Abstract
The vast majority of mitochondrial proteins are imported from the cytosol. For matrix-localized proteins, the final step of translocation across the inner membrane is mediated by the mitochondrial translocation motor, of which mhsp70 is a key component. The ATP-dependent function of mhsp70 is regulated by a complex, composed of a J-protein (called Pam18 or Tim14) and a J-like protein (called Pam16 or Tim16), and the nucleotide exchange factor Mge1. In this study, we investigated the structural properties of a recombinant purified Pam18/Tim14–Pam16/Tim16 complex using cross-linking with the bifunctional reagent DSS and CD-spectroscopy. The results of the study show that both Pam18/Tim14 and Pam16/Tim16 are thermally unstable proteins that unfold at very low temperatures (Tm values of 16.5°C and 29°C, respectively). Upon mixing the proteins in vitro, or when both proteins are co-overexpressed in bacteria, Pam18/Tim14 and Pam16/Tim16 form a heterodimer that is thermally more stable than the individual proteins (Tm = 41°C). Analysis of the properties of the complex in GdnHCl shows that dissociation of the heterodimer is the limiting step in achieving full denaturation.
Keywords: mitochondrial import, translocation motor, Pam18/Tim14, Pam16/Tim16
The import of nuclear-encoded proteins into the mitochondria is a multistep process that is mediated by the coordinated action of translocation machineries localized in both the outer and inner mitochondrial membranes. In the outer membrane, the multimeric TOM complex serves as both a receptor for recognition of mitochondrial precursor proteins and a main portal of entry for proteins into mitochondria (Pfanner and Chacinska 2002; Mokranjac and Neupert 2005; Rapaport 2005). On their way to the matrix, proteins that contain cleavable N-terminal targeting signals are transferred from the TOM complex to the TIM23 preprotein translocase. This complex is composed of a core of two multispanning integral inner membrane proteins: Tim17 and Tim23. The latter forms a channel, which allows proteins to be integrated into or to cross the inner membrane. A third protein, Tim50, seems to serve as a sorting receptor in the mitochondrial intermembrane space and regulates the pore formed by Tim23 (Meinecke et al. 2006). Import of proteins into the matrix requires cooperation between the TIM23 and TOM complexes, which form the so-called translocation contact sites. Tim23 and Tim50 were proposed to be involved in the formation of these contact sites, and recently it was suggested that a new component of the TIM23 complex, Tim21, may also contribute to their formation (Endo et al. 2003; Chacinska et al. 2005; Mokranjac et al. 2005).
A precursor protein in transit across the Tim23 channel requires additional help in order to be imported completely into the mitochondrial matrix. This step is catalyzed by the translocation motor that is formed by the ATP-hydrolyzing 70-kDa heat-shock protein (mhsp70), its J-domain-containing cochaperones Tim14 (also called Pam18) and Tim16 (also called Pam16), and the nucleotide exchange factor Mge1 (Bolliger et al. 1994; Westermann et al. 1995). A further component, Tim44, is suggested to associate transiently with the TIM23 complex and to recruit mhsp70 to the import channel in a nucleotide-dependent manner (Kronidou et al. 1994; Rassow et al. 1994; Berthold et al. 1995; Weiss et al. 1999). Mge1 and the Tim14(Pam18)/Tim16(Pam16) complex most likely modulate the ATP-dependent function of the motor (Stojanovski et al. 2006).
Both Pam18/Tim14 and Pam16/Tim16 are essential for the proper import of proteins into the mitochondrial matrix. Consequently, deletion of either protein is lethal in yeast (D'Silva et al. 2003; Mokranjac et al. 2003; Frazier et al. 2004; Kozany et al. 2004). Studies in isolated mitochondria demonstrated that Pam18/Tim14 and Pam16/Tim16 associate in the mitochondria to form a hetero-oligomer, which is detectable using cross-linking and immunoprecipitation (Frazier et al. 2004; Kozany et al. 2004). Both Pam18/Tim14 and Pam16/Tim16 contain an N-terminal hydrophobic domain that is suggested to play a role in their association with the inner membrane. Additionally, both proteins expose large parts of their structure to the matrix side of the mitochondria (D'Silva et al. 2003; Mokranjac et al. 2003; Frazier et al. 2004; Kozany et al. 2004). The matrix-localized domain of Pam18/Tim14 is a J domain, which plays a role in its association with mhsp70. It is suggested that the role of the J domain is to regulate the function of mhsp70 (Mokranjac et al. 2003; Truscott et al. 2003). Pam16/Tim16 contains a J-like domain that is suggested to play an antagonistic role in Pam18/Tim14 function (Li et al. 2004). Further in vitro studies using the purified recombinant matrix-localized domains of both Pam18/Tim14 and Pam16/Tim16 demonstrated that both proteins form heterodimeric oligomers in solution. It was also demonstrated that the formation of the heterodimers is essential for the function of both proteins in vivo (D'Silva et al. 2005).
