Abstract
Oncolytic viruses represent a novel cancer treatment strategy. Despite their promising preclinical data, however, corresponding clinical trials have disappointed. To aid preclinical analyses, we hypothesized that three-dimensional tumor cell clusters or spheroids might provide an assay system superior to conventional monolayer cell cultures. Spheroids show viral infection, replication and oncolytic patterns distinct from conventional monolayer assays. Therefore, viral tumor penetration and oncolysis measurements may be improved with such three-dimensional models. Also, preclinical analyses of oncolytic viruses frequently measure mitochondrial activity, but more accurate measures of oncolysis might involve quantitation of intracellular protein release. Therefore, we measured luciferase released from luciferase-expressing spheroids and found unique patterns that maintained consistency with various viruses and doses. The relative variations between viruses and doses may represent temporal differences in oncolysis dynamics. Analysis of five recombinant replicative adenoviruses with promise for clinical application showed that Ad5/3-Δ24 produced the most luciferase release 1 week after infection and achieved the earliest and highest peak luciferase release level. Ad5/3-Δ24 also effected the earliest subtotal spheroid cell death. These findings closely parallel monolayer oncolysis assays with these agents. Therefore, the luciferase-expressing tumor spheroid assay represents a promising three-dimensional model for preclinical analysis of replicative oncolytic agents.
Keywords: adenovirus, oncolytic virus, ovarian cancer, replication-competent viruses, spheroid
Introduction
Cancer patients with advanced stage disease still suffer unacceptably high mortality rates, and most patients with metastastic solid tumors cannot be cured. These poor outcomes demand new therapeutic strategies. Gene therapy with oncolytic replicative agents1 represent a novel approach for cancer treatment. These agents include mostly viruses, such as adenovirus2 (Ad), but other microbes3 have also been investigated. As replicative agents multiply their own genome, this amplification may overcome the common problem of low gene transfer.4 Although featuring augmented gene transfer, some replication-competent agents also have shown the ability to penetrate tumors.5 The viral replication cycle itself can cause oncolysis with subsequent release of newly generated virions. In turn, these viral progeny extend infection to neighboring tumor cells.6 Thus, this added antitumor effect does not require transgene expression but only active replication. As replication of therapeutic agents must be mitigated in nontumorous tissues, two methods to limit replication in Ads have been developed.
One method of replication control involves manipulation of genes essential for viral replication, such as the viral early gene E1, with tissue- or tumor-specific promoters.7 As promoters are activated by transcription factors preferentially present in particular tissues or tumors, viral promoter activation can be limited to selected tissues by incorporation of specific promoters, which are only active in target tissue, to drive AdE1gene expression. One conditionally replicative Ad (CRAd) RGDCRADcox-2R employs the cyclooxygenase-2 (Cox-2) promoter.8 As Cox-2 has been shown to be highly expressed in a number of epithelial tumors, RGDCRADcox-2R seems promising also for ovarian cancer.9 CRAd application of another tissue-specific promoter vascular endothelial growth factor (VEGF) exploits the need of cancers for neovascularization. To grow and metastasize, many malignancies including ovarian cancer express abundant VEGF, which binds to endothelial cell VEGF receptors to stimulate angiogenesis. The recently developed Ad5VEGFE1 and Ad5/3VEGFE1 incorporate the VEGF promoter.5
A second means to control viral replication involves deletions in the viral genome that require specific cellular factors to compensate for these alterations. The two CRAds Ad5-Δ24RGD10 and Ad5/3-Δ2411 each incorporate a 24 base pair deletion in the E1A gene. This gene’s product is responsible for binding the retinoblastoma (Rb) protein, which allows Ads to induce S-phase entry. Because of the deletion, this CRAd has limited ability to overcome the G1–S checkpoint in noncycling normal cells, but can replicate efficiently in tumor cells where this interaction is not necessary. As the Rb-p16 pathway is defective in almost all human tumors, CRAds with this E1A 24 base pair deletion replicate preferentially in tumor cells relative to normal cells. Both Ad5-Δ24RGD and Ad5/3-Δ24 have undergone preclinical evaluation for ovarian cancer12,13 and are now in clinical development for glioma and ovarian cancer.6
Despite the potential of replicative agents such as CRAds for effective gene transfer and tumor penetration, their transition to clinical application is limited by assay substrates. Although preliminary clinical results have substantiated their efficacy, corresponding clinical trials have yielded relatively few successes.14 For improved preclinical assays, primary tumor tissue allows more accurate evaluation of Ad agents than use of tumor cell lines.5 Therefore, we have developed three-dimensional aggregates or spheroids of unpassaged and purified ovarian cancer cells as a means for prolonging primary tumor cell viability.15 Spheroids also provide a three-dimensional in vitro model for replicative viral infection.15 As viral penetration of solid tumor proceeds in three dimensions, conventional two-dimensional cell culture assays might produce data that poorly represent these in vivo events. Spheroids, however, are often greater than 1 mm in diameter16 and therefore allow viral spread through a solid rather than planar substrate. We have also shown that replicative viruses exhibit orderly spread in spheroids from their outer to inner layers.16 These infections also cause a marked reduction in spheroid size.16 As cancer cell spheroids should provide an assay substrate with greater morphologic similarity to in vivo tumor, measurements of the penetration speed and oncolysis of replicative agents should be more accurate with these three-dimensional assays.
As a first goal, we examine differences between two-and three-dimensional assays for viral infection, replication and oncolysis. Of these three functions, the oncolytic potency of prototype CRAds represents the most relevant parameter for preclinical study. As a second goal, therefore, we endeavor to develop a three-dimensional assay for quantitation of CRAd-mediated oncolysis. In a previous study, we measured firefly luciferase released from human cancer cell spheroids to quantify tumor cell killing by a replicative Ad with a luciferase transgene expression cassette.15 As Ads kill infected cells by effecting membrane disruption, detection of intracellular luciferase protein17 indicates cellular membrane disruption and consequent cell death. This method, however, can only test CRAds that incorporate the luciferase gene, but clinical therapeutic agents rarely accommodate such reporter genes. Also, levels of luciferase released may poorly estimate levels of actual cytotoxicity but rather depend on variables related to Ad gene expression. A superior assay could implement tumor cells that already express luciferase. To this end, we employ the ovarian cancer cell line SK-OV-3luc that has been stably transfected with the firefly luciferase gene. In this report, we investigate its luciferase expression in the supernatant and cells of these spheroids both uninfected and infected by replicative Ads. We hypothesize that CRAd-infected SK-OV-3luc spheroids will release luciferase from these cells to allow measurements of viral-mediated oncolysis (Figure 1). Photomicrographs of ovarian cancer spheroids, including those undergoing Ad replication-mediated oncolysis, have been recently reported.15,16
Figure 1.
