Abstract
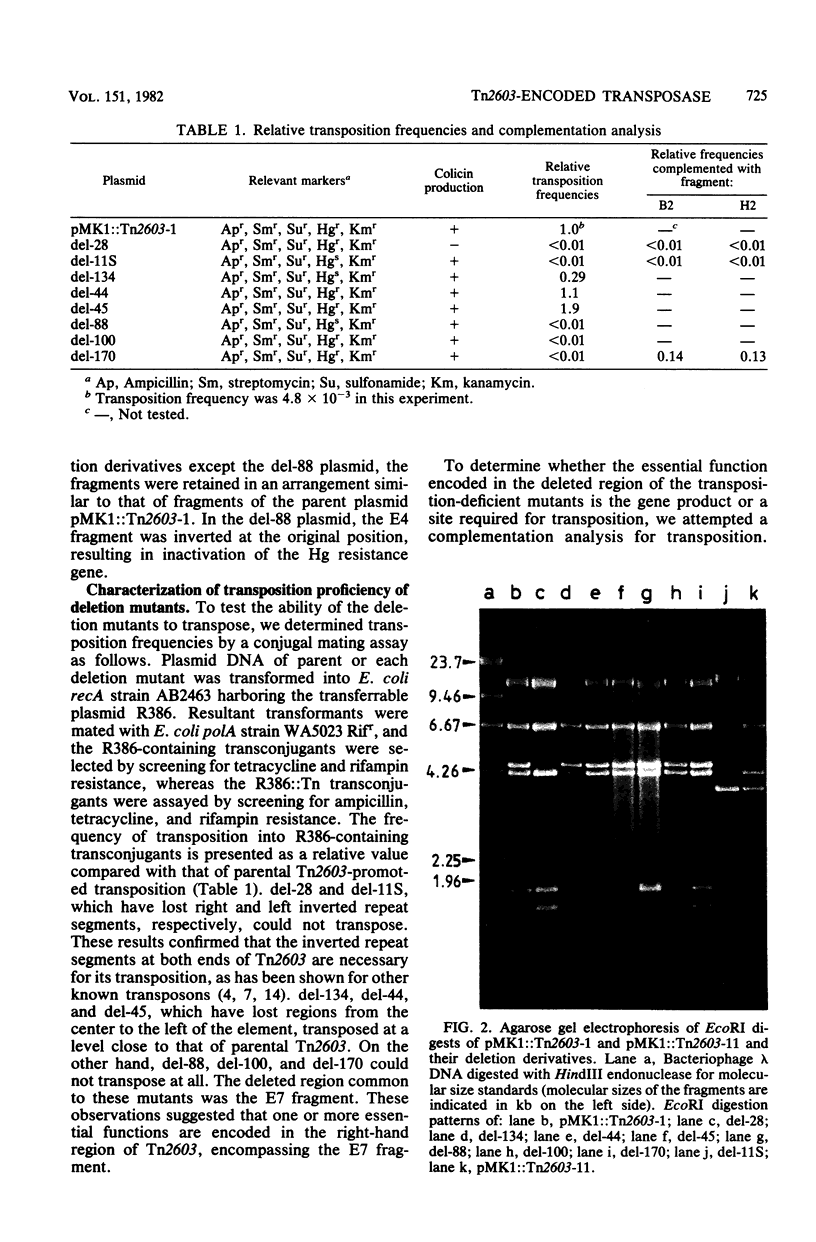
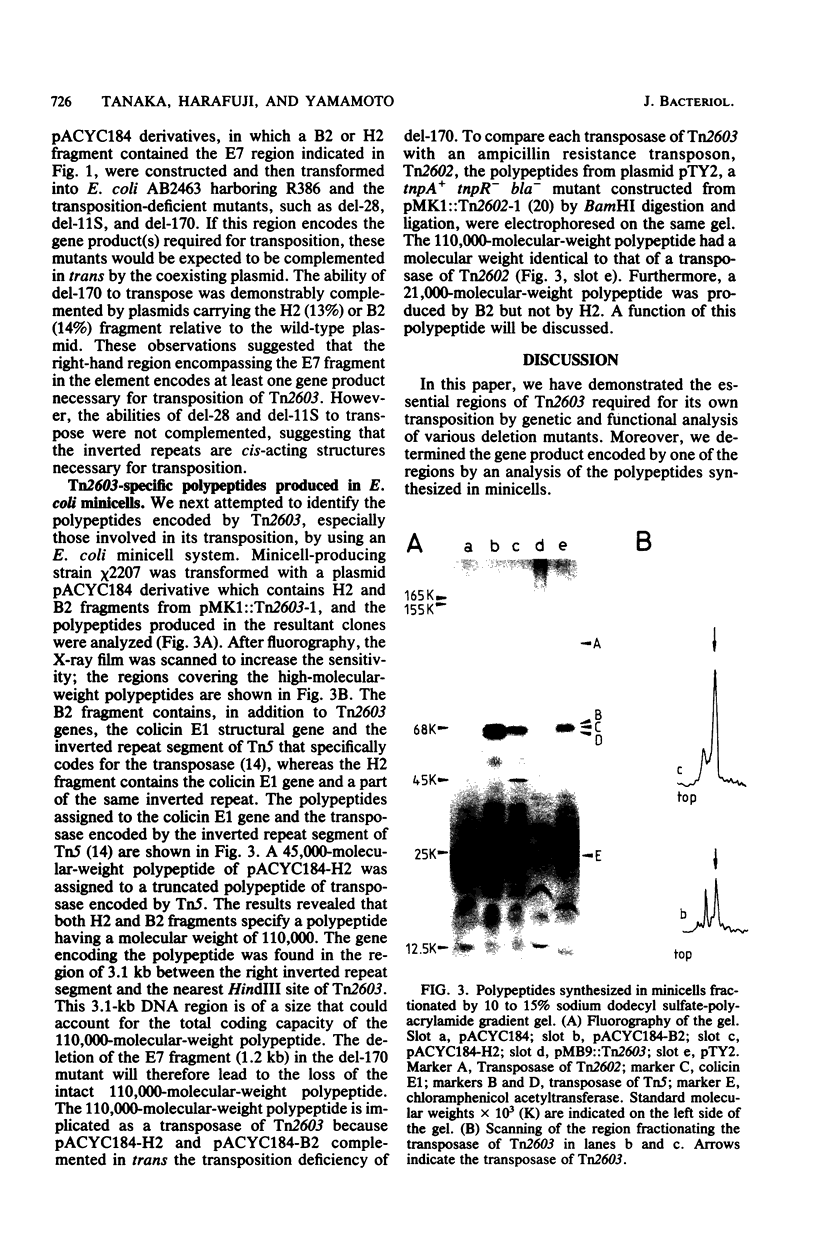
Tn2603 is a multiple-resistance transposon encoding resistance to ampicillin, streptomycin, sulfonamide, and mercury and having a molecular size of 20 kilobase pairs, with 200-base-pair inverted repeats at both ends. The essential sites and functions of Tn2603 which are required for its transposition were determined through construction and characterization of various deletion mutants affecting the efficiency of transposition. Deletions were introduced in plasmid pMK1::Tn2603 by partial digestion with restriction endonuclease EcoRI in vitro. Analysis of deletion mutants showed that the inverted repeat segments at both ends of the trans-acting diffusible product(s) encoded in the right-hand side of the central portion were required for the transposition of Tn2603. An essential gene product was revealed as a protein having a molecular weight of 110,000 by analysis of polypeptides synthesized in Escherichia coli minicells. This protein was assumed to be the so-called transposase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Davis M. A., Roberts D. E., Takeshita K., Kleckner N. Genetic organization of transposon Tn10. Cell. 1981 Jan;23(1):201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Chandler M. On the molecular mechanisms of transposition. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4858–4862. doi: 10.1073/pnas.78.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R. E., Heffron F., Falkow S. Identification of the protein encoded by the transposable element Tn3 which is required for its transposition. Nature. 1979 Dec 20;282(5741):797–801. doi: 10.1038/282797a0. