Abstract
Rcd-1, a protein highly conserved across eukaryotes, was initially identified as a factor essential for nitrogen starvation-invoked differentiation in fission yeast, and its Saccharomyces cerevisiae homolog, CAF40, has been identified as part of the CCR4–NOT transcription complex, where it interacts with the NOT1 protein. Mammalian homologs are involved in various cellular differentiation processes including retinoic acid-induced differentiation and hematopoetic cell development. Here, we present the 2.2 Å X-ray structure of the highly conserved region of human Rcd-1 and investigate possible functional abilities of this and the full-length protein. The monomer is made up of six armadillo repeats forming a solvent-accessible, positively-charged cleft 21–22 Å wide that, in contrast to other armadillo proteins, stays fully exposed in the dimer. Prompted by this finding, we established that Rcd-1 can bind to single- and double-stranded oligonucleotides in vitro with the affinity of G/C/T ≫ A. Mutation of an arginine residue within the cleft strongly reduced or abolished oligonucleotide binding. Rcd-1’s ability to bind to nucleic acids, in addition to the previously reported protein–protein interaction with NOT1, suggests a new feature in Rcd-1’s role in regulation of overall cellular differentiation processes.
Keywords: Rcd-1, transcription, X-ray crystallography, structure
Cellular differentiation is an essential characteristic of cells required for the formation of multicellular organisms. However, many unicellular organisms, such as yeast, also differentiate in order to survive in hostile environments. Recent studies suggest that the mechanisms and factors involved in differentiation are not very cell type specific, let alone organism specific (Yamamoto et al. 1999; Hiroi et al. 2002).
Schizosaccharomyces pombe, a fission yeast, resembles higher eukaryotes in several ways including genetic structure, cell cycle control, and various signaling pathways. It also undergoes cellular differentiation during sexual development involving conjugation, meiosis, and sporulation. Sexual differentiation is controlled primarily through two signals: nutrient starvation and the presence of mating pheromone. Transcriptional factor Ste11 is required and activates several genes involved in conjugation and meiosis (Kelly et al. 1988; Watanabe et al. 1988; Hughes et al. 1990; Sugimoto et al. 1991; Willer et al. 1995). Included are the mei2, rep1, and ste genes (including ste11), and other mating-type genes (Kelly et al. 1988; Watanabe et al. 1988; Hughes et al. 1990; Sugimoto et al. 1991; Miyamoto et al. 1994; Sugiyama et al. 1994; Willer et al. 1995).
Nitrogen starvation as well as carbon starvation are two major nutrient-depletion signals triggering sexual development in budding yeast. Pat1/Ran1 kinase plays a unique role in the regulation of sexual development, primarily blocking the onset of meiosis until conjugation occurs by inactivating a key triggering factor, Mei2 (Beach et al. 1985; Iino and Yamamoto 1985a,b; Watanabe et al. 1997). Inactivation of Pat1 in haploid cells induces lethal meiosis that can be suppressed through inactivation of the mei2 gene. As Ste11 is required for the expression of mei2 (Sugimoto et al. 1991), factors inhibiting ste11 expression or function would rescue the pat1 inactivated cells.
Rcd1 (required for cell differentiation) was identified as a differentiation-controlling factor crucial for nitrogen starvation-induced sexual development in S. pombe by screening a genetic library for suppressors of pat1 lethality (Okazaki et al. 1998). Surprisingly, cells with nonfunctional rcd1 were also shown to be sterile, despite initial assumptions that these strains would have been highly proficient for sexual development. Rcd-1 is essential for nitrogen starvation-invoked induction of ste11 (Okazaki et al. 1998). Paradoxically, overexpression of Rcd-1 rescued pat1 inactivated cells, supposedly through the inhibiting activity of a dominant active mutant of Mei2 (Y. Watanabe and M. Yamamoto, unpubl.).
