Abstract
Carbohydrate-deficient glycoprotein syndrome (CDGS) represents a class of genetic diseases characterized by abnormal N-linked glycosylation. CDGS patients show a large number of glycoprotein abnormalities resulting in dysmorphy, encephalopathy, and other organ disorders. The majority of CDGSs described to date are related to an impaired biosynthesis of dolichyl pyrophosphate-linked Glc3Man9GlcNAc2 in the endoplasmic reticulum. Recently, we identified in four related patients a novel type of CDGS characterized by an accumulation of dolichyl pyrophosphate-linked Man9GlcNAc2. Elaborating on the analogy of this finding with the phenotype of alg5 and alg6 Saccharomyces cerevisiae strains, we have cloned and analyzed the human orthologs to the ALG5 dolichyl phosphate glucosyltransferase and ALG6 dolichyl pyrophosphate Man9GlcNAc2 α1,3-glucosyltransferase in four novel CDGS patients. Although ALG5 was not altered in the patients, a C→T transition was detected in ALG6 cDNA of all four CDGS patients. The mutation cosegregated with the disease in a Mendelian recessive manner. Expression of the human ALG5 and ALG6 cDNA could partially complement the respective S. cerevisiae alg5 and alg6 deficiency. By contrast, the mutant ALG6 cDNA of CDGS patients failed to revert the hypoglycosylation observed in alg6 yeasts, thereby proving a functional relationship between the alanine to valine substitution introduced by the C→T transition and the CDGS phenotype. The mutation in the ALG6 α1,3-glucosyltransferase gene defines an additional type of CDGS, which we propose to refer to as CDGS type-Ic.
Within the last 20 years, various alterations of asparagine-linked glycosylation have been identified and classified as subtypes of carbohydrate-deficient glycoprotein syndromes (CDGS) (1). Besides characteristic clinical features, such as dysmorphism, psychomotor retardation, and stroke-like episodes, CDGS patients may present on an individual basis more specific symptoms, such as coagulation disorders or cerebellar ataxia (2). The constellation and gravity of the symptoms vary among CDGS patients, which likely reflect different enzymatic defects along the glycosylation pathways. Mutations in the phosphomannomutase-2 gene (3), which affect the availability of mannose for oligosaccharide synthesis, are the most frequent causes of CDGS. A second type of CDGS characterized by truncated complex type N-linked glycans has been found to originate from a mutation in the N-acetylglucosaminyltransferase-II gene (4). Although no specific therapy exists to correct the above-mentioned defects, it has been found that CDGS type-Ib patients bearing an inactive phosphomannose isomerase enzyme can be treated effectively by oral mannose supplementation (5).
A further type of CDGS has recently been documented that is characterized by an accumulation of dolichyl pyrophosphate-linked Man9GlcNAc2 (Man9GlcNAc2-PP-Dol) within the cells of affected patients (6, 7). This feature was directly related to the hypoglycosylation of secreted proteins detected in the corresponding CDGS patients. An analogous accumulation of Man9GlcNAc2-PP-Dol has been described in alg5 and alg6 Saccharomyces cerevisiae mutant strains (8–10). The ALG5 and ALG6 genes, which respectively code for the dolichyl phosphate glucosyltransferase and the Man9GlcNAc2-PP-Dol α1,3-glucosyltransferase enzymes, participate in the glucosylation of the oligomannose core. The addition of three glucose residues to the oligomannose core is critical for optimal substrate recognition by the oligosaccharyltransferase complex and therefore is necessary to ensure the efficient transfer of the oligomannose core to nascent glycoproteins (11, 12). Because the biosynthetic pathway of N-linked core glycosylation is highly conserved in eukaryotic cells, we have taken advantage of the knowledge gathered in yeast to determine the molecular basis of the glucosylation defect identified in CDGS patients.
MATERIALS AND METHODS
Cloning of Human ALG5 and ALG6 cDNAs.