In this study, we have purified the recombinant soluble domains of Pam18/Tim14, Pam16/Tim16, and the Pam18/Tim14–Pam16/Tim16 complex after their overexpression in bacteria. The purified proteins were studied using cross-linking and circular dichroism (CD) spectroscopy. We show that Pam18/Tim14 and Pam16/Tim16, individually, are only marginally stable proteins that denature readily at low temperatures. However, the complex formed between the two proteins is much more stable. We also show that dissociation of the Pam18/Tim14–Pam16/Tim16 complex is the limiting step for denaturation of the hetero-oligomer.
Results
Protein purification
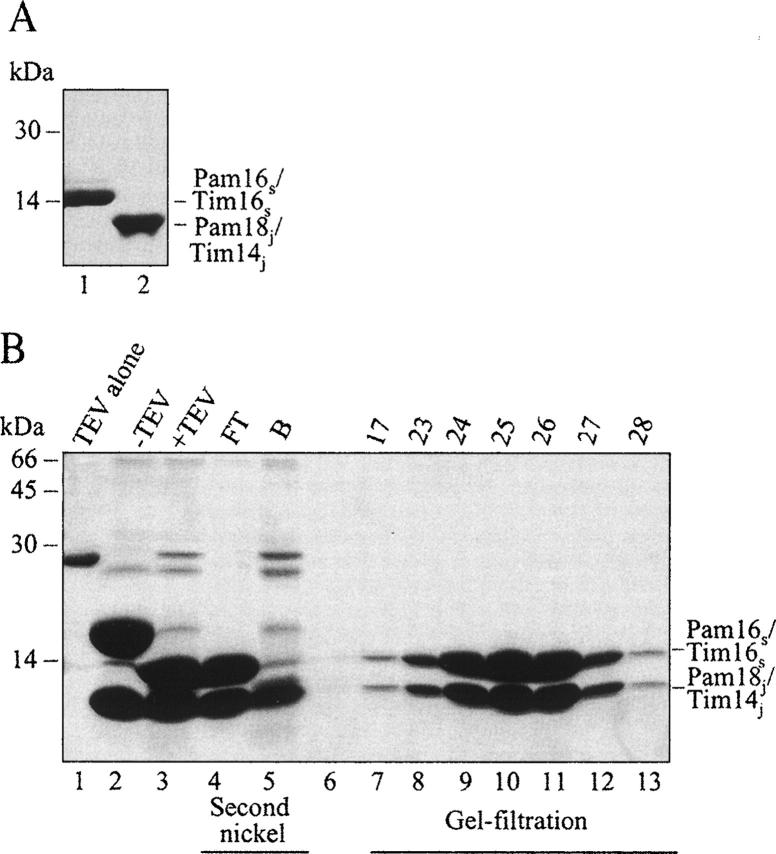
For the purpose of this study, we sought to purify the soluble domains of Pam18/Tim14 and Pam16/Tim16 that are relevant to their interaction (D'Silva et al. 2005). The hydrophobic regions of these proteins may play a role in the correct positioning of the complex next to the import channel and are not expected to play a role in complex formation (D'Silva et al. 2005). To this end, we have cloned Pam16/Tim16, lacking the first 24 amino acids, which are highly hydrophobic (Li et al. 2004), and the 19-amino acid C-terminal segment, which seems to constitute a low-complexity region. The latter is predicted to form an unordered segment in the secondary structure of the protein (Wootton and Federhen 1996). This recombinant protein is termed Pam16s/Tim16s. The putative J domain of Pam18/Tim14 (Pam18j/Tim14j) was also cloned. The latter contains the soluble matrix-localized domain of Pam18/Tim14 (amino acids 84–169). Both proteins were also overexpressed in bacteria, either individually or together in the same bacteria. All proteins contain an octa histidine tag, which enabled a first step of purification on Ni-agarose. As a second step, the histidine tag was removed using the TEV protease, and this step was followed by a second Ni column. Finally, the proteins were purified by gel filtration. As demonstrated in the representative gel (Fig. 1), the proteins are >95% pure.
Figure 1.
SDS-PAGE analysis of purified recombinant proteins. Purified proteins were analyzed using 16% SDS-PAGE and stained with Coomassie blue; 2.5 μg of the purified recombinant Pam16s/Tim16s (A, lane 1) and Pam18j/Tim14j (A, lane 2) were analyzed in each lane. (B) Purification steps of the Pam18j/Tim14j–Pam16s/Tim16s complex. (Lane 2) Proteins eluted after first Ni-agarose column. (Lane 3) The purified complex after digestion with the TEV protease. (Lane 4) Material that did not bind a second nickel column (primarily the Pam18j/Tim14j–Pam16s/Tim16s complex). (Lane 5) Proteins that bound to second Ni column: TEV and uncleaved proteins still containing the histidine tag. (Lanes 7–13) Fractions (4 mL each) eluted from the Superdex200 gel filtration column. The fraction number is indicated at the top of the gel. The Mw of the molecular-weight markers are indicated to the left of the gel.