Homologs of the rcd1 gene from S. pombe are quite conserved throughout eukaryotic genomes (Fig. 1A; Okazaki et al. 1998). One homolog, CAF40, was identified as a component of the CCR4–NOT transcription complex in Saccharomyces cerevisiae (Chen et al. 2001). This complex is evolutionarily conserved in eukaryotes and is involved in initiation (Denis and Malvar 1990; Sakai et al. 1992; Collart and Struhl 1993), elongation (Denis et al. 2001), degradation (Tucker et al. 2001), and deadenylation (Tucker et al. 2001) of mRNA, as well as regulation of TFIID activity (Chen et al. 2002). In the CCR4–NOT complex, only NOT1 is essential (Collart and Struhl 1993; Maillet et al. 2000), although it has no identifiable functional domains, apparently acting as a scaffold. CAF40 associates with NOT1 through the latter's N-terminal 1100 residues, although its presence was not required for CCR4–NOT complex formation (Chen et al. 2001). As CAF40 has also been shown to interact with human NOT1, this suggests that hRcd-1 would be a part of the human CCR4–NOT complex (Chen et al. 2001).
Figure 1.

Rcd-1 sequences. (A) Alignment of the sequences of Rcd-1 from various organisms as indicated (GenBank accession nos.: Homo sapiens, NP_005435; S. pombe, NP_594984; Neurospora crassa, XP_327357; Rattus norvegicus, NM_001009357; Mus musculus, NP_067358; Gallus gallus, NP_001006521; Drosophila melanogaster, NP_728284; Caenorhabditis elegans, NP_498048; Arabidopsis thaliana, NP_196802; and S. cerevisiae, NP_014111). Identical residues across sequences are shaded in dark gray, while conserved residues are shaded in light gray (criteria defined by Clustal W). Hyphens indicate gaps. Secondary structural elements determined in the crystal structure are aligned with the corresponding sequence. (B) The sequences of the six armadillo repeat-like regions within human Rcd-1 are aligned according to their secondary structural elements. Conserved residues across species are shown in blue. The consensus sequence from β-catenin is shown at the bottom.
Mammalian Rcd-1 was investigated for its functional similarity to the yeast homologs using F9 mouse teratocarcinoma cell differentiation, which is regulated by retinoic acid (RA) (Hiroi et al. 2002). F9 cells differentiate into primitive endoderm upon treatment with RA (Hogan et al. 1981; Edwards and McBurney 1983). Inhibition of mouse Rcd-1 (mRcd-1) production through the use of antisense oligonucleotides effectively blocked RA-induced differentiation (Hiroi et al. 2002). As well, overexpression of mRcd-1-induced spontaneous differentiation, apparently sensitizing the F9 cells to rather low levels of RA (Hiroi et al. 2002).
Rcd-1 is highly expressed in various mammalian tissues. Antibodies against Rcd-1 were used to probe various tissues from adult and embryonic rat, showing the highest levels of expression in the lung, spleen, testes, and thymus of adult rats, while expressed in virtually all tissues of the early rat embryo (Hiroi et al. 2002). Northern blots using human Rcd-1 as a probe showed significant expression in human testes, ovaries, and thymus (Okazaki et al. 1998). mrcd1 was also isolated as an erythropoietin-responsive gene potentially involved in hematopoetic cell development (Gregory et al. 2000). A recent finding reports that mRcd-1 interacts with the c-Myb protein (Haas et al. 2004), which, in turn, is required for the generation of monocytes from a myeloid progenitor (for review, see Friedman 2002). These results are consistent with Rcd-1’s proposed involvement in differentiation control.
Despite the increasing knowledge about the role Rcd-1 plays in cellular differentiation, little is known about the mechanisms by which Rcd-1 may exert its influence. Structural information could provide more insight regarding its actual functioning within cells. We now report the crystal structure of the highly conserved region of human Rcd-1 (a region spanning 90% of the full-length protein). The armadillo-repeat-like structure in conjunction with interesting surface features and charge clustering suggest possible means by which Rcd-1 can mediate its effects on cellular differentiation-related gene control.