Expressed sequence tag (EST) fragments similar to the S. cerevisiae ALG5 sequence were retrieved from the EST division of GenBank by using the tblastx algorithm (13). The human ALG5 cDNA was isolated from a T-cell cDNA library (14) by using a 602-bp fragment derived from the assembled AA425251 and AA478430 ESTs as a probe. The probing fragment was generated by PCR by using 50 ng of human T cell cDNA as template with the primers 5′-CACCTACCAAACAACTTTCTGTCG-3′ and 5′-TACATCAAATGCCCATCGTTCAAC-3′ for 35 cycles at 94°C for 45 s, 55°C for 30 s, and 72°C for 60 s. A fragment of the human ALG6 cDNA was amplified from T-cell cDNA by using the degenerate oligonucleotides 5′-GCKCARAGACATTGGATGGAAAT-3′ and 5′-AGGATAGTYTTYTCATGTACKTG-3′ designed according to the yeast ALG6 gene sequence. The PCR conditions were three cycles at 94°C for 45 s, 40°C for 30 s, and 72°C for 60 s, followed by 5 cycles at 94°C for 45 s, 45°C for 30 s, and 72°C for 60 s and 30 cycles at 94°C for 45 s, 50°C for 30 s, and 72°C for 60 s. The resulting 908-bp fragment was phosphorylated and subcloned into the SmaI site of pBluescript II SK(+) (Stratagene). The 908-bp ALG6 fragment was used as a hybridization probe to isolate full-length cDNA from a human T-cell cDNA library.
Northern Blotting.
The human ALG5 and ALG6 mRNAs were detected by Northern blot analysis by using commercially available multiple tissues poly(A)+ RNA blots (CLONTECH). The ALG5 1,018-bp EcoRI-BbsI and ALG6 1,432-bp ScaI-BamHI fragments were prepared from the human cDNAs and were labeled with [α-32P]CTP (Hartmann Analytics, Braunschweig, Germany) by random priming. Blots were hybridized to the probes for 16 h at 42°C, were washed in 0.1 × standard saline citrate (SSC) and 0.1% SDS up to 60°C, and were exposed for 5 days (ALG6) and 2 days (ALG5) between intensifying screens at −70°C.
Cell Culture, DNA, RNA Extraction.
Human primary fibroblasts from CDGS patients and healthy controls were cultivated in DMEM F12 high glucose medium with 10% fetal calf serum (Life Technologies, Paisley, Scotland). Cells (2 × 107) were used for either genomic DNA or RNA isolation. Genomic DNA was extracted following standard protocols (15), and total RNA was isolated by the procedure of Chomczynski and Sacchi (16).
Reverse Transcription (RT)–PCR Amplification.
RT was performed with 100 units of Moloney murine leukemia virus reverse transcriptase (New England Biolabs) using 8 μg of total RNA and a (dT)15-oligonucleotide as primer in a volume of 100 μl for 2 h at 37°C. ALG5 and ALG6 cDNA were amplified by PCR with the respective primer pairs 5′-GAGGGCTGCCACGGCATGGAG-3′, 5′-GAAGCATAAGAACACAACTGA-AGACTG-3′and 5′-TTAAAAGTACTCTGGCACTGGTG-3′, and 5′-CTTTCAGCACTGTTCACATTTTC-3′ by using 5 μl of RT product as template. The cycling conditions were 35 cycles at 94°C for 45 s, 53°C for 30 s, and 72°C for 1.5 min (ALG5) and 35 cycles at 94°C for 45 s, 58°C for 30 s, and 72°C for 2 min (ALG6). The corresponding ALG5 and ALG6 fragments were purified by GeneClean (Bio 101) and were directly sequenced (14).
ALG6 Genotyping.
PCR amplification with the ALG6 intronic primer 5′-GGTTTTAAATTTAAGTTGTCTGAGCTTCCAGGG-3′ and the ALG6 exonic primer 5′-AAGAGAATGGATTTTTCATGTACTTGGAAAGAA-3′ was performed on 100 ng of genomic DNA for 35 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 45 s. The allelic discrimination was performed by digesting the PCR products with HhaI for 1 h at 37°C followed by electrophoretic separation in 3% NuSieve 3:1 agarose (FMC) gels.
Yeast Strains and Media.
S. cerevisiae strains YG 91: Matα ade2–101 his3Δ200 ura3–52 Δalg5∷HIS3, YG 355: Matα ade2–101 his3Δ200 ura3–52 Δalg5∷HIS3 wbp1–2, YG592: Matα ade2–101 his3Δ200 ura3–52 lys2–801 Δalg6∷HIS3, YG36: Matα ade2–101 his3Δ200 ura3–52 alg6 wbp1–2 were used in this study. Standard yeast media and genetic techniques were applied (17).
Complementation of S. cerevisiae Strains.
