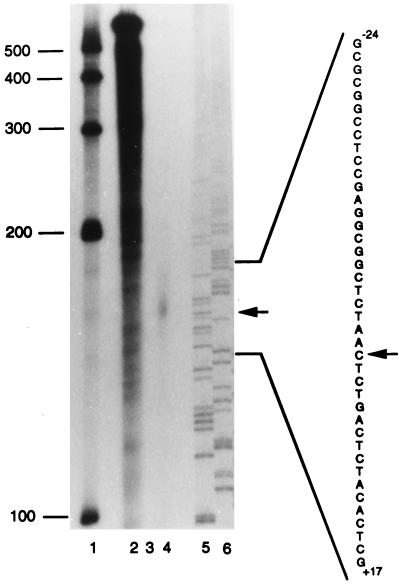
Figure 2.
Determination of the transcription start site of murine SOCS-3 mRNA by RNase protection assay. As described in Materials and Methods, a 32P-labeled antisense probe spanning nucleotides +160 to −273 of the murine SOCS-3 gene was hybridized at 42°C with 10 μg yeast RNA (lanes 2 and 3) or 10 μg total RNA derived from LIF-stimulated AtT-20 cells (lane 4). Lane 2 shows the undigested full-length probe. RNase A/RNase T1 mix was added for digestion of unprotected fragments to samples of lane 3 (negative control) and lane 4. Lanes 5 and 6 show 33P-sequencing of A and C with antisense primer (+160 to +143). Arrows indicate a protected band in lane 4 and the corresponding nucleotide sequence.
Effect of Different Cytokines on SOCS-3 Promoter Activity and Gene Expression.
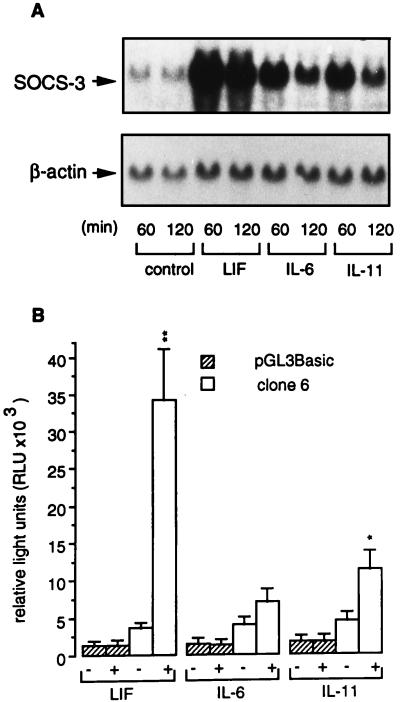
AtT-20 cells were either untreated or stimulated with 0.5 × 10−9 M LIF, IL-6, and IL-11 for 60 or 120 min. Northern blot analysis showed a SOCS-3-specific signal of uniform transcript size of ≈2.8 kb (Fig. 3A). LIF was the most potent inducer of SOCS-3 mRNA expression. Although less potent stimuli of SOCS-3 gene expression, IL-6 and IL-11 showed a similar pattern of SOCS-3 mRNA induction (Fig. 3A). For measurement of SOCS-3 promoter activity, transient transfections of AtT-20 cells were performed either with pGL3Basic alone or clone 6, a construct containing −2757 to +929 5′ genomic region of murine SOCS-3 linked to the luciferase reporter gene in pGL3Basic vector. AtT-20 cells transfected with clone 6 showed a significantly higher basal luciferase activity than cells transfected with pGL3Basic alone (4043 ± 443 vs. 1611 ± 398 relative light units/sec; P < 0.001). Stimulation with 0.5 × 10−9 M LIF, IL-6, or IL-11 caused no further increase of luciferase activity in control AtT-20 cells transfected with pGL3Basic alone. However, AtT-20 cells transfected with clone 6 showed an approximately 10-fold (P < 0.01), 2-fold (not significant), and 3-fold (P < 0.05) stimulation of luciferase activity in comparison to untreated cells, after stimulation with LIF, IL-6, and IL-11, respectively (Fig. 3B).
Figure 3.