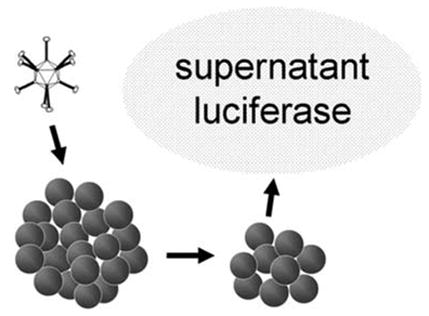
Adenoviral infection of tumor cell spheroids leads to cell membrane lysis and subsequent release of intracellular enzymes that can be quantified as a measure of cell killing.
Materials and methods
Cell culture
The human ovarian adenocarcinoma cell lines SKO-V3.ip1 and SK-OV-3luc were generous gifts from Dr Janet Price (University of Texas MD Anderson Cancer Center) and Dr Robert Negrin (Stanford University), respectively. The 293 and A549 cell lines were purchased from Microbix (Toronto, Canada) and the American Type Culture Collection (Manassas, VA), respectively. Transformed 911 human embryonic retinoblasts were kindly provided by Dr Alex van der Eb (Leiden University Medical Center, The Netherlands). SKO-V3.ip1 cells were cultured in Dulbecco’s modified Eagle’s medium/Ham’s F-12 (50:50 mixture) growth medium. SK-OV-3luc cells were cultured in McCoy’s growth medium. All other cells were cultured in the growth media recommended by their manufacturers. All growth media were supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 IU/ml penicillin and 25 μg/ml streptomycin. All media used for infection were supplemented with 2% fetal bovine serum. All cells were cultured at 37°C in a 5% CO2 atmosphere.
Culture of spheroids
Spheroids were cultured as described previously.15,16 Briefly, tumor cells were suspended with growth medium and added to Costar Ultra Low Attachment Cluster 96- or 24-well plates (Corning Incorporated, Corning, NY) and incubated on a rocking platform. The wells in these plates have a hydrophilic coating that prevents cell adhesion as occurs normally in commonly used treated flasks or plates. Most tumor cells cultured in these containers preferentially adhere to each other to form clusters or spheroids within a week.
Recombinant Ads
As E1-deleted Ads generally have mitigated- replication competency, we employed RGDTKSSTR for such a control virus role. RGDTKSSTR,18,19 RGDCRADcox-2R,8 Ad5luc1,20 AdVEGFLuc, Ad5VEGFE1, Ad5/3VEGFE1,16 Ad5-Δ24RGD,10,12 Ad5/3-Δ24,11 Ad5-Δ24RGDGFP16 and the wild-type serotype 5 Ad Ad300wt (American Type Culture Collection, Manassas, VA) have been described previously. AdVEGFLuc, Ad5VEGFE1 and Ad5/3VEGFE1 were propagated on 911 cells. Ad5-Δ24RGD and Ad5-Δ24RGD were propagated on A549 cells. The other viruses were propagated on 293 cells and all were purified on cesium chloride gradients. The vector particle (vp) concentration determined at 260 nm was 2.82 × 1011, 8.7 × 1011, 1.28 × 1011, 1.01 × 1012 3.41 × 1012, 6.33 × 1012, 6.3 × 1011, 2.6 × 1012 and 4.5 × 1012 vp/ml for RGDTKSSTR, Ad300wt, Ad5luc1, AdVEGFLuc, Ad5VEGFE1, Ad5/3VEGFE1, RGDCRADcox-2R, Ad5-Δ24RGD and Ad5/3-Δ24, respectively. To determine the functional titer of the viruses, we performed plaque assays and counted plaque forming units (PFU) after overnight infection periods to allow entry of all functional virions without confounding by receptor density. The RGDTKSSTR, Ad300wt, Ad5luc1, AdVEGFLuc, Ad5VEGFE1, Ad5/3VEGFE1, RGDCRADcox-2R, Ad5-Δ24RGD and Ad5/3-Δ24 viruses had titers of 9.98 × 108, 5.7 × 1010, 4.7 × 1010, 1.5 × 1010 1.75 × 1010, 1.3 × 1010, 1.3 × 1010, 5.2 × 1010 and 9.5 × 1011 PFU/ml, respectively.
Comparison of viral infection in monolayers and spheroids
To investigate differences in infectivity of monolayers and spheroids, SKOV3.ip1 cells were cultured as monolayers and spheroids with 5 × 105 cells/well. Either 0, 10, 100 or 1000 vp/cell of Ad5luc1, a recombinant Ad that incorporates a luciferase gene expression cassette, were added to each well in quadruplicate. Forty hours after infection, luciferase expression was measured with the luciferase assay system (Promega Corporation, Madison, WI) as recommended.
Comparison of viral replication in monolayers and spheroids
To compare viral replication in SKOV3.ip1 monolayers and spheroids (1 × 105 cells/well), both were infected with 0 or 100 vp/cell of Ad300wt. Every other day until 18 days after infection, cells from one monolayer and one spheroid well were frozen at −20°C. Samples from mock-infected wells were similarly frozen. DNA was purified from each sample with the DNeasy Tissue Kit (Qiagen Inc., Valencia, CA). Quantitative PCR was performed on these DNA samples with a Roche Light-cycler (Roche Diagnostics Corporation, Indianapolis, IN) as previously described.15,16 The probe used was specific for the Ad-E4 gene. Viral DNA copies detected were normalized to the number of cells by simultaneous measurement of actin copies. Mock values were subtracted from experimental values.
Comparison of viral-mediated oncolysis in monolayers and spheroids
To analyze differences in viral-induced oncolysis in one- and two-dimensional cell cultures, SKOV3.ip1 cells were cultured with 1 × 104 cells/well as monolayers and spheroids. Both were infected with 0 or 100 vp/cell of Ad300wt. Daily measurements of cell killing were performed with lactate dehydrogenase (LDH) cytotoxicity assay (Roche Applied Science, Indianapolis, IN). LDH release was detected as an indicator of cytolysis by this colorimetric assay that measures light absorption at 490 nm. Mock infection values were subtracted from all data.