The oligomeric state of recombinant Pam16s/Tim16s, Pam18j/Tim14j, and their complex
A previous study carried out using gel filtration concluded that the soluble domains of Pam18/Tim14 and Pam16/Tim16 individually can each form homodimers, while together Pam18/Tim14 with Pam16/Tim16 will prefer to assemble into a heterodimer. We therefore, first examined the oligomeric state of the purified proteins using cross-linking with the bifunctional reagent DSS (Fig. 2). Pam18j/Tim14j was exposed to DSS for increasing times, and the cross-linking products were analyzed using SDS-PAGE. When Pam18j/Tim14j was exposed to DSS for increasing times, one main cross-linking product was observed (Fig. 2A). This product migrates faster than the 30-kDa molecular-weight marker, suggesting that this product is a homodimer of Pam18j/Tim14j. Interestingly, at longer exposure times, the intensity of the stain on the monomer and dimer decreased (1 h and 2 h) (Fig. 2A). The decrease in the intensity of the bands suggests that the oligomer of Pam18j/Tim14j has a tendency to aggregate even to forms that do not enter the gel. Overall, the cross-linking pattern presented in Figure 2A is typical of a protein that is present in solution as a mixture of monomers and dimers, which tend to aggregate.
Figure 2.
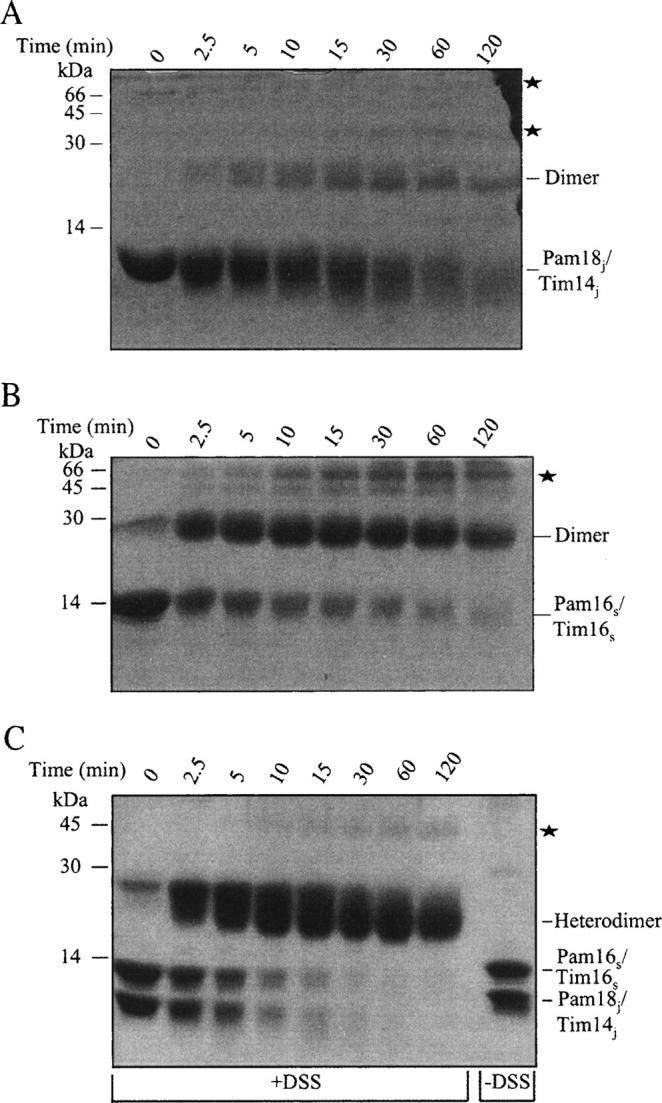
Analysis of the oligomeric state of the purified proteins using cross-linking. The proteins (1 mg/mL) were subjected to cross-linking at room temperature, using 1 mM of DSS, for the times indicated at the top of the gel. The cross-linking products (10 μg in each lane) were analyzed by 16% SDS-PAGE and stained with Coomassie blue. (A) Cross-linking products of Pam18j/Tim14j. (B) Cross-linking products of Pam16s/Tim16s. (C) Cross-linking products of the Pam18j/Tim14j–Pam16s/Tim16s complex. ★ indicates cross-linking products with mobility lower than that of a dimer (these present probably aggregates that entered the gel).
We also subjected Pam16s/Tim16s to cross-linking using DSS (Fig. 2B). The cross-linking pattern is similar to that of Pam18j/Tim14j, with some obvious differences as follows: (1) The intensity of the dimer is stronger for the Pam16s/Tim16s, and (2) the decrease in dimer intensity over time is less pronounced for Pam16s/Tim16s compared with Pam18j/Tim14j. We conclude that Pam16s/Tim16s forms mainly dimers in solution, which have less of a tendency to aggregate compared with Pam18j/Tim14j.
The oligomeric state of the purified Pam18j/Tim14j–Pam16s/Tim16s complex was also studied using cross-linking (Fig. 2C). Exposure of the complex to DSS leads to the formation of a cross-linking product that migrates faster than the 30-kDa marker. This result confirms the previously reported results that Pam18j/Tim14j forms heterodimers upon association with Pam16s/Tim16s (D'Silva et al. 2005). In contrast to the individual proteins (Fig. 2A,B), less protein aggregated when the complex was cross-linked, as judged from the almost constant intensity of the stain in the dimeric bands at the longer exposure times to cross-linker (Fig. 2C).