Stimulation of murine SOCS-3 mRNA and SOCS-3 reporter gene activity by different cytokines in corticotroph AtT-20 cells. (A) AtT-20 cells were treated with 0.5 × 10−9 M LIF, IL-6, or IL-11 for 60 and 120 min. Northern blot analysis was performed with 25 μg total RNA per lane. (Upper) SOCS-3 mRNA. (Lower) β-actin mRNA. (B) Luciferase activity of pGL3Basic alone and a −2757/+929 murine SOCS-3 promoter-pGL3Basic construct (clone 6) was measured in AtT-20 cells. Cells were treated with 0.5 × 10−9 M LIF, IL-6, or IL-11. Relative light units were calculated from four independently performed experiments. Each experiment was performed with n = 3 wells per group. * indicate in-group significance of untreated (−) vs. treated (+); ∗, P < 0.05; ∗∗, P < 0.01.
Effect of Overexpressed Dominant Negative STAT-3 Mutants or wt SOCS-3 on LIF-Induced SOCS-3 Gene Expression and Promoter Activity.
AtT-20 cells overexpressing wt STAT-3 (AtT-20W) showed a 5.4 ± 0.7-fold increase of SOCS-3 mRNA levels after 45-min stimulation with 0.5 × 10−9 M LIF. In comparison, AtT-20 cells overexpressing the dominant negative mutants STAT-3F (AtT-20F) and STAT-3D (AtT-20D) showed a diminished 3.4 ± 0.4 (P = 0.07) and 2.6 ± 0.1-fold (P < 0.02) induction of SOCS-3 mRNA after stimulation with LIF (Fig. 4 A and B). Similarly, transient transfection experiments with clone 6 showed stimulation of luciferase activity by LIF (6.9 ± 0.5-fold) in AtT-20W cells, whereas only a 5.4 ± 0.5-fold (P = 0.09) and 3.4 ± 0.4-fold (P < 0.01) stimulation was observed in AtT-20F and AtT-20D cells, respectively (Fig. 4C).
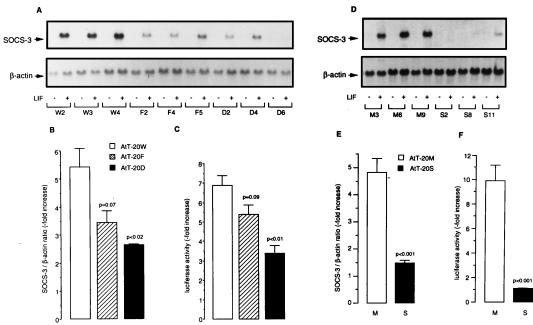
Figure 4.
Effect of overexpressed dominant negative STAT-3 mutants or wt SOCS-3 on LIF-induced SOCS-3 promoter activity and gene expression. Corticotroph AtT-20 cells overexpressing wt STAT-3 (AtT-20W) or dominant negative STAT-3 mutants (AtT-20F and AtT-20D), as well as wt SOCS-3 (AtT-20S) and mock-transfected (AtT-20M), were isolated after stable transfection, as described (15, 28). (A and D) Cells were treated with 0.5 × 10−9 M LIF for 45 min. Northern blot analysis was performed with 15 μg total RNA per lane; shown is a representative experiment. (Upper) SOCS-3 mRNA. (Lower) β-actin mRNA. (B and E) Northern blot signals for SOCS-3 mRNA were analyzed by quantitative densitometry and normalized for β-actin. The relative increase of LIF-induced SOCS-3 mRNA was calculated from three independently performed experiments. Each experiment was performed with three different clones per group. (C and F) Luciferase activity of a −2757/+929 murine SOCS-3 promoter-pGL3Basic construct (clone 6) was measured in different cell clones and treated with 0.5 × 10−9 M LIF. LIF-induced luciferase activity was normalized to the untreated control for each clone. Relative induction of luciferase activity after stimulation was calculated from three independently performed experiments. Each experiment was performed with three independent clones per group.