Validation of luciferase expression by SK-OV-3luc cells
To validate the luciferase-expressing characteristics of SK-OV-3luc, we first compared its luciferase expression with cell mass. Quadruplicate monolayer and spheroid cultures of 103, 104, 105 and 106 cells/well were established. One day after monolayers were seeded, and 1 week after spheroids were seeded, cellular and supernatant luciferase was measured in all samples with the Bright-Glo Luciferase Assay System (Promega Corporation, Madison, WI).
Validation of luciferase release by viral-infected SK-OV-3luc cells
To measure viral-induced oncolysis in SK-OV-3luc spheroids, 5 × 105 SK-OV-3luc cells were added to each well to form spheroids and a week later infected in quadruplicate with either 0 or 100 vp/cell of Ad300wt. Supernatant luciferase was measured daily after infection for 15 days after infection by removing 100 μl aliquots of growth medium from each well for assay with the Bright-Glo Luciferase Assay System. All media removed was immediately replaced with equal volumes of fresh growth medium.
Measuring released luciferase from SK-OV-3luc spheroids infected by multiple doses of virus
To observe differences in luciferase release from spheroids infected with different doses of virus, SK-OV-3luc spheroids (5 × 105 cells/well) were infected with 0, 0.1, 1, 10, 100 and 1000 vp/cell of Ad300wt. Four replicates were assayed per dose. For the following 18 days, 100 μl aliquots of growth medium from each well (500 μl per well) were daily extracted for luciferase assays. Each aliquot removed was immediately replaced with an identical volume of fresh media. Mock-infection values were subtracted from all data.
Comparison of oncolysis by five CRAds with SK-OV-3luc spheroids
To compare the oncolytic potency of several CRAds with high clinical potential, SK-OV-3luc spheroids (5 × 105 cells/well) were infected in quadruplicate with 0 or 100 vp/cell of RGDTKSSTR (E1-deleted control), RGDCRADcox-2R, Ad5VEGFE1, Ad5/3VEGFE1, Ad5-Δ24RGD and Ad5/3-Δ24. For the next 18 days, 100 μl aliquots were daily removed from the 500 μl of growth medium in each well for luciferase assays. With each assay, the removed supernatant was replaced in each well with fresh growth medium.
Statistical analysis
Differences in tumor cell monolayers and spheroids were compared at three doses by Student’s t-test. A generalized linear model was fit to the data to examine the effect of serial viral replication with days as a continuous variable and group (monolayers vs spheroid) as a treatment effect. A quadratic term was added for days to model the curvature in the data. The analysis of viral-induced oncolysis over time was performed with a repeated measures model with PROC MIXED (SAS Ver. 9.0), which treated the within-group effect of time as a continuous variable and the treatment group as a fixed effect. The observed curvature in the data was included in the model as a quadratic term. The effects of treatment group, time and the interaction of treatment group and time were evaluated by F-tests. The differences in predicted treatment means averaged over time were compared with a t-test. Validation of luciferase expression by spheroids was evaluated by a generalized linear model and the correlation coefficient. Time–dependent changes in luciferase concentration between infected and uninfected spheroids were examined by a mixed model as described above, taking into account the replicates in each system. Similarly, changes in luciferase release from infected spheroids were modeled for different doses of wild-type Ad by a mixed model with planned comparisons for the 0.1 vs 10, 0.1 vs 1000 and 10 vs 1000 vp/cell. Cumulative luciferase over time between dose groups and five- CRAds with spheroids was compared by linear regression with a quadratic term. The rate of change was compared to the referent group (0.1 vp/cell or RGDTKSSTR). For all analyses a two-sided P-value of <0.05 was deemed statistically significant.
Results
Monolayer and spheroid models differ with regard to viral infectivity, replication and oncolysis
To compare viral infectivity in two- and three-dimensional model systems, tumor cell monolayers and spheroids were infected with equal amounts of Ad5luc1. Forty hours after infection, transgene expression was measured to quantify viral infection. At all three doses, monolayers supported more viral infection than that observed in spheroids (Figure 2a). Transgene expression was 6.4-, 1.8- and fourfold higher in monolayers than spheroids. Overall, viral infectivity was 406% greater in monolayers with significantly higher infectivity than spheroids at 10 vp/cell (P = 0.02). There was no statistical difference at the 100 and 1000 vp/cell doses (P = 0.27 and 0.75, respectively).
Figure 2.
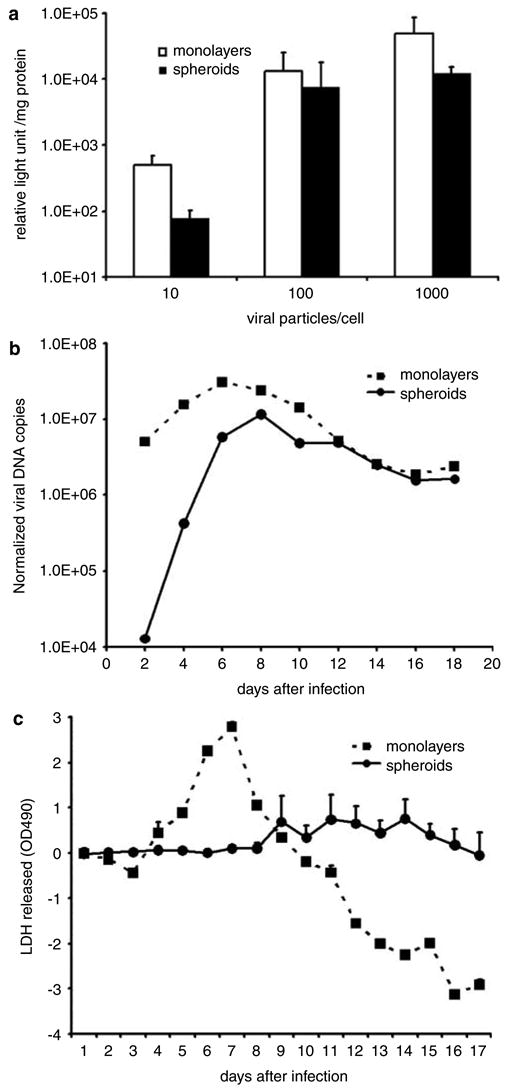
Monolayer and spheroid models differ in their assessment of viral infection, replication and oncolysis. (a) Luciferase expression from tumor cells infected with Ad5luc1 show more viral infectivity in monolayer than spheroid assay systems (all error bars represent one standard deviation). (b) Quantitation of viral DNA in tumor cells infected with wild-type Ad shows more viral replication in monolayers than spheroids. (c) An LDH-based cytotoxicity assay of tumor cells infected with wild-type Ad shows more LDH release in monolayers than spheroids (values from uninfected cells have been subtracted from infected cells).