Results
Determination of the oligomerization state of the full-length and truncated versions of human and mouse Rcd-1
Following purification, recombinant human Rcd-1 (hRcd-1), hRcd-1 ΔN (residues 18–299), hRcd-1 ΔC (residues 1–285), hRcd-1 ΔNC (residues 18–285), and mouse Rcd-1 (mRcd-1) proteins were loaded in separate experiments onto a Superdex 200 column. hRcd-1 and mRcd-1 eluted within a single peak of ∼70 kDa, slightly larger than the estimated sizes of hRcd-1 and mRcd-1 dimers. On reducing SDS-PAGE gels, hRcd-1 and mRcd-1 ran as monomers showing bands ∼34 kDa in size, and no contaminating bands could be detected by Coomassie blue staining. hRcd-1 ΔN and hRcd-1 ΔC eluted from the column in fractions corresponding to a molecular mass of ∼63 kDa, close to the estimated size of the hRcd-1 ΔN or the hRcd-1 ΔC dimer. On reducing SDS-PAGE gels, peak fractions ran as monomers ∼32 kDa in size. hRcd-1 ΔNC eluted from the column in fractions corresponding to a molecular mass of ∼52.5 kDa, slightly smaller than the estimated size of an hRcd-1ΔNC dimer. On a reducing SDS-PAGE gel, peak fractions ran as monomers ∼29 kDa in size (data not shown). These results indicate that the stable solution forms of both full-length as well as truncated human and mouse Rcd-1 are dimers.
As an independent and more direct method to determine oligomerization state, analytical ultracentrifugation runs were performed on purified hRcd-1 and hRcd-1 ΔNC at three different concentrations. They, too, showed that both hRcd-1 and hRcd-1 ΔNC form dimers in solution (see Supplemental Fig. 1). The calculated molecular masses were 69.8 kDa and 62.0 kDa for hRcd-1 and hRcd-1 ΔNC, respectively. This is well within experimental error of the theoretical sizes of the respective dimers, 67.3 kDa for hRcd-1 and 61.0 kDa for hRcd-1 ΔNC.
During purification, the two R227E mutant versions of hRcd-1 and hRcd-1 ΔNC behaved identically to the corresponding wild-type versions, especially, their CD spectra and elution profiles from a gel chromatography column were identical (data not shown). This suggests that the point mutations had no effect on the overall structure.
Crystallization of native and Se-Met derivatives of hRcd-1, hRcd-1 ΔN, hRcd-1 ΔNC, and mRcd-1
Initial crystallization conditions were deduced from conditions found in the Hampton Crystal Screens I, II, and the Index Screen. In general, crystals formed in the hanging drops containing slightly acidic conditions in the presence of PEG 3350 and a divalent cation such as Ca2+, Mg2+, or Mn2+. Human and murine Rcd-1 crystals as well as their truncated versions were reproducibly grown in 100 mM MES (pH 5.7), 16%–18% PEG 3350, 115 mM MnSO4, and 1% glucose. Crystals typically reached dimensions of 0.4–0.8 × 0.2 × 0.1 mm in 3 d. The crystals were strongly birefringent but tended to grow in clusters or as visible twins.
Native hRcd-1 crystals belonged to space group P1 with cell axes a = 68.90 Å, b = 68.65 Å, c = 125.42 Å, α = 90.02°, β = 90.10°, and γ = 84.12° with an estimated eight monomers per asymmetric unit. However, data collected from these crystals proved problematic because of the poor quality of the diffraction patterns (limited resolution, anisotropic diffraction).
Native hRcd-1 ΔN crystals tended to be extremely clustered and twinned and therefore did not permit meaningful data collection.
Crystals of hRcd-1 ΔNC belonged to space group P1 with unit cell parameters a = 68.51 Å, b = 68.96 Å, c = 69.36 Å, α = 95.16°, β = 117.26°, and γ = 91.91° with four monomers per asymmetric unit. Table 1 summarizes the data collection statistics. The region comprising hRcd-1 ΔNC is identical for murine and human homologs. Se-Met-derivatized hRcd-1 ΔNC crystallized under the same conditions as native protein and also adopted space group P1. The crystal cell parameters were similar to those of the native hRcd-1 ΔNC: a = 68.55 Å, b = 69.98 Å, c = 69.43 Å, α = 94.57°, β = 117.11°, and γ = 92.39° with four monomers per asymmetric unit.
Table 1.