The 1,164-bp human ALG5 cDNA ScaI-PstI fragment and the 1,781-bp human ALG6 cDNA ScaI-ScaI fragments were blunted and subcloned into the YEp352-GAL1–10 (18, 19) vector opened with XbaI-HindIII and blunted. Yeast strains were grown overnight in 5 ml of liquid yeast extract/peptone/dextrose medium. Series of 1:10 dilutions starting with 2 × 106 cells were made in yeast extract/peptone/dextrose medium, and 5 μl of each dilution were spotted on YPgalactose plates and were incubated at 25°C. A reduced growth rate is reflected by a reduced cell density at a given dilution. Western blot analysis was performed as described (20) using anti-carboxypeptidase Y (CPY) specific antibodies.
RESULTS
The phenotypic accumulation of Man9GlcNAc2-PP-Dol oligosaccharide has been detected in four related CDGS patients, who presented signs of psychomotor retardation and epilepsy (S. Grünewald, personal communication). The isoelectric focusing of serum transferrin showed increased levels of the disialylated form (6), which is typical of CDGS. The analogy of the Man9GlcNAc2-PP-Dol accumulation to the alg5 and alg6 phenotypes of S. cerevisiae prompted us to investigate the integrity of these genes in the CDGS patients.
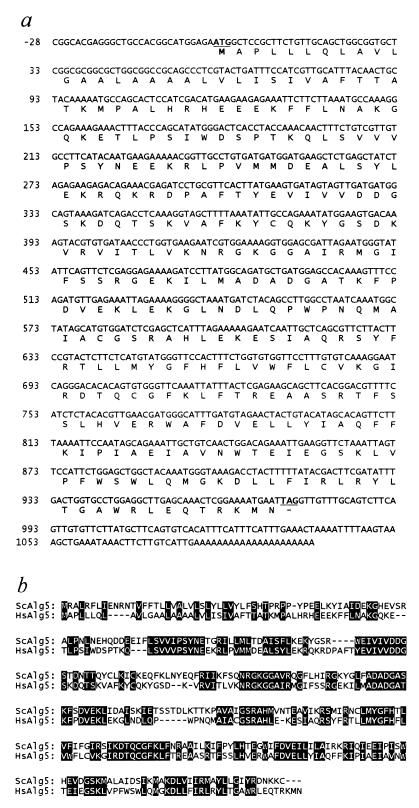
We have cloned the human orthologs of the ALG5 and ALG6 genes by blast (13) searching of EST databases and low-stringency PCR amplification. Several human EST fragments similar to the yeast ALG5 were retrieved from GenBank, and a putative ORF of 644 bp was constructed by assembling the ESTs AA425251 and AA478430. Using this fragment as a probe, we have isolated a full-length ALG5 cDNA from a human T-lymphocyte cDNA library. The 1,126-bp human ALG5 cDNA included an ORF of 729 bp encoding a polypeptide of 242 amino acids (Fig. 1a). The human ALG5 gene showed 37% identity and 58% similarity to the S. cerevisiae ortholog (Fig. 1b).
Figure 1.
Sequence of the human ALG5 dolichyl phosphate glucosyltransferase cDNA. (a) Primary structure and predicted amino acid sequence of the human ALG5 cDNA. The ATG codon and stop codon are underlined. (b) Protein alignment of the S. cerevisiae (sc)Alg5 and the Homo sapiens (hs)Alg5 protein sequences. Conserved residues are shaded.
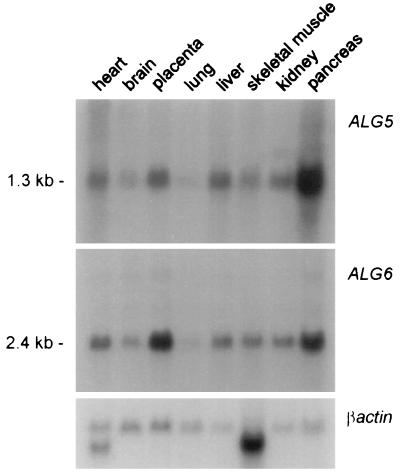
An EST search using the yeast ALG6 gene as query failed because only EST fragments similar to the closely related yeast ALG8 gene (21) emerged at the time the search was performed. As an alternate approach, we designed a series of degenerate oligonucleotides within regions of the S. cerevisiae ALG6 sequence that were dissimilar to the yeast ALG8 sequence. A PCR performed on human T-cell cDNA using two degenerate oligonucleotides as primers yielded a 908-bp fragment that was similar to the yeast ALG6 as determined by sequencing. This 908-bp fragment permitted the isolation of a complete human cDNA, which contained a 1,524-bp-long ORF coding for a protein of 507 amino acids (Fig. 2a). Though shorter by 37 amino acids, the human Alg6 protein shared several conserved regions with its S. cerevisiae ortholog (Fig. 2b). The overall identity and similarity between the two orthologs was 32% and 51%, respectively. The human Alg6 contains several hydrophobic regions, compatible with the transmembrane topology attributed to the yeast Alg6 (10). The human ALG5 and ALG6 genes were found to be expressed in all tissues examined by Northern blotting (Fig. 3). The intensity of the hybridization signal varied among tissues, showing the same pattern for both ALG5 and ALG6, indicating a possible coordinated expression.