To observe differences in viral replication by two- and three-dimensional model systems, tumor cell monolayers and spheroids were infected with 100 vp/cell of wild-type Ad. Serial measurements of viral DNA in both models showed more virus in the monolayers during the first week (Figure 2b). Although more virus was detected in monolayers during the first 4 days after infection, viral replication in spheroids surpassed that in monolayers later in the first week. Amounts of virus in both monolayers and spheroids evened 12 days after infection. Overall, significantly higher normalized viral DNA copies were detected in the monolayer system relative to the spheroid system (P = 0.04) from a linear model. The cumulative amount of viral DNA in monolayers was more than three-fold that in spheroids.
To compare viral-induced oncolysis in two- and three-dimensional cell cultures, tumor cell monolayers and spheroids were infected with 100 vp/cell of wild-type Ad. Daily assay of cell killing by measurement of LDH release showed a prominent rise reaching a peak of 2.79 7 days after infection (Figure 2c). The subsequent decline produced negative values because mock-infected group values were subtracted from all data. Spheroid oncolysis increased at a much slower rate reaching a much lower peak of 0.76 14 days after infection. A repeated measures model showed that spheroid oncolysis was significantly lower compared to monolayer (95% CI difference, −1.16, −0.08, P = 0.04).
Validation of luciferase expression by SK-OV-3luc spheroids
SK-OV-3luc cells readily formed spheroids within a week. The size and shape of these spheroids were consistently uniform between cultures. To validate the luciferase-expressing characteristics of these cells in monolayers and spheroids, we compared their luciferase expression with cell mass. In monolayers, luciferase expression correlated well with cell number (Figure 3). Quadruplicate 1 × 102, 1 × 103, 1 × 104, 1 × 105 and 1 × 106 cell populations of SK-OV3luc expressed means of 4337, 67 343, 728 089, 4 893 177 and 17 393 995 relative light units (RLU)/cell, respectively. In spheroids, the same numbers of cells expressed 150 195, 240 517, 1 510 610, 9 247 323 and 19 070 203 RLU/cell, respectively. Luciferase expression correlated significantly with cell mass in both monolayers and spheroids (P = 0.001, r2 = 0.75 from a linear model) and did not differ between the model systems. In monolayer SK-OV-3luc cultures, the luciferase detected in the supernatants represented 7.30, 5.44 and 12.98% of that measured in the cellular fraction (data not shown). As spheroids, their supernatant luciferase was 0.41%, 0.51% and 0.67% of that in the cells (data not shown), respectively.
Figure 3.
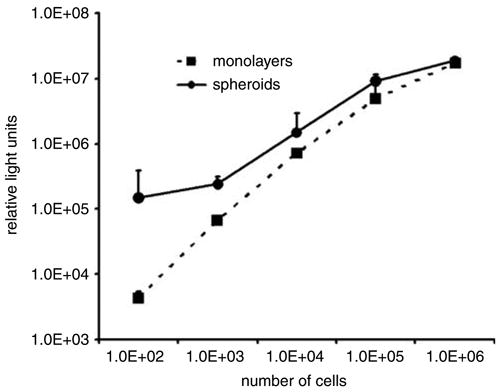
Cellular luciferase expression from SK-OV-3luc cells cultured as both monolayers and spheroids show adequate correlation with cell mass. This correlation shows that SK-OV-3luc spheroids possess amounts of luciferase proportional to their cell mass.
Validation of luciferase release by viral-infected SK-OV-3luc spheroids
Oncolysis of SK-OV-3luc spheroids infected by wild-type Ad was analyzed by measurements of supernatant luciferase daily for 15 days after infection. Except for the first day during this period, the uninfected spheroids released amounts of luciferase that were comparable to those observed in the previous experiment and ranged from 0.0097 to 0.0825% of the cellular luciferase. On the first day after infection, the supernatant luciferase was 2.4% that of the cellular luciferase. In infected spheroids, however, supernatant luciferase levels were significantly higher than those of uninfected spheroids (P = 0.04, repeated measure analysis) and progressively increased to 4315 RLU, compared to a peak of 990 RLU in the uninfected group (Figure 4). Luciferase levels from the infected spheroids reached their peak 7 days after infection, then gradually declined and then showed an apparent second smaller peak 2 weeks after infection.
Figure 4.
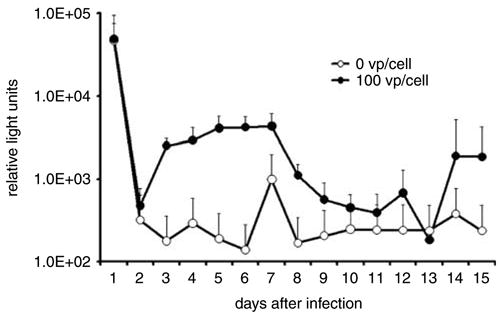
Supernatant luciferase released from infected SK-OV-3luc spheroids is markedly higher than levels from uninfected spheroids The difference in these levels should represent the amount of viral-induced oncolysis.
Luciferase from SK-OV-3luc spheroids infected by multiple doses of virus
SK-OV-3luc spheroids were infected with different doses of wild-type Ad. Figure 5a shows evidence of a trend: higher doses led to earlier peaks in luciferase release. The 0.1, 1, 10, 100 and1000 vp/cell doses produced peaks in cell killing at 6, 12, 8, 10 and 6 days after infection, respectively. These luciferase levels reached peaks of 75 339, 107 403, 140 184, 85 475 and 75 339 RLU, respectively. A repeated measures model showed that luciferase release was significantly higher at the 10 vp/cell dose compared to the 0.1 vp/cell dose (95% CI 1604.2, 32 523; P = 0.03), in the 1000 vp/cell dose compared to the 0.1 vp/cell dose (95% CI 16 448, 47 367; P = 0.0002), and of borderline significance between the 1000 cell dose compared to the 10 vp/cell dose (95% CI −615.2, 30 303; P = 0.059). Although all doses produced comparable peak levels of luciferase release, lower doses tended to produce later peaks. The luciferase release from spheroids infected with 1000 vp/cell at 14 days was not recorded.
Figure 5.