The thermal stability of purified recombinant Pam16s/Tim16s and Pam18j/Tim14j
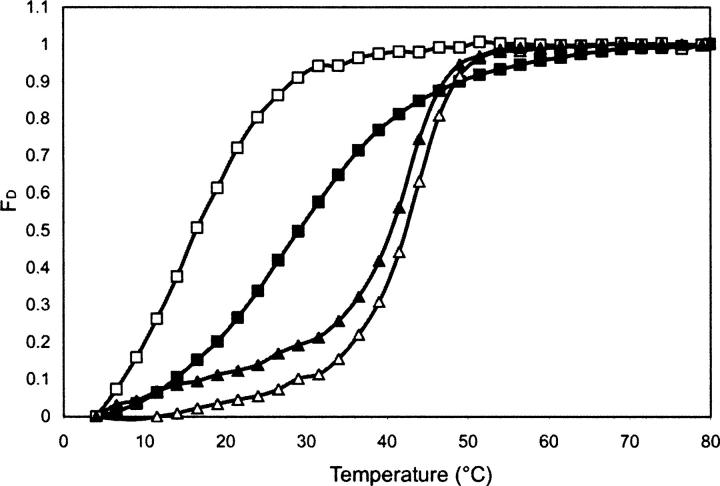
The cross-linking results presented above raise the possibility that the individual proteins are much less stable than the heterologous complex. To examine this possibility, we studied the thermal stability of the purified proteins using CD spectroscopy (Fig. 3). Thermal unfolding midpoints were determined by monitoring changes in secondary structure content as detected by CD spectroscopy at 222 nm. We monitored the ellipticity at 222 nm, since a characteristic helical minimum was observed at this wavelength for both the individual proteins and the heterologous complex (Fig. 4; data not shown). Notably, recombinant Pam18j/Tim14j and Pam16s/Tim16s start to denature at a relatively low temperature (∼10°C) with unfolding midpoints of 16.5°C and 29°C, respectively (Fig. 3). In contrast, the purified Pam18j/Tim14j–Pam16s/Tim16s complex that was obtained by co-overexpression of the two proteins together in bacteria was much more stable than were the individual proteins, with a Tm of ∼41°C. Similarly, significant stability (Tm = 40°C) was also observed when the complex was formed by mixing the individual proteins as described previously (D'Silva et al. 2005). A Tm of 41°C was also determined for the complex of the full-length Pam18/Tim14–Pam16/Tim16, which suggests that removal of the N-terminal hydrophobic segments does not affect the stability of the dimers (data not shown).
Figure 3.
Examination of the thermal stability of purified proteins using CD spectroscopy. The fraction of denatured protein (FD) was obtained by following the changes in the ellipticity at 222 nm at various temperatures. 0.1 mg/mL Pam18j/Tim14j (□), 0.1 mg/mL Pam16s/Tim16s (▪), and Pam18j/Tim14j at 0.05 mg/mL (▴) was mixed with Pam16s/Tim16s at 0.05 mg/mL and incubated for 30 min at 4°C prior to analysis. (▵) Pam18j/Tim14j–Pam16s/Tim16s complex at 0.1 mg/mL.
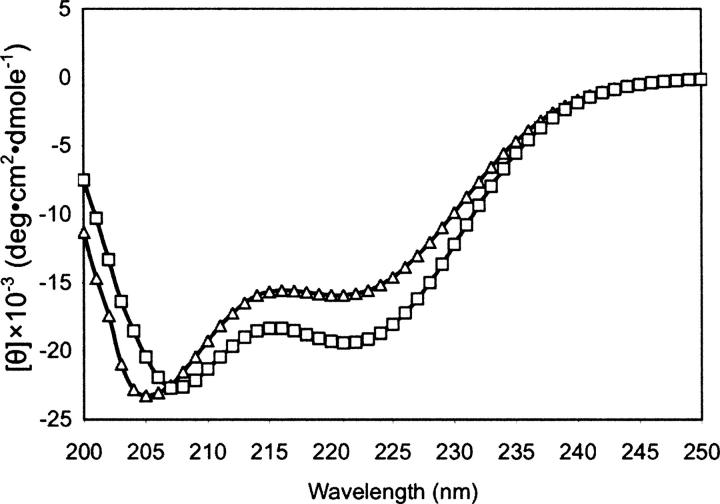
Figure 4.
Changes in dichroism upon formation of a complex between Pam18j/Tim14j and Pam16s/Tim16s. Mean residue ellipticities (θ) were plotted as a function of wavelength (nm). Buffer containing 0.025 mg/mL Pam16s/Tim16s was placed in one compartment of a tandem cuvette, and buffer containing a similar concentration of Pam18j/Tim14j was placed in the other. CD spectra were collected before (▵) and after (□) mixing the components of the two compartments.