Figure 2.
Sequence of the human ALG6 dolichyl pyrophosphate Man9GlcNAc2 α1,3-glucosyltransferase cDNA. (a) Nucleotide and deduced amino acid sequence of the human ALG6 cDNA. The ATG codon and stop codon are underlined. The nucleotide position 998 is shaded. (b) Alignment of the S. cerevisiae (sc)Alg6 and the H. sapiens (hs)Alg6 protein sequences. Conserved residues are shaded. The amino acid substitution A333V found in CDGS is indicated by a subscript V.
Figure 3.
Expression pattern of the ALG5 and ALG6 genes in adult human tissues as determined by Northern blot analysis. Each lane represents ≈2 μg of poly(A)+ RNA. The bottom panel shows the βactin hybridization signal as a loading control. At the left, the size of the transcripts is indicated in kilobases.
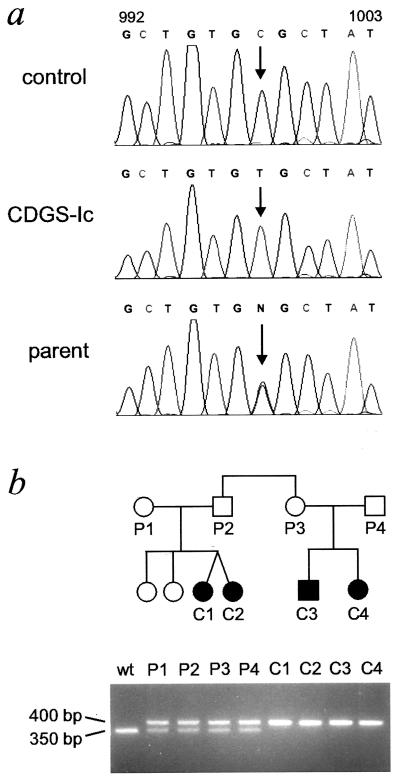
Total RNA was isolated from fibroblasts obtained from the four CDGS patients harboring the Man9GlcNAc2-PP-Dol accumulation phenotype. The integrity of the ALG5 and ALG6 cDNA sequence in these patients was investigated by RT-PCR performed on total RNA. The intensity of the RT-PCR signals obtained indicated similar expression levels for the ALG5 and ALG6 cDNAs in healthy control and in the CDGS fibroblasts. Direct sequencing of the PCR products obtained from the four patients revealed the absence of any mutation in the ALG5 sequence as compared with the sequence obtained from cDNA derived from unaffected individuals (data not shown). By contrast, a C→T transition was detected at the nucleotide position 998 of the ALG6 cDNA in all four CDGS patients. This C→T transition indicated an amino acid substitution from alanine to valine at position 333 of the Alg6 protein. The 998C/T mutation was confirmed at the genomic level by sequencing the ALG6 region surrounding the mutation as effected by PCR amplification using intron- and exon-specific primers (Fig. 4a). A similar analysis performed on genomic DNA isolated from the parents of CDGS-Ic patients showed the heterozygosity of the parents for the mutant ALG6 allele (Fig. 4a). The 998C/T transition introduced a restriction-fragment-length polymorphism in the ALG6 gene sequence attributable to the loss of an HhaI cleavage site, thereby permitting a simple genotyping of the CDGS patients and their relatives (Fig. 4b). By using this test, 134 ALG6 alleles from geographically matched control subjects have been investigated. This study revealed a strict segregation of the 998C/T mutation with the CDGS-Ic phenotype.