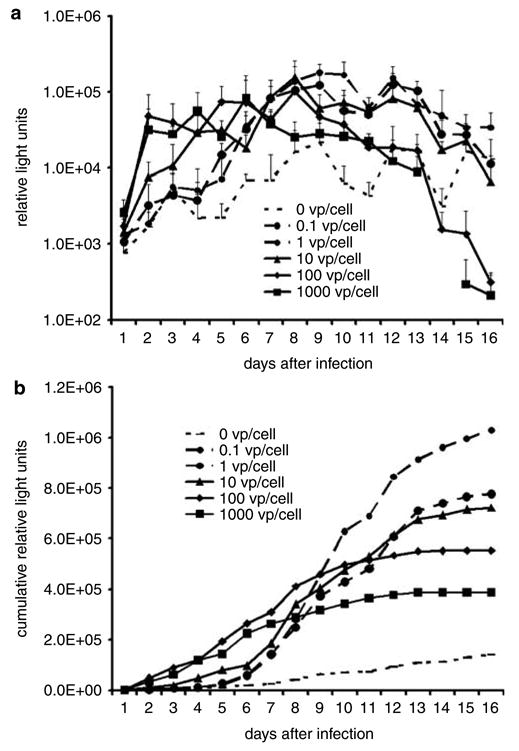
Supernatant luciferase release from infected SK-OV-3luc spheroids show patterns that reflect differences in viral doses. (a) Higher doses generally led to earlier and higher peaks in luciferase release. (b) When displayed as cumulative values, these data show that because more cells survive longer, lower doses of virus allow a more prolonged luciferase release and apparently greater total luciferase release than higher doses do.
As even modest variations in luciferase release limit precise comparison of groups, Figure 5b presents an alternative view of the same data. Displayed as cumulative data, each data point represents the particular day’s value added to the sum of values from all previous days. This view shows that lower doses of virus allow more prolonged luciferase release and apparently greater total luciferase production. The 100 and 1000 vp/cell produced significantly higher cumulative luciferase release over time compared to the 0.1 vp/cell group. There were no significant differences in luciferase release among the 0.1, 1 and 10 vp/cell over time.
Comparison of oncolysis by five CRAds with SK-OV-3luc spheroids
Five CRAds with high potential for clinical application were analyzed by our new three-dimensional oncolysis assay. These were compared with an E1-deleted non-replicative control, RGDTKSSTR. Luciferase released into the supernatant surrounding infected SK-OV-3luc spheroids was measured daily as a measure of tumor cell lysis. These data are presented as cumulative data where each day’s value equals the luciferase measurement for that time added to the sum of all previous values (Figure 6). Compared to the E1-deleted control group, the cumulative luciferase release rate progressively decreased in the RGDCRADcox-2R, AD5VEGFE1 and AD5/3VEGFE1 groups and increased in the wild-type AD5 and Ad5/3-Δ24 groups. Ad5/3-Δ24 resulted in the most luciferase release during the first week after infection. It was followed in order by Ad5-Δ24RGD, Ad300wt, RGDCRADcox-2R, RGDTKSSTR, mock, Ad5/3VEGFE1 and Ad5VEGFE1. The mock, RGDTKSSTR, Ad300wt, RGDCRADcox-2R, Ad5-VEGFE1, Ad5/3VEGFE1, Ad5-Δ24RGD and Ad5/3-Δ24 groups reached peak supernatant luciferase levels at 10, 6, 10, 6, 6, 10, 6 and 3 days after infection, respectively. These peaks amounted to 135 892, 103 123, 108 651, 74 684, 36 240, 44 124, 99 754 and 174 411 RLU, respectively. The mock, RGDTKSSTR, Ad300wt, RGDCRADcox-2R, Ad5VEGFE1, Ad5/3VEGFE1, Ad5-Δ24RGD and Ad5/3-Δ24 groups reached cessation of oncolysis at >18, >18, 16, 13, 15, 13, 16 and 13 days after infection, respectively. Cessation of oncolysis was defined as less than 10 000 RLU with no subsequent peaks above this level for the particular CRAd.
Figure 6.
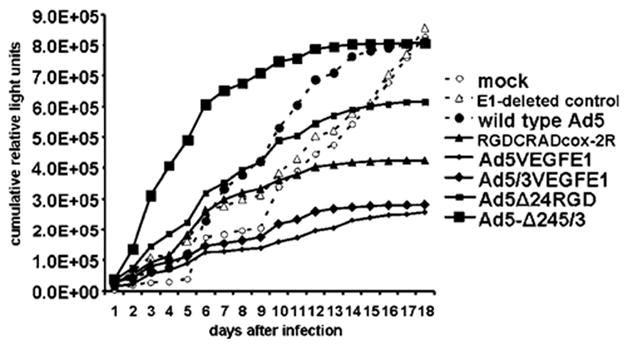
Oncolysis by five CRAds with high clinical potential was evaluated with the SK-OV-3luc spheroid assay. Ad5/3-Δ24 produced the most luciferase release during the first week after infection. It also achieved the earliest (3 days after infection) and highest peak level of luciferase release. Ad5/3-Δ24 was also fastest (day 13) in causing near-complete spheroid death (increase in luciferase of less than 10 000 RLU/measurement).
Discussion
As solid tumors in vivo are three-dimensional, models for analysis of replication competent oncolytic agents should also have a similar morphology. Discrepancies between two- and three-dimensional assays may introduce error into analysis of antitumor agents. A difference between oncolysis of spheroids differing in sizes was illustrated in earlier studies.15 As smaller spheroids present targets little more than two cells thick, infection should show a spread of transgene expression similar to that in monolayers. As expected, progression of gene expression by a replicative vector proceeded slower and for a longer period through the larger spheroids. In this study, we show evidence that two- and three-dimensional luciferase-expressing model systems show a similar difference in their assessment of viral infection, replication and oncolysis.
With regard to viral infectivity, a particular number of cells possess more surface area as a monolayer culture than as a three-dimensional cluster. Greater area for contact between cells and virus enhances opportunity for viral binding and internalization.21 Therefore, monolayers should allow greater viral infectivity by their relatively high surface area relative to spheroids. Indeed, our findings show that infectivity was more than fourfold greater in the monolayers than in spheroids (Figure 2a).