To gain more insight into this phenomenon, we analyzed the secondary structures of Pam16s/Tim16s and Pam18j/Tim14j alone and after complex formation by using CD spectroscopy. Pam16s/Tim16s was placed in one cell of a tandem cuvette, while an equivalent concentration of Pam18j/Tim14j was placed in the second. A spectrum was obtained before and after mixing the contents of the two cells, allowing detection of changes in spectra that resulted from molecular interactions between the components of both chambers. A significant difference between the spectra was observed before and after mixing the chambers (Fig. 4). The difference is reflected by an increase in the negative ellipticity of the signal at 222 nm, which is characteristic of an increase in α-helical content. We conclude that both Pam18/Tim14 and Pam16/Tim16 are thermally very unstable proteins, which upon forming heterodimers undergo a conformational change that renders them considerably more stable.
The pattern of GdnHCl-induced unfolding of the Pam18j/Tim14j–Pam16s/Tim16s heterodimer
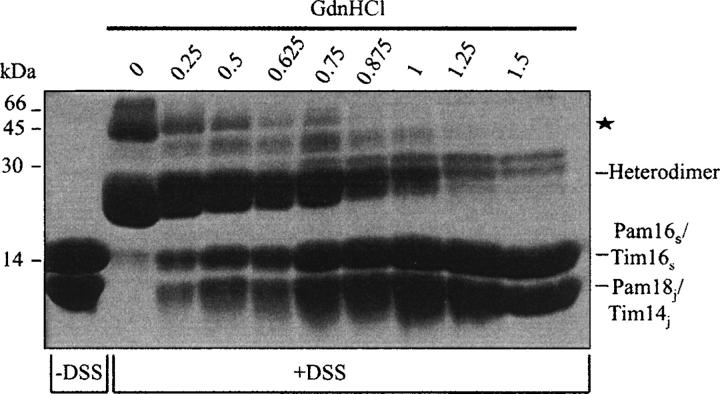
In order to obtain more information on the stabilizing effect of dimer formation, we investigated the pattern of unfolding of the complex following treatment with GdnHCl. For this purpose, we examined the ellipticity at 222 nm after incubation of the Pam18j/Tim14j–Pam16s/Tim16s complex with increasing GdnHCl concentrations. A biphasic change in the ellipticity of the complex is observed with increasing GdnHCl concentrations (Fig. 5A). The first phase (∼20% unfolding) is slight and is observed at a GdnHCl concentration of up to 0.5 M. When the concentration of GdnHCl is increased above 0.5 M, there is a profound increase in ellipticity that reaches its maximum at 1.25 M GdnHCl. This suggests that a major unfolding event occurs at GdnHCl concentrations of >0.5 M. We asked whether this unfolding event is limited by the dissociation of the dimers. To answer this question, we carried out a cross-linking analysis of the oligomeric state of the complex in the presence of increasing concentrations of GdnHCl (Fig. 6). An inverse correlation was obtained between the GdnHCl-dependent unfolding and the presence of dimers, as detected by cross-linking. At low GdnHCl concentrations, little dissociation of the Pam18j/Tim14j–Pam16s/Tim16s complex is observed, and most of the protein exists in heteromeric form. At GdnHCl concentrations >0.5 M, we observe a strong dissociation of the dimers. A maximal and almost complete dissociation of the complex is seen at GdnHCl concentrations >1 M. At these concentrations, significant unfolding of the proteins was also observed. These results suggest that the dissociation of the dimer is the limiting step for the unfolding process of the Pam18j/Tim14j–Pam16s/Tim16s complex. Thus, the Pam18j/Tim14j–Pam16s/Tim16s dimer undergoes a two-state denaturation process. Upon treatment of the dimer with 2.5 M GdnHCl and subsequent dilution of the denaturant to 0.5 M, the signal at 222 nm was fully restored.
Figure 5.
Denaturation of the Pam18j/Tim14j–Pam16s/Tim16s complex as a function of GdnHCl concentration. (A) Denaturation was monitored by far-UV CD spectroscopy at 222 (nm). CD spectra were recorded at 20°C following a 1-h incubation of the complex (1 mg/mL) at room temperature in buffer containing the indicated GdnHCl concentration. (B) As in A in the presence of the indicated urea concentrations.
Figure 6.
Cross-linking of the Pam18j/Tim14j–Pam16s/Tim16s complex in the presence of increasing concentrations of GdnHCl. Complex (1 mg/mL) was incubated at room temperature for 1 h in PBS containing the indicated GdnHCl concentrations and then subjected to cross-linking with 1 mM DSS for 30 min. −DSS indicates complex without cross-linker; ★ indicates cross-linking products with mobility lower than that of a dimer.
The denaturation of the complex was also examined in the presence of increasing urea concentrations (Fig. 5B). The complex was fully denatured at 3 M urea. When the Pam18j/Tim14j–Pam16s/Tim16s complex was incubated with 4 M urea, and subsequently diluted to 0.8 M, the signal at 222 nm was restored. Thus the denaturation of the complex in both GdnHCl and urea is reversible. The ΔG for the denaturation of the complex in the presence of GdnHCl and urea was determined as described by Need and Timm (1994), and was found to be 9.1 and 10.8 kcal/mol, respectively. ΔG values for the conformational stability of dimeric proteins that undergo a two-state denaturation were estimated to be in the range of 9.5–27.8 kcal/mol (Need and Timm 1994). Thus, despite profound thermal stabilization of the Pam18/Tim14–Pam16/Tim16 complex relative to the individual proteins, its conformational stability in the presence of GdnHCl and urea is still low compared with other dimeric proteins (Need and Timm 1994).