Figure 4.
Mutation at the ALG6 allele in CDGS patients. (a) Electropherograms of the genomic ALG6 sequence obtained from a control subject, a CDGS type-Ic patient (CDGS-Ic), and a parent of a CDGS type-Ic patient. The region surrounding the 998C/T base transition is shown. (b) Pedigree of CDGS type-Ic family. CDGS patients are indicated in black. The bottom panel shows the bands obtained after HhaI digestion of genomic PCR ALG6 fragments effected on parent (P1–P4) and patient (C1–C4) DNA. The mutant allele remains uncut and runs at 400 bp whereas the wild-type allele is cleaved by HhaI and yields two fragments of 350 bp and 50 bp, but only the larger fragment (350 bp) is visible.
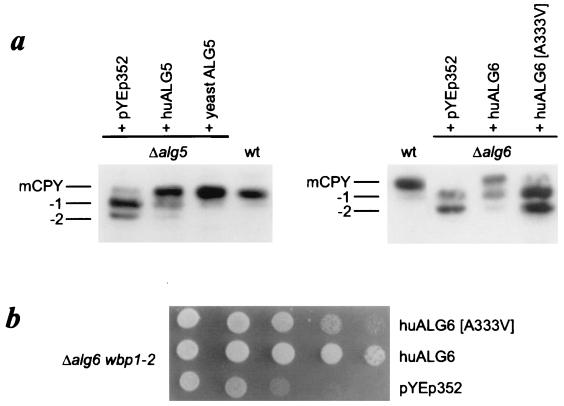
In yeasts, the alg5 and alg6 mutations lead to the hypoglycosylation of secreted proteins (9, 10). To investigate the functional similarities of the human and yeast orthologs, the human ALG5 and both wild-type and mutant ALG6 cDNA were subcloned into the yeast YEp352 expression vector (18). Expression was controlled by the inducible/repressible GAL1–10 promoter (19). After transformation into the alg5 and alg6 yeast strains, we ascertained the processing of the CPY protein. This vacuolar protein carries four N-linked oligosaccharides. A deficiency in glycosylation of the lipid-linked oligosaccharide results in the incomplete glycosylation of glycoprotein because of the decreased affinity of the oligosaccharyltransferase toward the incomplete substrate. The reduced glycosylation caused by the alg5 or alg6 deletion in the yeast strains was visualized by the appearance of CPY glycoforms lacking one or two oligosaccharide chains. Expression of the human ALG5 and ALG6 cDNA partially complemented the respective yeast mutations, as shown by the improved glycosylation of CPY (Fig. 5a). By contrast, the human ALG6 cDNA bearing the 998C/T mutation failed to correct the hypoglycosylation phenotype, demonstrating the adverse effect of the A333V substitution onto Alg6 function (Fig. 5a). The combination of the alg6 mutation with the wbp1–2 allele, encoding a mutant form of the oligosaccharyltransferase subunit Wbp1, results in a synthetic growth defect caused by the accumulation of incomplete Man9GlcNAc2-PP-Dol and reduced oligosaccharyltransferase activity (10, 21). While the human ALG6 cDNA was able to correct the growth defect of alg6wbp1–2 yeasts, the ALG6[A333V] cDNA was only capable of partially restoring growth of the yeast alg6wbp1–2 strain (Fig. 5b).
Figure 5.
Complementation of S. cerevisiae mutant strains by human ALG5 and ALG6. (a) Western blot analysis of carboxypeptidase Y (mCPY) showing the glycoforms lacking one (−1) or two (−2) oligosaccharide chains. In the left panel, alg5 yeasts transformed with the vector (pYEp352), with pYEp352-huALG5 (huALG5), and with pYEp352-yeastALG5 (yeastALG5) are indicated. The right panel shows CPY glycoforms in alg6 yeasts transformed with pYEp352 alone, with pYEp352-huALG6, and with pYEp352-huALG6[A333V]. (b) Growth of alg6wbp1–2 yeasts transformed with pYEp352, pYEp352-huALG6, and pYEp352-huALG6[A333V] constructs. Tenfold dilutions starting from the left with the same cell number were plated on YPgalactose plates and were incubated at 25°C for 5 days.