Data collection and refinement statistics for hRcd-1 ΔNC
Structure of hRcd-1 ΔNC
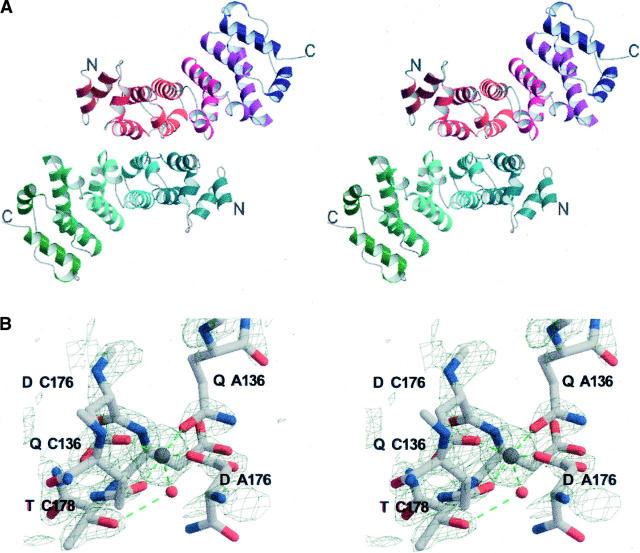
The crystal structure of human Rcd-1 truncated at its N and C termini (hRcd-1 ΔNC; residues 18–285) was derived using the Se-Met-based MAD technique (Fig. 2). The overall fold of the hRcd-1 ΔNC monomer is an 18 α-helix bundle with helices packed in a clockwise spiraling manner (Fig. 2A). The tertiary arrangement of helices shows high similarity with the armadillo (arm) repeat-containing karyopherin α (DALI Z-score of 18; Holm and Sander 1994). After refinement, the average RMSD between Cα atoms from each monomer is 1.18 Å (calculated using DeepView/Swiss-Pdb Viewer 3.7; Guex and Peitsch 1997). This is a rather large value for subunits of an oligomeric protein, pointing to flexibility within the structure. This finding could also explain why all crystals obtained were in space group P1. Also, when the Cα atoms of hRcd-1 ΔNC are superimposed on the closest fitting parts of the other structurally known arm-like proteins, the results are surprising. Aligning hRcd-1 (residues 120–252) with karyopherin α (residues 131–263), the RMSD between Cα atoms is 7.77 Å; the sequence similarity within this region spanning roughly three arm repeats is 0.060. Comparing similarly spanning regions in β-catenin (residues 272–404) to karyopherin α (residues 252–384) yields an RMSD of 3.34 Å, while plakophilin (residues 539–671) to karyopherin α (residues 260–392) has an RMSD of 7.56 Å; sequence similarities for these pairings were 0.170 and 0.174, respectively. Aligning roughly one arm repeat of hRcd-1 (residues 153–194) with karyopherin α (residues 164–205), the RMSD between Cα atoms is 1.13 Å; the sequence identity and similarity within this region are both 0.20. Taken together, this suggests that while the geometry of a single arm repeat is quite conserved, the pitch of the resultant superhelices generated by multiple arm repeat-containing proteins can vary by differing amounts as a result of the flexibility of this domain. As well, primary sequence similarities for particular arm repeat regions can vary widely, even within the same protein.
Figure 2.
Structure of hRcd-1 ΔNC. (A) Stereo ribbon diagram of the dimer. The twofold axis runs approximately horizontal to the page. Monomer A is colored from dark red to indigo (for helices), while monomer B is colored from dark blue to dark green. Individual arm three-helix repeats in each monomer are identified by a different color. N and C termini are labeled. (B) Stereo diagram showing the electron density (2F o-F c) around the Mn2+ ion found at the interface between the dimers within the unit cell; a 3σ cutoff is used to emphasize the metal density and tight coordination of the liganding atoms. The residue type and position from adjacent monomers are labeled. The electron density is shown in light gray. Residues from one of the dimers make up the left side of the image, while residues from the other dimer make up the right side of the image. An approximate twofold axis runs nearly vertically in the plane of the paper intersecting both the Mn2+ ion (gray sphere) and water molecule (red sphere).