In measuring viral replication, monolayer cultures should have higher initial amounts of virus because of their relatively greater surface area and subsequent high proportion of initially infected cells. Spheroids hold in their interior a reserve of viable cells protected from initial infection.22 Accordingly, greater amounts of virus were detected in the monolayers during the first week (Figure 2b), but as virus reached the interior of the spheroids, viral replication accelerated more rapidly than that of monolayers until about 2 weeks after infection, when amounts of viruses in both models evened. In addition to the difference in viral replication rates, summation of all viral DNA measurements showed more overall replication in monolayers. In the 18-day period after infection, cumulative viral DNA measurements in monolayers were more than threefold greater than in spheroids. This difference concurs with knowledge that greater initial infection leads to greater viral-gene expression and virus replication.11,23–26 Therefore, monolayers may support higher overall viral replication than spheroids because of greater initial opportunity for viral infection. Also, monolayer cultures may allow more rapid cellular regeneration during the experiment. In contrast, spheroids incorporate a higher proportion of dormant cells just as clinical tumors do.22
Levels of oncolysis were initially higher in tumor cell monolayers, with a peak in cell killing 1 week after infection, compared to the peak at 2 weeks in spheroids (Figure 2c). Also, the peak in cell killing for monolayers was over three times higher than for spheroids. As mock-infected values were subtracted from all data, the negative values in monolayers during the second week after infection indicate greater cell death relative to uninfected groups. Simultaneously, the positive values in spheroids indicate that viruses were still actively killing tumor cells. The three-dimensional spheroids, therefore, allow a more prolonged analysis of viral-induced oncolysis and may model complexities viruses encounter in a three-dimensional environment.
Altogether, these data show marked differences in viral-mediated oncolysis between two- and three-dimensional assays. Therefore, any analyses of agent propagation over an extended period of time in monolayer cultures may be flawed. Spheroids, however, provide a three-dimensional assay substrate that may more accurately simulate agent penetration in human solid tumors. Their unique morphology and sustained cellular viability also allow extended longitudinal analyses.
For preclinical analyses of replicative viral agents, oncolysis may be a parameter even more important than infection or replicativity. Tumor cell killing can be indirectly measured by cell-viability assays, such as the mean transit time (MTT) assay (ATCC, Manassas, VA), [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] (MTS) assays (Promega) and WST-1 assays (Roche) that quantify mitochondrial activity. The experience of ours and others27,28 with applying these assays to spheroids shows that they require exceptionally prolonged incubation periods to obtain reasonable results. These deviations from the manufacturers’ recommendations may be necessitated by the inadequate reagent penetration of spheroids. Furthermore, viral infection induces host cell metabolic activity that increases mitochondrial activity and confounds results. Therefore, we sought to explore other means of quantifying viral-induced oncolysis. Exclusion -cell counts with Trypan blue staining identifies tumor cells with disrupted membranes, such as those killed by virus. This method, however, was technically difficult to apply (data not shown), as tumor cell spheroids are very difficult to disrupt into individual cells. Moreover, both metabolic and staining assays require killing of the assayed tumor cultures for measurements. Convenience and reduced variability could be achieved by an assay that measures one sample through several time points.
One such assay that also measures cell membrane integrity is the 51Cr-release assay, but this test requires preinfection labeling of the tumor cells with a radioactive reagent. Other nonradioactive assays can also measure progressive release of cytoplasmic substances such as LDH. LDH is detectable in glioma29 and other cancer cells,30 and its measurement has correlated with the MTT assay.30 This assay also allows continuous supernatant testing and consequent preservation of a spheroid sample. Importantly, it measures actual cell killing in contrast to cell viability assays that depend indirectly on mitochondrial activity (see Figure 1).
Assays for quantitative measurement of transduced reporter enzymes released from lysed tumor cells would apply the same principle as the established LDH based oncolysis assay. Expression of enzymes not naturally occurring in mammalian cells, such as firefly luciferase, allows highly sensitive detection and a large dynamic range.17 Current luciferase detection assays such as the Promega Bright-Glo Luciferase Assay System can detect concentrations of luciferase as low as 10−20 mol.31 Furthermore, as luciferase has only an approximate 30 min half-life in growth medium,17 its assay should provide a measure that is temporally precise.
As monolayers, luciferase expression from SK-OV-3luc cells closely correlated with the number of cells (Figure 3). SK-OV-3luc spheroid cultures, however, showed that larger spheroids generally have relatively lower luciferase expression. This difference may be explained by the knowledge that both larger spheroids and human solid tumor hold fewer viable cells at their cores.22 The proportion of supernatant luciferase in SK-OV-3luc monolayer cultures is at least one order of magnitude more than that in spheroid cultures. The greater luciferase release in uninfected cells may represent a confounding factor in measuring viral-mediated oncolysis. Therefore, the lower amount of luciferase release from uninfected SK-OV-3luc spheroids is preferable.
Our experiments infecting SK-OV-3luc spheroids with wild-type Ad showed that infected spheroids consistently released more luciferase than uninfected spheroids did (Figure 4). Infected spheroids produced a peak at 1 week after infection, which is more than fourfold greater than that from uninfected spheroids. The relatively high initial luciferase release 1 day after infection may be due to the daily growth medium exchanges performed only after day 1. It is unclear why the mock-infected group has an outstanding increase in luciferase release on day 7.
To investigate whether different doses of virus used for infection would create any variations in results, we infected SK-OV-3luc spheroids with viral doses that ranged over four orders of magnitude. In general, all supernatant luciferase measurements closely paralleled each other, but higher doses led to earlier peaks in luciferase release (Figure 5a). These temporal differences in luciferase release seen with varying doses agree with our previous observations in spheroids infected by multiple doses of a replication-competent Ad with expression of a luciferase transgene.15 In these earlier studies, higher doses also led to earlier peaks and shorter durations of luciferase release. In this report, some of the later time points at the highest dose showed more luciferase release from noninfected cells than infected cells. These data may represent total cell death in the infected cells. When the data are viewed as cumulative data, Figure 5b shows that lower doses of virus allow more prolonged luciferase release and apparently greater total luciferase release than higher doses do. These findings suggest that this spheroid assay might allow sufficient accuracy even with a wide range of viral doses.
We attempted to correlate luciferase release with LDH release from SK-OV-3luc spheroids infected by wild-type Ad (data not shown). Despite infection of spheroids in quadruplicates, the LDH assay suffered unacceptably high variability on all 18 days after infection. The luciferase assay, however, showed more consistent measurements at nearly all time points. Although the correlation between the two assays was poor (Pearson’s correlation coefficient 0.482), the SK-OV-3luc assay shows greater precision. The SK-OV-3luc assay also showed insufficient correlation with protein assay, quantitative PCR measurements of Ad DNA copies and an MTS cell viability assay (data not shown). The protein and DNA assays, however, are not well accepted methods for quantification of viral-mediated oncolysis quantification. As discussed earlier, the unacceptably long incubation times and viral alteration of cellular metabolism impair cell viability assays, such as the MTS assay. Therefore, utilization of SK-OV-3luc cells and measuring supernatant luciferase is our current method-of-choice for comparing replication-competent oncolytic viruses.