DISCUSSION
CDGS type-Ia and type-Ib are caused by a shortage of mannose incorporation into N-glycans consecutive to mutations in the phosphomannomutase-2 and phosphomannose isomerase genes, respectively. The decreased availability in mannose impairs the biosynthesis of the lipid-linked oligomannose core, which results in a reduced N-linked glycosylation of proteins. In the present study, we show that the altered glucosylation of the oligomannose core caused by a mutation in the ALG6 α1,3-glucosyltransferase gene also leads to hypoglycosylation of proteins, thereby defining an additional type of CDGS. We propose to refer to the form of CDGS caused by ALG6 mutation as type-Ic because it represents a defect in glycosylation localized in the endoplasmic reticulum, proximally from the transfer of the oligomannose core to nascent proteins. This nomenclature simplifies the classification of CDGS forms between pre-Golgi (type-I) and Golgi-localized defects (type-II). This system also prevents any confusion with CDGS type-III and type-IV, which represent clinically defined forms that are as yet uncharacterized at the molecular level (22, 23). Körner et al. recently have described a CDGS type-V that is related to a deficiency in Alg6 α1,3-glucosyltransferase activity (7). The similarity of this finding with the CDGS type-Ic reported here suggests that both types may represent the same entity. However, the genetic cause of CDGS type-V remains unclear because the integrity of the ALG6 gene was not determined in the corresponding CDGS patient (7). In addition, several clinical features attributed to the single CDGS-V patient, such as recurrent infections, ataxia, and cerebellar atrophy, were not observed in the four CDGS-Ic patients.
The effect of the A333V substitution on the properties of the Alg6 protein is yet unclear. The replacement of an alanine by a more bulky amino acid like valine has been shown in various cases to be linked to profound alterations of protein functions, such as those observed for superoxide dismutase-1 in familial amyotrophic lateral sclerosis (24) and for the follicle-stimulating hormone receptor in hereditary hypergonadotropic ovarian failure (25). The proximity of the A333V mutation to a large domain of the Alg6 protein that is conserved between yeast and man suggests a possible alteration of the catalytic properties of Alg6. However, the A333V mutation does not inactivate the glucosyltransferase as deduced from the partial growth restoration observed in the alg6 yeast transfected with the mutant human ALG6[A333V] cDNA (Fig. 5b).
The 998C/T missense mutation in the ALG6 gene has only been identified in the four CDGS type-Ic patients but never in control healthy subjects, indicating that this mutation does not reflect a polymorphism unrelated to this entity. Accordingly, the CDGS phenotype correlated with the genotype for the mutant ALG6 allele because only patients were homozygous whereas their parents were heterozygous. Therefore, we conclude that the 998C/T mutation in ALG6 is at the origin of the accumulation of Man9GlcNAc2-PP-Dol in the endoplasmic reticulum, which leads to hypoglycosylation of proteins and, hence, to CDGS.
With the exception of the N-acetylglucosaminyltransferase-II deficiency, all types of CDGS characterized to date have been attributed to mutations in the biosynthesis pathway of the dolichol-bound oligomannose core. As demonstrated in the present study, conservation of the early N-linked glycosylation pathway among eukaryotes enables the application of yeast glycosylation mutants as tools to elucidate the molecular basis of CDGS.
Acknowledgments
We thank the patients and their families for contributing to this study and J. Jaeken for introducing us to these CDGS cases. We express our gratitude to G. Matthijs and H.W. van Kerkwijk for providing us with biological specimens from the CDGS families. Our thanks also go to Drs. J. de Rijk-van Andel, J. B. C. de Klerk, and H. Stroink, who diagnosed CDGS in these patients. This work was supported by the Stiftung für wissenschaftliche Forschung an der Universität Zürich and by the Swiss National Science Foundation (Grant 3100-46836.96 to E.G.B. and T.H. and Grant 3100-040350.94 to M.A.).