Discussion
Pam18/Tim14 and Pam16/Tim16 are two essential proteins for the function of the mitochondrial protein import translocation motor. By use of immunoprecipitation and in organelle cross-linking, it was shown that these proteins form a hetero-oligomer in the matrix of mitochondria (Frazier et al. 2004; Kozany et al. 2004). In vitro studies using purified recombinant Pam18j/Tim14j and Pam16s/Tim16s showed that the purified proteins form a heterodimer when they are mixed in solution (D'Silva et al. 2005). In this work, we used chemical cross-linking and CD spectroscopy to study the structural stability of Pam18j/Tim14j and Pam16s/Tim16s both individually and in complex. Using cross-linking with DSS, we confirmed that a heterodimer is formed when Pam18/Tim14 and Pam16/Tim16 associate. CD analysis showed that, in contrast to the great instability of the individual proteins (Tm of 16.5°C and 29°C for Pam18j/Tim14j and Pam16s/Tim16s, respectively), the Pam18/Tim14–Pam16/Tim16 complex is much more stable (Tm ∼ 41°C). This finding was also supported by the observation that the GdnHCl-induced loss of complex secondary structure (CD) correlated with dissociation of heterodimers. Pam18/Tim14 and Pam16/Tim16 represent a unique case in which the individual proteins can form homodimers but unfold readily at physiological temperatures. When both proteins are mixed, the equilibrium is shifted toward the formation of very thermally stable heterodimers. Interestingly, despite profound thermal stabilization upon the formation of the Pam18/Tim14–Pam16/Tim16 complex, the conformational stability of the complex as calculated by equilibrium denaturation (9.1 and 10.8 kcal/mol in GdnHCl and urea, respectively) is in the low range of ΔG that is known for dimeric proteins (9.5–27.8 kcal/mol) (Need and Timm 1994).
The normal growth temperature of yeast is 30°C, and higher temperatures are considered to be heat shock. Thus, any dissociation of the Pam18/Tim14–Pam16/Tim16 complex in vivo would theoretically lead to their denaturation. It is therefore tempting to speculate that the Pam18/Tim14–Pam16/Tim16 complex prefers the formation of stable hetero-oligomers in vivo and exerts a regulatory function on the translocation motor through a conformational change, rather than by dissociation to monomers.
Materials and methods
Cloning of Pam18j/Tim14j
The nucleotide sequence encoding amino acid residues 84–169 of Saccharomyces cerevisiae Pam18/Tim14 was cloned by PCR (for primers used, see Table 1) from a yeast DNA genomic library, between restriction sites for BamHI (forward primer-I) and XhoI (backward primer-II). The PCR product was subcloned into the pGEM-T easy plasmid and sequenced to rule out PCR-induced errors. The resulting construct was cut and cloned into a modified pET21d that contained a TEV protease site between the protein and the histidine tag. The engineered plasmid overexpresses Pam18j/Tim14j containing an octa histidine tag at its N terminus that can be removed after treatment with TEV protease. The BL21 tuner Escherichia coli strain (Novagen) was used as an overexpression host in all constructs used in this study.
Table 1.
Primers used in this study
Cloning of Pam16s/Tim16s
The nucleotide sequence encoding amino acid residues 25–130 of S. cerevisiae Pam16/Tim16 was cloned by PCR between BamHI (forward primer-III) and NotI (backward primer-IV) restriction sites. The rest of the procedure was similar to the Pam18j/Tim14j cloning procedure described above.
Cloning of the Pam18j/Tim14j–Pam16s/Tim16s complex
The nucleotide sequence encoding amino acid residues 84–169 of S. cerevisiae Pam18/Tim14 was cloned by PCR from a yeast DNA genomic library between NdeI (forward primer-V) and XhoI (backward primer-II) restriction sites. The nucleotide sequence encoding amino acid residues 25–130 of S. cerevisiae Pam16/Tim16 was cloned to contain BamHI and NotI restriction sites as described in the previous section. Each of the PCR products was subcloned in pGEM-T easy and sequenced to rule out PCR-induced sequence errors. The resulting two constructs were cut and ligated into a modified pETDuet vector. The resulting plasmid co-overexpresses Pam16s/Tim16s, containing an octa hisitidine tag at its N terminus, and Pam18j/Tim14j. The histidine tag is removable by digestion with TEV protease.