Mock-transfected AtT-20 cells (AtT-20M) showed an approximately 5-fold increase of SOCS-3 mRNA levels after 45-min stimulation with 0.5 × 10−9 M LIF, whereas AtT-20 cells overexpressing wt SOCS-3 (AtT-20S) showed a significant inhibition of LIF-induced SOCS-3 mRNA expression (Fig. 4 D and E). Similarly, transient transfection experiments with clone 6 revealed luciferase activity to be stimulated by LIF (9.9 ± 1.3-fold) in AtT-20M cells, whereas LIF-induced luciferase activity in AtT-20S cells was abrogated and did not differ substantially from luciferase activity in untreated AtT-20S cells (Fig. 4F).
Functional Analysis of Different SOCS-3 5′ Region-Luciferase Constructs.
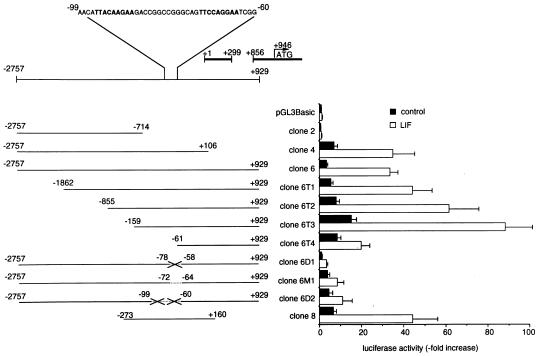
Clone 6 is the −2757 to +929 5′ genomic region of murine SOCS-3 linked to the luciferase reporter in pGL3Basic vector. 3′ Truncations of clone 6 are: clone 4 (nucleotides −2757 to +106) and clone 2 (nucleotides −2757 to −714). 5′ Truncations of clone 6 are: clone 6T1 (nucleotides −1862 to +929), clone 6T2 (nucleotides −855 to +929), clone 6T3 (nucleotides −159 to +929), and clone 6T4 (nucleotides −61 to +929). Analysis of clone 6 sequence with Mat Inspector V2.2 (33) revealed potential STAT binding sites containing the consensus binding sequence TT(N)5AA (34–36) located at nucleotides −95 to −87 and nucleotides −72 to −64, as well as at nucleotides −345 to −337 and −1400 to −1392, respectively. However, only the STAT binding site from nucleotides −72 to −64 showed the more specific sequence TTCCAGGAA (34–36), indicating a potential binding site for STAT-1 and STAT-3. Therefore, in subsequent experiments, we focused on the STAT binding site at nucleotides −72 to −64, constituting part of the tandem STAT binding region pair nucleotides −95 to −87 and nucleotides −72 to −64. By using overlap extension PCR, we deleted the complete tandem STAT binding region from nucleotides −99 to −60 (clone 6D2), or only the 3′ located STAT binding element from nucleotides −78 to −58 (clone 6D1). In clone 6M1, the 3′ located STAT binding element from nucleotides −72 to −64 was not deleted, but mutated to ATCGACGAT, thus destroying the specific binding sequence TTCCAGGAA. Clone 8 is a minimal −273 to +160 5′ genomic region of murine SOCS-3 linked to the luciferase reporter in pGL3Basic vector. Basal and LIF-induced luciferase activity were assayed after transient transfection of corticotroph AtT-20 cells with the different constructs (Fig. 5).
Figure 5.
Luciferase activity of different constructs of the genomic 5′ region of murine SOCS-3. Different constructs were obtained as described in Materials and Methods. After transient transfection, luciferase activity of each construct was measured in untreated (filled bars) and LIF-stimulated (empty bars) AtT-20 cells. Relative luciferase activity was normalized to the activity of pGL3Basic alone in untreated AtT-20 cells, which was defined as 1.0. Crossed lines indicate deletions of STAT binding elements in clones 6D1 and 6D2, in between the named nucleotides. Dotted line indicates a mutation of the wt STAT binding sequence (5′-TTCCAGGAA-3′) with mutant (5′-ATCGACGAT-3′) in clone 6M1.