ABBREVIATIONS
- CDGS
carbohydrate-deficient glycoprotein syndrome
- Man9GlcNAc2-PP-Dol
dolichyl pyrophosphate-linked Man9GlcNAc2
- EST
expressed sequence tag
- CPY
carboxypeptidase Y
- RT
reverse transcription
Footnotes
References
- 1.Jaeken J, Carchon H, Stibler H. Glycobiology. 1993;3:423–428. doi: 10.1093/glycob/3.5.423. [DOI] [PubMed] [Google Scholar]
- 2.Krasnewich D, Gahl W A. Adv Pediatr. 1997;44:109–140. [PubMed] [Google Scholar]
- 3.Matthijs G, Schollen E, Pardon E, Veiga-Da-Cunha M, Jaeken J, Cassiman J J, Van Schaftingen E. Nat Genet. 1997;16:88–92. doi: 10.1038/ng0597-88. [DOI] [PubMed] [Google Scholar]
- 4.Tan J, Dunn J, Jaeken J, Schachter H. Am J Hum Genet. 1996;59:810–817. [PMC free article] [PubMed] [Google Scholar]
- 5.Niehues R, Hasilik M, Alton G, Korner C, Schiebe-Sukumar M, Koch H G, Zimmer K P, Wu R, Harms E, Reiter K, et al. J Clin Invest. 1998;101:1414–1420. doi: 10.1172/JCI2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burda P, Borsig L, de Rijk-van Andel J, Wevers R, Jaeken J, Carchon H, Berger E G, Aebi M. J Clin Invest. 1998;102:647–652. doi: 10.1172/JCI2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Körner C, Knauer R, Holzbach U, Hanefeld F, Lehle L, von Figura K. Proc Natl Acad Sci USA. 1998;95:13200–13205. doi: 10.1073/pnas.95.22.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Runge K W, Huffaker T C, Robbins P W. J Biol Chem. 1984;259:412–417. [PubMed] [Google Scholar]
- 9.te Heesen S, Lehle L, Weissmann A, Aebi M. Eur J Biochem. 1994;224:71–79. doi: 10.1111/j.1432-1033.1994.tb19996.x. [DOI] [PubMed] [Google Scholar]
- 10.Reiss G, te Heesen S, Zimmerman J, Robbins P W, Aebi M. Glycobiology. 1996;6:493–498. doi: 10.1093/glycob/6.5.493. [DOI] [PubMed] [Google Scholar]
- 11.Murphy L A, Spiro R G. J Biol Chem. 1981;256:7487–7494. [PubMed] [Google Scholar]
- 12.Burda P, Aebi M. Glycobiology. 1998;8:455–462. doi: 10.1093/glycob/8.5.455. [DOI] [PubMed] [Google Scholar]
- 13.Altschul S F, Boguski M S, Gish W, Wootton J C. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 14.Hennet T, Dinter A, Kuhnert P, Mattu T S, Rudd P M, Berger E G. J Biol Chem. 1998;273:58–65. doi: 10.1074/jbc.273.1.58. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 9.16–9.19. [Google Scholar]
- 16.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 17.Berlin V, Brill J A, Trueheart J, Boeke J D, Fink G R. Methods Enzymol. 1991;194:774–792. doi: 10.1016/0076-6879(91)94058-k. [DOI] [PubMed] [Google Scholar]
- 18.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 19.Fagan M C, Scott J F. Gene. 1985;40:217–229. doi: 10.1016/0378-1119(85)90044-7. [DOI] [PubMed] [Google Scholar]
- 20.Burda P, te Heesen S, Brachat A, Wach A, Dusterhoft A, Aebi M. Proc Natl Acad Sci USA. 1996;93:7160–7165. doi: 10.1073/pnas.93.14.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stagljar I, te Heesen S, Aebi M. Proc Natl Acad Sci USA. 1994;91:5977–5981. doi: 10.1073/pnas.91.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stibler H, Westerberg B, Hanefeld F, Hagberg B. Neuropediatrics. 1993;24:51–52. doi: 10.1055/s-2008-1071513. [DOI] [PubMed] [Google Scholar]
- 23.Stibler H, Stephani U, Kutsch U. Neuropediatrics. 1995;26:235–237. doi: 10.1055/s-2007-979762. [DOI] [PubMed] [Google Scholar]
- 24.Rosen D R, Bowling A C, Patterson D, Usdin T B, Sapp P, Mezey E, McKenna-Yasek D, O’Regan J, Rahmani Z, Ferrante R J, et al. Hum Mol Genet. 1994;3:981–987. doi: 10.1093/hmg/3.6.981. [DOI] [PubMed] [Google Scholar]
- 25.Aittomaki K, Lucena J L, Pakarinen P, Sistonen P, Tapanainen J, Gromoll J, Kaskikari R, Sankila E M, Lehvaslaiho H, Engel A R, et al. Cell. 1995;82:959–968. doi: 10.1016/0092-8674(95)90275-9. [DOI] [PubMed] [Google Scholar]