Purification of Pam18j/Tim14j and, Pam16s/Tim16s individually and as a complexs
Bacteria were grown for 3–4 h at 37°C in 3 L of LB media, containing 100 μg/mL ampicillin and 34 μg/mL chloramphenicol. Upon reaching an A600 of 0.3, the temperature was lowered to 16°C, and growth was continued until the culture reached an A600 of 0.6. Then, protein expression was induced with 1 mM of IPTG. Cells were harvested after 16 h and stored at −20°C until purification. About 12 g of cells were suspended in 120 mL of buffer A (buffer A: 400 mM NaCl, 1% Triton X-100, 20 mM imidazole, 50 mM Tris-HCl at pH 7.4) containing 45 mg/mL of DNAase, 2 mM phenylmethylsulfonyl fluoride, and three tablets of complete EDTA-free protease inhibitor cocktail (Roche). After lysis of cells by a microfluidizer, debris was removed by centrifugation for 1 h at 15,000 rpm in a Sorvall SS-34 rotor. The soluble fraction was loaded on a Ni-NTA-agarose column (∼20 mL) pre-equilibrated with buffer A. The column was washed with three volumes of buffer A without Triton until a stable baseline was achieved. Fractions were eluted with a linear gradient (90 mL) of buffer B (buffer A containing 1 M imidazole). Fractions enriched with the protein were pooled and digested with TEV protease (TEV:protein 1:50 w/w) for 12 h at 4°C, in dialysis tubular membrane against 3 L of buffer A. After the digestion, the protein was passed over a second 5 mL nickel column to remove (1) the TEV protease, which itself contains an uncleavable octa histidine tag; (2) uncleaved proteins still containing a histidine tag; and (3) nonspecifically bound proteins. Fractions enriched with the desired protein were concentrated and loaded onto a HiLoad 16/60 Superdex 200 column (120 mL total bead volume, Amersham) pre-equilibrated with PBS buffer (pH 7.4) at a flow rate of 1 mL/min (or 400 mM NaCl and 50 mM Tris-HCl at pH 7.4) for cross-linking of proteins as in Fig. 2. The fractions were collected and concentrated using Vivaspin protein concentrators (5000 Mw cutoff, Vivascience), divided into aliquots, and flash frozen in liquid N2. All purification procedures were carried out at 4°C.
Cross-linking of proteins
Cross-linking of the individual proteins and of the complex (Fig. 2) was carried out with 1 mM of DSS at room temperature in 20 mM of Na-HEPES (pH 7.4), containing 100 mM of KCl, at a protein concentration of 0.5 mg/mL. The cross-linking reaction was stopped at different times by the addition of SDS-containing sample buffer and boiling for 5 min. The cross-linking products (20 μL) were analyzed by 16% SDS-PAGE.
Cross-linking in the presence of GdnHCl
Cross-linking was carried out essentially as described (Guo et al. 2004) with minor modifications. Pam18j/Tim14j–Pam16s/Tim16s complex, 1 mg/mL, was incubated for 1 h at room temperature in PBS buffer containing varying concentrations of GdnHCl. Cross-linking was initiated by the addition of 1 mM DSS, and the sample was incubated for 30 min. Cross-linking was stopped by the addition of 96 μL of 200 mM glycine and incubated for 5 min on ice. Then, 200 μL of DDW, 9 μL of 10% deoxycholic acid solution, and 21.6 μL of 100% TCA were added, followed by an additional 5-min incubation. Samples were centrifuged at 14,000 rpm for 10 min, and the pellet was redissolved in 50 μL of SDS-containing sample buffer. When it was required, TRIS base was added to neutralize the acidic pH. Cross-linking products were analyzed by 14% SDS-PAGE.
Circular dichroism
All CD measurements were performed with an Aviv CD spectrometer model 202. Each spectrum is an average of five scans. The raw data were corrected by subtracting the contribution of the buffer to the CD signal. Data were smoothed and converted to molar ellipticity units.
For chemical denaturation experiments, all spectra were measured between 260 and 190 nm following a 1-h incubation with GdnHCl. For all measurements, a cell with a 0.1-mm path length was used. The measurements were taken at a constant temperature of 20°C and a protein concentration of 1 mg/mL.
Temperature-dependent denaturation studies were performed in a 1-mm path-length quartz cuvette at a sample concentration of 0.1 mg/mL. Melting data were collected at 2.5°C intervals, with a 10-sec averaging time and a 2.5-min equilibration time per data point.
For binding (mixing) experiments (Fig. 4), spectra were measured in tandem quartz cells with path lengths of 2 × 4.37 mm. Buffer containing 0.025 mg/mL protein was placed in each compartment. Measurements were taken at 4°C. The samples were equilibrated for 15 min after mixing, to let the folding processes take place.
Miscellaneous
The concentrations of Pam16s/Tim16s and of the Pam18j/Tim14j–Pam16s/Tim16s complex were obtained using the predicted extinction coefficient of the proteins at 280 nm. For both, the molar extinction coefficient (M−1cm−1) at 280 nm was 11,460. The concentration of Pam18j/Tim14j, which lacks Trp, Tyr, or Cys residues, was determined using the Bicinchoninic Acid Protein Assay (Sigma B9643) with BSA as a standard.
Acknowledgments
We thank Dr. Celeste Weiss for useful discussions. This work is supported by the German-Israeli Foundation (grant no. 753/181) and the German-Israeli Project Cooperation (DIP, F5.1).
Footnotes
Reprint requests to: Abdussalam Azem, Department of Biochemistry, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv 69778, Israel; e-mail: Azema@tauex.tau.ac.il; fax: 972-3-640-6834.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062459607.