Relative luciferase activities were calculated in comparison to basal luciferase activity of pGL3Basic alone, which was normalized as 1.0. Basal luciferase activity of clone 2 did not differ from pGL3Basic, and neither clone showed induction of luciferase activity by LIF. However, clones 4 and 6 showed 7- and 4-fold higher basal as well as ≈ 35-fold higher LIF-stimulated luciferase activity than pGL3Basic (P < 0.001). Increasing 5′ truncations of clone 6 up to nucleotide −159 caused a gradual increase of basal and LIF-stimulated luciferase activity, with clone 6T2 (P < 0.05) and clone 6T3 (P < 0.001) showing significantly higher basal and LIF-induced luciferase activities than clone 6, respectively. Clone 6T3 revealed highest basal and LIF-induced luciferase activities, being 15-fold and 88-fold elevated in comparison to basal pGL3Basic (P < 0.001). Further 5′ truncation to nucleotide −61 in clone 6T4 caused a decrease of basal, and more markedly, LIF-inducible promoter activity. Mutated clone 6D1 (P < 0.001) and clone 6M1 (P < 0.01) showed reduced LIF-induced luciferase activity in comparison to wt clone 6. Further deletions in clone 6D2 did not result in altered luciferase activity in comparison to clone 6D1. Clone 8 showed basal and LIF-induced luciferase activity comparable to clone 6 (Fig. 5).
EMSA.
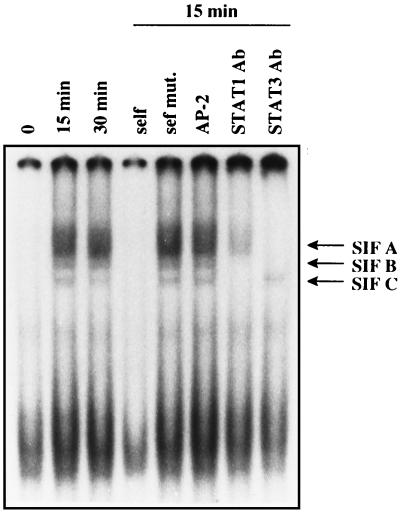
EMSA showed specific binding of nuclear extracts from LIF-induced AtT-20 cells to a ds oligonucleotide probe spanning nucleotides −75 to −55 (STAT oligo), including the STAT-1/STAT-3 binding element from −72 to −64 (Fig. 6). Although nuclear extracts from unstimulated AtT-20 cells did not form specific complexes with the oligoprobe, nuclear extracts from LIF-stimulated AtT-20 cells formed three specific complexes, compatible with STAT-3 homodimer, STAT-1/STAT-3 heterodimer, and STAT-1 homodimer (34–36). The three complexes were not evident during self-competition with 100-fold excess of unlabeled ds STAT oligo, whereas the same ds oligo mutated at positions −72, −69, −67, and −64 (see Materials and Methods), or a nonspecific ds AP-2 oligo had no effect. An apparent nonspecific band was present at the bottom of each lane and was not competed by unlabeled ds STAT oligo. Incubation with a specific antibody directed against STAT-1 abolished the two bands representing STAT1 homodimer and STAT-1/STAT-3 heterodimer. Similarly, incubation with a specific antibody directed against STAT-3 abolished the two bands representing STAT-3 homodimer and STAT-1/STAT-3 heterodimer.
Figure 6.
Gel shift analysis with nuclear cell extracts (20 μg) from AtT-20 cells. A 32P-labeled ds oligonucleotide (STAT oligo), containing the STAT binding consensus sequence (sense 5′-−75CAGTTCCAGGAATCGGGGGGC−55-3′) was used as a probe. Cells were either untreated or treated with 1 nM LIF for 15 min or 30 min. By using nuclear cell extract from 15-min LIF-treated AtT-20 cells, competition of the probe with a 100-fold excess of the unlabeled STAT oligo or the unlabeled STAT oligo mutated at positions −72, −69, −67, and −64, or an unrelated AP-2 oligo could be demonstrated. A presumably unspecific band, unaltered by competition with unlabeled STAT oligo, is shown at the bottom of each lane. Incubation with a STAT-1 antibody or with a STAT-3 antibody abolished DNA binding of specific complexes as evidenced by the absence of specific bands.