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Kopecko D. J., Brevet J., Cohen S. N. Involvement of multiple translocating DNA segments and recombinational hotspots in the structural evolution of bacterial plasmids. J Mol Biol. 1976 Dec;108(2):333–360. doi: 10.1016/s0022-2836(76)80124-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McCormick M., Wishart W., Ohtsubo H., Heffron F., Ohtsubo E. Plasmid cointegrates and their resolution mediated by transposon Tn3 mutants. Gene. 1981 Nov;15(2-3):103–118. doi: 10.1016/0378-1119(81)90120-7. [DOI] [PubMed] [Google Scholar]
- Reed R. R. Resolution of cointegrates between transposons gamma delta and Tn3 defines the recombination site. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3428–3432. doi: 10.1073/pnas.78.6.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein S. J., Jorgensen R. A., Postle K., Reznikoff W. S. The inverted repeats of Tn5 are functionally different. Cell. 1980 Mar;19(3):795–805. doi: 10.1016/s0092-8674(80)80055-9. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltkamp E., Barendsen W., Nijkamp H. J. Influence of protein and ribonucleic acid synthesis on the replication of the bacteriocinogenic factor Clo DF13 in Escherichia coli cells and minicells. J Bacteriol. 1974 Apr;118(1):165–174. doi: 10.1128/jb.118.1.165-174.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Baba R., Yamagishi S. Physical and functional mapping of Tn2603, a transposon encoding ampicillin, streptomycin, sulfonamide, and mercury resistance. Mol Gen Genet. 1981;181(4):464–469. doi: 10.1007/BF00428737. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Nohara C., Fukunaga Y., Yamagishi S. Transposition of the oxacillin-hydrolyzing penicillinase gene. J Bacteriol. 1981 Feb;145(2):808–813. doi: 10.1128/jb.145.2.808-813.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Yamagata S., Horii K., Yamagishi S. Comparison of transcription of beta-lactamase genes specified by various ampicillin transposons. J Bacteriol. 1982 Apr;150(1):269–276. doi: 10.1128/jb.150.1.269-276.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]