References
- Berthold, J., Bauer, M.F., Schneider, H.C., Klaus, C., Dietmeier, K., Neupert, W., and Brunner, M. 1995. The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ATP driving system. Cell 81: 1085–1093. [DOI] [PubMed] [Google Scholar]
- Bolliger, L., Deloche, O., Glick, B.S., Georgopoulos, C., Jeno, P., Kronidou, N., Horst, M., Morishima, N., and Schatz, G. 1994. A mitochondrial homolog of bacterial GrpE interacts with mitochondrial hsp70 and is essential for viability. EMBO J. 13: 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska, A., Lind, M., Frazier, A.E., Dudek, J., Meisinger, C., Geissler, A., Sickmann, A., Meyer, H.E., Truscott, K.N., and Guiard, B., et al. 2005. Mitochondrial presequence translocase: Switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120: 817–829. [DOI] [PubMed] [Google Scholar]
- D'Silva, P.D., Schilke, B., Walter, W., Andrew, A., and Craig, E.A. 2003. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. 100: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva, P.R., Schilke, B., Walter, W., and Craig, E.A. 2005. Role of Pam16’s degenerate J domain in protein import across the mitochondrial inner membrane. Proc. Natl. Acad. Sci. 102: 12419–12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, T., Yamamoto, H., and Esaki, M. 2003. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J. Cell Sci. 116: 3259–3267. [DOI] [PubMed] [Google Scholar]
- Frazier, A.E., Dudek, J., Guiard, B., Voos, W., Li, Y., Lind, M., Meisinger, C., Geissler, A., Sickmann, A., and Meyer, H.E., et al. 2004. Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 11: 226–233. [DOI] [PubMed] [Google Scholar]
- Guo, Q., Zhao, F., Guo, Z., and Wang, X. 2004. Intermediates in the inactivation and unfolding of dimeric arginine kinase induced by GdnHCl. J. Biochem. 136: 49–56. [DOI] [PubMed] [Google Scholar]
- Kozany, C., Mokranjac, D., Sichting, M., Neupert, W., and Hell, K. 2004. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 11: 234–241. [DOI] [PubMed] [Google Scholar]
- Kronidou, N.G., Oppliger, W., Bolliger, L., Hannavy, K., Glick, B.S., Schatz, G., and Horst, M. 1994. Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc. Natl. Acad. Sci. 91: 12818–12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Dudek, J., Guiard, B., Pfanner, N., Rehling, P., and Voos, W. 2004. The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J. Biol. Chem. 279: 38047–38054. [DOI] [PubMed] [Google Scholar]
- Meinecke, M., Wagner, R., Kovermann, P., Guiard, B., Mick, D.U., Hutu, D.P., Voos, W., Truscott, K.N., Chacinska, A., and Pfanner, N., et al. 2006. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312: 1523–1526. [DOI] [PubMed] [Google Scholar]
- Mokranjac, D. and Neupert, W. 2005. Protein import into mitochondria. Biochem. Soc. Trans. 33: 1019–1023. [DOI] [PubMed] [Google Scholar]
- Mokranjac, D., Sichting, M., Neupert, W., and Hell, K. 2003. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 22: 4945–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac, D., Popov-Celeketic, D., Hell, K., and Neupert, W. 2005. Role of Tim21 in mitochondrial translocation contact sites. J. Biol. Chem. 280: 23437–23440. [DOI] [PubMed] [Google Scholar]
- Need, K.E. and Timm, D.E. 1994. Conformational stability of dimeric proteins: Quantitative studies by equilibrium denatuaration. Protein Sci. 3: 2167–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner, N. and Chacinska, A. 2002. The mitochondrial import machinery: Preprotein-conducting channels with binding sites for presequences. Biochim. Biophys. Acta 1592: 15–24. [DOI] [PubMed] [Google Scholar]
- Rapaport, D. 2005. How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane? J. Cell Biol. 171: 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow, J., Maarse, A.C., Krainer, E., Kubrich, M., Muller, H., Meijer, M., Craig, E.A., and Pfanner, N. 1994. Mitochondrial protein import: Biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol. 127: 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski, D., Rissler, M., Pfanner, N., and Meisinger, C. 2006. Mitochondrial morphology and protein import-A tight connection? Biochim. Biophys. Acta 1763: 414–421. [DOI] [PubMed] [Google Scholar]
- Truscott, K.N., Voos, W., Frazier, A.E., Lind, M., Li, Y., Geissler, A., Dudek, J., Muller, H., Sickmann, A., and Meyer, H.E., et al. 2003. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, C., Oppliger, W., Vergeres, G., Demel, R., Jeno, P., Horst, M., de Kruijff, B., Schatz, G., and Azem, A. 1999. Domain structure and lipid interaction of recombinant yeast Tim44. Proc. Natl. Acad. Sci. 96: 8890–8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, B., Prip-Buus, C., Neupert, W., and Schwarz, E. 1995. The role of the GrpE homologue, Mge1p, in mediating protein import and protein folding in mitochondria. EMBO J. 14: 3452–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton, J.C. and Federhen, S. 1996. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 266: 554–571. [DOI] [PubMed] [Google Scholar]