DISCUSSION
Cloning of the SOCS-protein family has provided important new insights into regulation of cytokine signaling and the Jak-STAT pathway (4–15). SOCS-3 is up-regulated by several cytokines in a tissue-specific manner (4, 15, 19, 37, 38), and so far has been demonstrated to act as an inhibitor of LIF (7, 9, 15), IL-11 (19), growth hormone (37), and leptin (38) signaling. We recently have shown SOCS-3 expression in the pituitary to be stimulated by LIF, whereas SOCS-3 inhibits LIF-induced POMC gene expression and ACTH secretion in a negative autoregulatory feedback (15). To further understand the regulation of SOCS-3 gene expression in the pituitary, we have cloned and characterized ≈ 3.8 kb of the genomic 5′ region of murine SOCS-3. We demonstrate that murine SOCS-3 promoter activity depends on a STAT-1/STAT-3 binding element, located at −72 to −64.
Northern blot analysis, 5′ RACE, and RNase protection assay revealed a single major transcript of murine SOCS-3 mRNA (≈ 2.8 kb). Previously, a slightly longer transcript size for SOCS-3 was reported, using Northern blot analysis (4). The major transcript form consists of at least two exons: a 5′ untranslated exon 1 (+1 to +299) and exon 2 (beginning at +856) that contains the intronless coding region of SOCS-3 (4). Although PCR-based methods suggested the existence of two longer splicing variants of murine SOCS-3 containing several 5′ untranslated exons and an alternative transcription start site region between −1193 and −1109, these splicing variants were not confirmed by RNase protection assay or Northern blot analysis. Therefore, the single 2.8-kb transcript of murine SOCS-3 mRNA seems to be the major transcript.
To demonstrate promoter activity in the 5′ genomic region of murine SOCS-3, we linked the full-length −2757 to +929 5′ genomic region of murine SOCS-3 to the luciferase reporter gene in pGL3Basic vector (clone 6). Transient transfection experiments with this construct in AtT-20 cells showed significant baseline promoter activity further enhanced by LIF, IL-6, and IL-11. In accordance, SOCS-3 mRNA expression was rapidly induced by these IL-6 receptor family cytokines. All cytokines of the IL-6 receptor family are known to use the common gp130 signaling subunit and to act through Jak2 activation and phosphorylation of STAT-3 and STAT-1, respectively (1, 17). Activation of SOCS-3 promoter activity and gene expression by LIF, IL-6, and IL-11 thus is concordant with our finding of a functionally important STAT-1/STAT-3 binding element in the murine SOCS-3 promoter region.
Luciferase activity of the different 5′ constructs was similar for both basal and LIF-induced luciferase activities of clone 4 and 6, indicating that the region from nucleotides +106 to +929 is likely not involved in promoter activity. The 5′ truncated clone 6T3 showing significantly higher basal and LIF-induced luciferase activity than clone 6 itself demonstrates that the region from −2757 to −159 contains apparent negative regulatory elements, but is not responsible for basal and LIF-induced SOCS-3 promoter activity. Further 5′ truncation to nucleotide −61 in clone 6T4 caused a decrease of basal and especially LIF-inducible promoter activity, which we hypothesized to be caused by loss of the specific STAT 1/STAT 3 binding element (TTCCAGGAA) at nucleotides −72 to −64. We therefore inactivated this STAT binding element, either by deletion (clone 6D1) or mutation of the consensus sequence (clone 6M1). Both clone 6D1 and clone 6M1 exhibited significantly reduced LIF-induced luciferase activity, in comparison to wt clone 6. Extending the deletion to the entire tandem STAT binding region in clone 6D2 showed no significant difference in the magnitude of basal vs. LIF-induced luciferase activity in comparison to clone 6D1. These results indicate that the specific STAT-1/STAT-3 binding element at −72 to −64 (TTCCAGGAA) mediates the LIF-induced rise in luciferase activity, whereas the more 5′ located STAT binding element at −95 to −87 (TTACAAGAA) does not significantly participate in this signal. The tested minimal promoter region from nucleotides −273 to +160 (clone 8) showed basal and LIF-induced luciferase activity comparable to clone 6, thus further demonstrating the functional importance of this region.