Five CRAds with potential for clinical application were compared with each other and a wild-type Ad control a nonreplicative control, and an uninfected control (Figure 6). Ad5/3-Δ24 produced the most luciferase release during the first week after infection. Ad5/3-Δ24 also produced the most rapid luciferase release by achieving the earliest (3 days after infection) and highest (174 411 RLU) peak levels. Ad5/3-Δ24 also boasted the fastest time (13 days) to reach a luciferase release level of 10 000 RLU or lower. This last parameter should represent cessation of luciferase release and consequent complete cell death for the tumor spheroid. This premise is based on previous observations, including microscopic examination, of CRAd-infected ovarian cancer spheroids over a similar duration.16
Some of these earlier studies employed cell viability studies16 that closely parallel results of this report and showed Ad5-Δ24RGD and Ad5/3-Δ24 CRAds to induce maximum tumor cell killing. The other CRAds with the Ad serotype 3 knob (Ad5/3VEGFE1 and Ad5/3-Δ24) effected more oncolysis than their counterparts with wild-type Ad5 or RGD-4C-modified knobs (Ad5VEGFE1 and Ad5-Δ24RGD). Earlier studies also showed that Ad5/3-Δ24 showed more DNA replication that the other CRAds tested in primary ovarian cancer spheroids.16 These results concur with the experiments of this report.
Unexpectedly, two of the CRAd-infected SK-OV-3luc spheroid groups actually showed levels of luciferase release lower than that from uninfected spheroids. It is unlikely that Ad infection prevents tumor cell lysis or preserves cell viability. As Ad infection decreases production of many host cellular proteins to support production of viral proteins, these viruses possibly suppressed cellular luciferase expression relatively more than they have effected oncolysis.
Low luciferase release values for any CRAd should not be absolutely interpreted as predictor of poor clinical efficacy. High specificity in cell killing also represents an important parameter in addition to oncolytic potency. Thus, these data should be evaluated together with complementary tests evaluating toxicity towards normal host tissue and other conventional tests. Ultimately, clinical data will be needed to confirm preclinical results. Tumor cell lines, such as SK-OV-3luc that express unique reporter proteins such as luciferase, may become useful in organotypic spheroids comprising heterogeneous mixtures of tumor cells and nontumor supportive cells. Such assays could measure oncolysis of replicative agents in a substrate even more similar to in vivo tumor and perhaps measure also the specificity of tumor vs normal cell killing. In summary, our results suggest that development of the SK-OV-3luc spheroid model might provide a sensitive three-dimensional preclinical tool for analysis and comparison of candidate clinical agents.
Acknowledgments
We thank Dr Robert S Negrin and Dr Christian Sheffold from the Division of Bone Marrow Transplantation at Stanford University Medical Center for use of the SK-OV-3luc cell line. This study was supported by NCI (R01 CA83821, P50 CA83591, P50 CA89019 and R01 CA93796), the Academy of Finland, Helsinki University Central Hospital Research Funds, University of Helsinki Internal Funds, Sohlberg Foundation, Sigrid Juselius Foundation, Finnish Cancer Society, Instrumentarium Research Fund, Emil Aaltonen Foundation, Finnish Oncology Association, Research and Science Foundation of Farmos.
References
- 1.Kirn D, Martuza RL, Zwiebel J. Replication-selective virotherapy for cancer: biological principles, risk management and future directions. Nat Med. 2001;7:781–787. doi: 10.1038/89901. [DOI] [PubMed] [Google Scholar]
- 2.Kanerva A, Hemminki A. Modified adenoviruses for cancer gene therapy. Int J Cancer. 2004;110:475–480. doi: 10.1002/ijc.20129. [DOI] [PubMed] [Google Scholar]
- 3.Theys J, Barbe S, Landuyt W, Nuyts S, Van Mellaert L, Wouters B, et al. Tumor-specific gene delivery using genetically engineered bacteria. Curr Gene Ther. 2003;3:207–221. doi: 10.2174/1566523034578357. [DOI] [PubMed] [Google Scholar]
- 4.Puumalainen AM, Vapalahti M, Agrawal RS, Kossila M, Laukkanen J, Lehtolainen P, et al. Beta-galactosidase gene transfer to human malignant glioma in vivo using replication-deficient retroviruses and adenoviruses. Hum Gene Ther. 1998;9:1769–1774. doi: 10.1089/hum.1998.9.12-1769. [DOI] [PubMed] [Google Scholar]
- 5.Lamfers ML, Hemminki A. Multicellular tumor spheroids in gene therapy and oncolytic virus therapy. Curr Opin Mol Ther. 2004;6:403–411. [PubMed] [Google Scholar]
- 6.Kanerva A, Hemminki A. Adenoviruses for treatment of cancer. Ann Med. 2005;37:33–43. doi: 10.1080/07853890410018934. [DOI] [PubMed] [Google Scholar]
- 7.Saukkonen K, Hemminki A. Tissue-specific promoters for cancer gene therapy. Expert Opin Biol Ther. 2004;4:683–696. doi: 10.1517/14712598.4.5.683. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto M, Davydova J, Wang M, Siegal GP, Krasnykh V, Vickers SM, et al. Infectivity enhanced, cyclooxygenase-2 promoter-based conditionally replicative adenovirus for pancreatic cancer. Gastroenterology. 2003;125:1203–1218. doi: 10.1016/s0016-5085(03)01196-x. [DOI] [PubMed] [Google Scholar]