Dominant negative STAT-3 mutants, isolated by substitution of a carboxyl-terminal tyrosine phosphorylation site Tyr705 to Phe705 (STAT-3F) or mutation at positions important for DNA binding (STAT-3D), recently have been described (39). Overexpression of these STAT-3 dominant negative mutants in corticotroph AtT-20 cells inhibits LIF-induced POMC gene expression and ACTH secretion (28). We now demonstrate that LIF-induced SOCS-3 promoter activity and gene expression also is decreased in these dominant negative STAT-3 mutant transfectants, thus being at least in part dependent on wt STAT-3. Overexpression of wt SOCS-3 in AtT-20 cells abrogated LIF-induced SOCS-3 promoter activity and gene expression, suggesting a negative autoregulatory feedback of SOCS-3 on its own cytokine-induced gene expression. SOCS-3 as well as SOCS-1 bind to Jak2 (9, 11) and block phosphorylation of gp130 and STAT3 (4, 7, 9, 11, 15). Our report of a functionally important STAT 1/STAT 3 binding site in the SOCS-3 promoter region at −72 to −64 provides a model for SOCS-3 acting as a negative autoregulatory inhibitor on its own gene expression (Fig. 7). Cytokine-induced gene expression of SOCS-1 also has been shown to be inhibited in cells overexpressing dominant negative STAT-3 mutants (6), whereas the promoter region of SOCS-1 has not yet been cloned. Thus, expression of SOCS-1 and SOCS-3 is stimulated by a STAT-3-dependent pathway, whereas they both behave as negative feedback regulators of STAT3 activation. Similarly, gene expression of another member of the SOCS protein family, namely CIS, is up-regulated by a STAT-5 dependent pathway (10, 40), whereas CIS is a negative feedback regulator of STAT-5 activation (8, 10). The human CIS gene promoter contains two functionally important pairs of STAT binding elements (10, 41). Structurally similar, the murine SOCS-3 gene promoter contains a pair of STAT binding elements TT(N5)AA (40–42) separated by only 14 nt. However, comparison of LIF-induced luciferase activity in AtT-20 cells transfected with clones 6, 6D1, 6M1, and 6D2 indicates that only the STAT-1/STAT-3 binding element at −72 to −64 is functionally important. EMSA with a specific ds oligonucleotide probe of this region revealed specific binding of three complexes, corresponding to STAT-3 homodimers, STAT-1/STAT-3 heterodimers, and STAT-1 homodimers (34). Specific inhibition with polyclonal STAT-3 and STAT-1 antibodies verified this observation.
Figure 7.
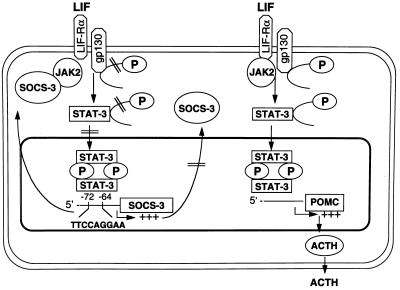
Model of interaction and negative autoregulatory feedback of SOCS-3 protein on POMC and SOCS-3 gene expression. The LIF-induced signaling cascade in the corticotroph cell envolves tyrosine phosphorylation of gp130, STAT-3, and STAT-1 (15, 23, 26, 27). LIF induces gene expression of POMC (28) and SOCS-3 in the corticotroph cell in a STAT-3 dependent manner. Although the region from nucleotides −173 to −160 in the rat POMC promoter is important for synergy of LIF and corticotropin-releasing hormone, it does not involve STAT protein binding (25); the putative STAT binding element in the POMC promoter has not yet been characterized. The murine SOCS-3 promoter has a functionally critical STAT-1/STAT-3 binding region at −72 to −64. SOCS-3 inhibits Jak2 activity by binding to its JH1 domain (9) and thus inhibits LIF-induced tyrosine phosphorylation of gp130 and STAT-3 in the corticotroph cell (15). By inhibiting Jak-STAT signaling, SOCS-3 negatively regulates LIF-induced POMC gene expression and ACTH secretion and also exerts a negative autoregulatory feedback on its own gene expression. This negative autoregulatory feedback of SOCS-3 on its own gene expression limits the accumulation of SOCS-3 protein in the corticotroph cell.
In conclusion, we have cloned and characterized the genomic 5′ region of murine SOCS-3. LIF-induced SOCS-3 gene expression is partially STAT-3 dependent, and the murine SOCS-3 promoter has a functionally important STAT-1/STAT-3 binding element at −72 to −64. As SOCS-3 is a potent inhibitor of the LIF-induced Jak-STAT signaling cascade in the corticotroph cell, these findings indicate a negative autoregulatory feedback of SOCS-3 on its own gene expression, thus limiting accumulation of SOCS-3 in the corticotroph cell (Fig. 7). The STAT-3 dependence and the negative autoregulatory feedback of SOCS-3 on its LIF-induced gene expression might explain the rapid and short-lived SOCS-3 mRNA peak after LIF stimulation (15). As LIF-induced POMC gene transcription and ACTH secretion are STAT-3 dependent (28) and SOCS-3 potently inhibits LIF-induced STAT-3 phosphorylation and subsequent POMC transcription and ACTH secretion (15), the suggested negative feedback of SOCS-3 on its own gene expression provides a mechanism enabling cortocotroph plasticity with fast “on” and “off” ACTH secretion in response to LIF and other neuro-immuno-endocrine stimuli.
Acknowledgments
We acknowledge the helpful technical advice of Drs. Lin Pei and Toni Prezant. This study was supported by a scholarship of the Deutsche Forschungsgemeinschaft (Au 139/1–1) and National Institutes of Health Grant DK 50238.
ABBREVIATIONS
- LIF
leukemia inhibitory factor
- POMC
proopiomelanocortin
- ACTH
adrenocorticotropic hormone
- Jak
Janus kinase
- STAT
signal transducers and activators of transcription
- RACE
rapid amplification of cDNA ends
- SOCS
suppressors of cytokine signaling
- CIS
cytokine-inducible SH2 containing protein
- wt
wild type
- EMSA
electromobility shift assay
- ds
double-stranded
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF117732).
References
- 1.Hirano T, Nakajima K, Hibi M. Cytokine Growth Factor Rev. 1997;8:241–252. doi: 10.1016/s1359-6101(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 2.Carter-Su C, Smit L S. Recent Prog Horm Res. 1998;53:61–82. [PubMed] [Google Scholar]
- 3.Haque S J, Wiliams B R G. Semin Oncol. 1998;25, Suppl. 1:14–22. [PubMed] [Google Scholar]
- 4.Starr R, Willson T A, Viney E M, Murray L J, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, Hilton D J. Nature (London) 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 5.Hilton D J, Richardson R T, Alexander W S, Viney E M, Willson T A, Sprigg N S, Starr R, Nicholson S E, Metcalf D, Nicola N A. Proc Natl Acad Sci USA. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et al. Nature (London) 1997;387:924–928. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 7.Minamoto S, Ikegame K, Ueno K, Narazaki M, Naka T, Yamamoto H, Matsumoto T, Saito H, Hosoe S, Kishimoto T. Biochem Biophys Res Commun. 1997;237:79–83. doi: 10.1006/bbrc.1997.7080. [DOI] [PubMed] [Google Scholar]
- 8.Yoshimura A, Ohkubo T, Kiguchi T, Jenkins N A, Gilbert D J, Copeland N G, Hara T, Miyajima A. EMBO J. 1995;14:2816–2826. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuhara M, Sakamoto H, Matsumoto A, Suzuki R, Yasukawa H, Mitsui K, Wakioka T, Tanimura S, Sasaki A, Misawa H, et al. Biochem Biophys Res Commun. 1997;239:439–446. doi: 10.1006/bbrc.1997.7484. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- 11.Endo T A, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, et al. Nature (London) 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto H, Yasukawa H, Masuhara M, Tanimura S, Sasaki A, Yuge K, Ohtsubo M, Ohtsuka A, Fujita T, Ohta T, et al. Blood. 1998;92:1668–1676. [PubMed] [Google Scholar]
- 13.Aman M J, Leonard W J. Curr Biol. 1997;7:R784–R788. doi: 10.1016/s0960-9822(06)00405-2. [DOI] [PubMed] [Google Scholar]