- 9.Kanerva A, Bauerschmitz GJ, Yamamoto M, Lam JT, Alvarez RD, Siegal GP, et al. A cyclooxygenase-2 promoter-based conditionally replicating adenovirus with enhanced infectivity for treatment of ovarian adenocarcinoma. Gene Therapy. 2004;11:552–559. doi: 10.1038/sj.gt.3302181. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki K, Fueyo J, Krasnykh V, Reynolds PN, Curiel DT, Alemany R. A conditionally replicative adenovirus with enhanced infectivity shows improved oncolytic potency. Clin Cancer Res. 2001;7:120–126. [PubMed] [Google Scholar]
- 11.Kanerva A, Zinn KR, Chaudhuri TR, Lam JT, Suzuki K, Uil TG, et al. Enhanced therapeutic efficacy for ovarian cancer with a serotype 3 receptor-targeted oncolytic adenovirus. Mol Ther. 2003;8:449–458. doi: 10.1016/s1525-0016(03)00200-4. [DOI] [PubMed] [Google Scholar]
- 12.Bauerschmitz GJ, Lam JT, Kanerva A, Suzuki K, Nettelbeck DM, Dmitriev I, et al. Treatment of ovarian cancer with a tropism modified oncolytic adenovirus. Cancer Res. 2002;62:1266–1270. [PubMed] [Google Scholar]
- 13.Kanerva A, Zinn KR, Peng KW, Ranki T, Kangasniemi L, Chaudhuri TR, et al. Noninvasive dual modality in vivo monitoring of the persistence and potency of a tumor targeted conditionally replicating adenovirus. Gene Therapy. 2005;12:87–94. doi: 10.1038/sj.gt.3302387. [DOI] [PubMed] [Google Scholar]
- 14.Kirn D. Clinical research results with dl1520 (Onyx-015), a replication-selective adenovirus for the treatment of cancer: what have we learned? Gene Therapy. 2001;8:89–98. doi: 10.1038/sj.gt.3301377. [DOI] [PubMed] [Google Scholar]
- 15.Lam JT, Bauerschmitz GJ, Kanerva A, Barker SD, Straughn JM, Wang M, et al. Replication of an integrin targeted conditionally replicating adenovirus on primary ovarian cancer spheroids. Cancer Gene Therapy. 2003;10:377–387. doi: 10.1038/sj.cgt.7700578. [DOI] [PubMed] [Google Scholar]
- 16.Lam JT, Kanerva A, Bauerschmitz GJ, Takayama K, Suzuki K, Yamamoto M, et al. Inter-patient variation in efficacy of five oncolytic adenovirus candidates for ovarian cancer therapy. J Gene Med. 2004;6:1333–1342. doi: 10.1002/jgm.635. [DOI] [PubMed] [Google Scholar]
- 17.Schafer H, Schafer A, Kiderlen AF, Masihi KN, Burger R. A highly sensitive cytotoxicity assay based on the release of reporter enzymes, from stably transfected cell lines. J Immunol Methods. 1997;204:89–98. doi: 10.1016/s0022-1759(97)00040-9. [DOI] [PubMed] [Google Scholar]
- 18.Hemminki A, Belousova N, Zinn KR, Liu B, Wang M, Chaudhuri TR, et al. An adenovirus with enhanced infectivity mediates molecular chemotherapy of ovarian cancer cells and allows imaging of gene expression. Mol Ther. 2001;4:223–231. doi: 10.1006/mthe.2001.0446. [DOI] [PubMed] [Google Scholar]
- 19.Hemminki A, Zinn KR, Liu B, Chaudhuri TR, Desmond RA, Rogers BE, et al. In vivo molecular chemotherapy and noninvasive imaging with an infectivity-enhanced adenovirus. J Natl Cancer Inst. 2002;94:741–749. doi: 10.1093/jnci/94.10.741. [DOI] [PubMed] [Google Scholar]
- 20.Krasnykh V, Belousova N, Korokhov N, Mikheeva G, Curiel DT. Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J Virol. 2001;75:4176–4183. doi: 10.1128/JVI.75.9.4176-4183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittereder N, March KL, Trapnell BC. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunz-Schughart LA, Kreutz M, Knuechel R. Multicellular spheroids: a three-dimensional in vitro culture system to study tumour biology. Int J Exp Pathol. 1998;79:1–23. doi: 10.1046/j.1365-2613.1998.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas JT, Kim M, Sumerel LA, Carey DE, Curiel DT. Efficient oncolysis by a replicating adenovirus (ad) in vivo is critically dependent on tumor expression of primary ad receptors. Cancer Res. 2001;61:813–817. [PubMed] [Google Scholar]
- 24.Hemminki A, Dmitriev I, Liu B, Desmond RA, Alemany R, Curiel DT. Targeting oncolytic adenoviral agents to the epidermal growth factor pathway with a secretory fusion molecule. Cancer Res. 2001;61:6377–6381. [PubMed] [Google Scholar]
- 25.Kanerva A, Mikheeva GV, Krasnykh V, Coolidge CJ, Lam JT, Mahasreshti PJ, et al. Targeting adenovirus to the serotype 3 receptor increases gene transfer efficiency to ovarian cancer cells. Clin Cancer Res. 2002;8:275–280. [PubMed] [Google Scholar]
- 26.Kanerva A, Wang M, Bauerschmitz GJ, Lam JT, Desmond RA, Bhoola SM, et al. Gene transfer to ovarian cancer versus normal tissues with fiber-modified adenoviruses. Mol Ther. 2002;5:695–704. doi: 10.1006/mthe.2002.0599. [DOI] [PubMed] [Google Scholar]
- 27.Grill J, Lamfers ML, van Beusechem VW, Dirven CM, Pherai DS, Kater M, et al. The organotypic multicellular spheroid is a relevant three-dimensional model to study adenovirus replication and penetration in human tumors in vitro. Mol Ther. 2002;6:609–614. [PubMed] [Google Scholar]
- 28.Lamfers ML, Grill J, Dirven CM, Van Beusechem VW, Geoerger B, Van Den Berg J, et al. Potential of the conditionally replicative adenovirus Ad5-Delta24RGD in the treatment of malignant gliomas and its enhanced effect with radiotherapy. Cancer Res. 2002;62:5736–5742. [PubMed] [Google Scholar]
- 29.Ryu R, Shin Y, Choi JW, Min W, Ryu H, Choi CR, et al. Depletion of intracellular glutathione mediates zinc-induced cell death in rat primary astrocytes. Exp Brain Res. 2002;143:257–263. doi: 10.1007/s00221-001-0991-7. [DOI] [PubMed] [Google Scholar]
- 30.Coley HM, Lewandowicz G, Sargent JM, Verrill MW. Chemosensitivity testing of fresh and continuous tumor cell cultures using lactate dehydrogenase. Anticancer Res. 1997;17:231–236. [PubMed] [Google Scholar]
- 31.Wood KV. Recent Advances and Prospects for Use of Beetle Luciferase as Genetic Reports. John Wiley and Sons; Chichester: 1991. p. 11. [Google Scholar]