- 14.Starr R, Hilton D J. Int J Biochem Cell Biol. 1998;30:1081–1085. doi: 10.1016/s1357-2725(98)00067-3. [DOI] [PubMed] [Google Scholar]
- 15.Auernhammer C J, Chesnokova V, Bousquet C, Melmed S. Mol Endocrinol. 1998;12:954–961. doi: 10.1210/mend.12.7.0140. [DOI] [PubMed] [Google Scholar]
- 16.Cheng J G, Pennica D, Patterson P H. J Neurochem. 1997;69:2278–2284. doi: 10.1046/j.1471-4159.1997.69062278.x. [DOI] [PubMed] [Google Scholar]
- 17.Melmed S. Trends Endocrinol Metab. 1997;8:391–397. doi: 10.1016/s1043-2760(97)00156-2. [DOI] [PubMed] [Google Scholar]
- 18.Bilezikjian L M, Turnbull A V, Corrigan A Z, Blount A L, Rivier C L, Vale W W. Endocrinology. 1998;139:3361–3364. doi: 10.1210/endo.139.7.6190. [DOI] [PubMed] [Google Scholar]
- 19.Auernhammer C J, Melmed S. Endocrinology. 1999;140:1559–1566. doi: 10.1210/endo.140.4.6636. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Ren S G, Melmed S. Endocrinology. 1996;137:2947–2953. doi: 10.1210/endo.137.7.8770918. [DOI] [PubMed] [Google Scholar]
- 21.Auernhammer C J, Chesnokova V, Melmed S. Endocrinology. 1998;139:2201–2208. doi: 10.1210/endo.139.5.6017. [DOI] [PubMed] [Google Scholar]
- 22.Chesnokova V, Auernhammer C J, Melmed S. Endocrinology. 1998;139:2201–2208. doi: 10.1210/endo.139.5.6017. [DOI] [PubMed] [Google Scholar]
- 23.Shimon I, Yan X, Ray D W, Melmed S. J Clin Invest. 1997;100:357–363. doi: 10.1172/JCI119541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akita S, Webster J, Ren S G, Takino H, Said J, Zand O, Melmed S. J Clin Invest. 1995;95:1288–1298. doi: 10.1172/JCI117779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bousquet C, Ray D W, Melmed S. J Biol Chem. 1997;272:10551–10557. doi: 10.1074/jbc.272.16.10551. [DOI] [PubMed] [Google Scholar]
- 26.Ray D W, Ren S G, Melmed S. J Clin Invest. 1996;97:1852–1859. doi: 10.1172/JCI118615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray D W, Ren S G, Melmed S. Ann NY Acad Sci USA. 1998;840:162–173. doi: 10.1111/j.1749-6632.1998.tb09560.x. [DOI] [PubMed] [Google Scholar]
- 28.Bousquet C, Melmed S. J Biol Chem. 1998;274:10723–10730. doi: 10.1074/jbc.274.16.10723. [DOI] [PubMed] [Google Scholar]
- 29.Chenchik A, Diachenko L, Moqadam F, Tarabykin V, Lukyanov S, Siebert P D. Biotechnology. 1996;3:526–534. doi: 10.2144/96213pf02. [DOI] [PubMed] [Google Scholar]
- 30.Siebert P D, Chenchik A, Kellogg D E, Lukyanov K A, Lukyanov S A. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henikoff S. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 32.Aiyar A, Xiang Y, Leis J. Methods Mol Biol. 1996;57:177–191. doi: 10.1385/0-89603-332-5:177. [DOI] [PubMed] [Google Scholar]
- 33.Quandt K, Frech K, Karas H, Wingender E, Werner T. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horvath C M, Wen Z, Darnell J E., Jr Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 35.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 36.Becker S, Groner B, Mueller C W. Nature (London) 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 37.Adams T E, Hansen J A, Starr R, Nicola N A, Hilton D J, Billestrup N. J Biol Chem. 1998;273:1285–1287. doi: 10.1074/jbc.273.3.1285. [DOI] [PubMed] [Google Scholar]
- 38.Bjorbaek C, Elmquist J K, Frantz J D, Shoelson S E, Flier J S. Mol Cell. 1998;1:619–625. doi: 10.1016/s1097-2765(00)80062-3. [DOI] [PubMed] [Google Scholar]
- 39.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle J N. Mol Cell Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verdier F, Rabionet R, Gouilleux F, Beisenherz-Huss C, Varlet P, Muller O, Mayeux P, Lacombe C, Gisselbrecht S, Chretien S. Mol Cell Biol. 1998;18:5852–5860. doi: 10.1128/mcb.18.10.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]