Arm motifs describe a general fold and have been found in many functionally unrelated proteins from β-catenin to karyopherin α (Peifer et al. 1994; Conti et al. 1998). All these proteins, however, bind other proteins (Coates 2003). The specific tertiary structures of the arm motif are generated through differences in the positions and lengths of the helices. Like β-catenin and karyopherin α, the tight, repetitive packing of the Rcd-1 helices creates a continuous hydrophobic core extending throughout the structure. While structurally similar, there is poor conservation of sequence between the arm repeats of hRcd-1 and those of β-catenin or karyopherin α (see Fig. 1B). This sequence divergence explains the failure of primary sequence-based structural predictions to identify any motifs for Rcd-1. The similarity in the fold between karyopherin α and hRcd-1 ΔNC only extends to the monomers; the quaternary structures of the dimers are rather different. Karyopherin α monomers interact with each other near the ends of the arm repeat region, creating a central cavity within the homodimer. hRcd-1 ΔNC shows no hole in the center of the dimer, with monomers interacting on the back side of the “C” formed by the arm repeats.
The hydrophobic part of the surface of each monomer forms the dimer interface including Ala64, Leu67, Val71, Tyr74, Pro75, Pro79, Ala112, Ala113, His114, Leu117, and Phe118 and their corresponding partners from the other monomer. Interestingly, no hydrogen bonding is evident within the ∼950 Å2 dimer interface. This area is surrounded by hydrophilic residues such as Gln68, Asn72, Thr160, Thr161, and Glu162, which, however, do not contribute to the interface. This interface resides at an approximate twofold symmetry axis between the monomers. The inherent variability of helix orientation in each monomer and between dimers prevented use of noncrystallographic symmetry during refinement.
hRcd-1 ΔNC dimers pack together in the unit cell with the assistance of a Mn2+ ion. During crystallization trials, it was found that divalent cations such as Mg2+, Ca2+, and Mn2+ all resulted in visually similar crystal forms; trials set up using MnSO4 yielded the highest quality crystals. The Mn2+ ion achieves octahedral coordination with oxygens from Gln152 and Asp192 from both dimers; the fifth and sixth coordination positions are occupied by water molecules, one of which is further stabilized by Thr194 from one of the monomers (Fig. 2A). At the interface between unit cells where the dimers from one unit cell pack against dimers from the next, no electron densities that could correspond to additional Mn2+ ions were found. Given that the presence of a divalent cation was essential for crystal formation, this arrangement likely reflects a crystallization artifact.
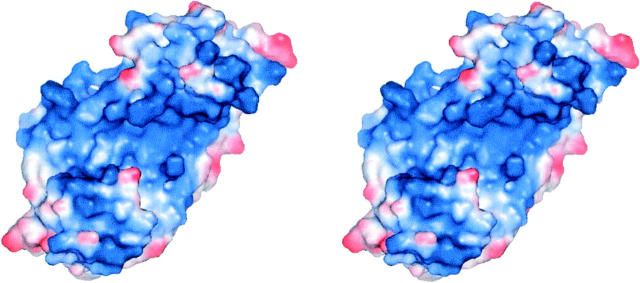
The most striking feature of hRcd-1 ΔNC dimers is their highly positively-charged cleft formed as a result of the consecutive arm motifs (Fig. 3). In this cleft, one finds Lys44, Lys45, Arg46, His85, His103, Arg107, Arg130, Lys148, Lys188, Lys230, and His231. Together with the three histidines, Tyr134, Phe184, and Trp275 form a sextet of aromatic amino acids. The majority of these residues are conserved across species (Fig. 1A). The diameter of the cleft is 20–21 Å wide (20 Å between the Cβ atoms of E47 and R227), roughly equivalent to the diameter of double-stranded DNA.
Figure 3.
Stereo representation of the electrostatic surface potential of a hRcd-1 ΔNC monomer. Values calculated using PyMOL. Positively-charged regions are shown in increasing blue, while negatively-charged regions are shown in increasing red. A highly positively-charged groove runs across the protein molecule.
hRcd-1 and hRcd-1 ΔNC bind to oligodeoxyribonucleic and ribonucleic acids in vitro; point mutation reduces and/or abolishes oligonucleotide binding
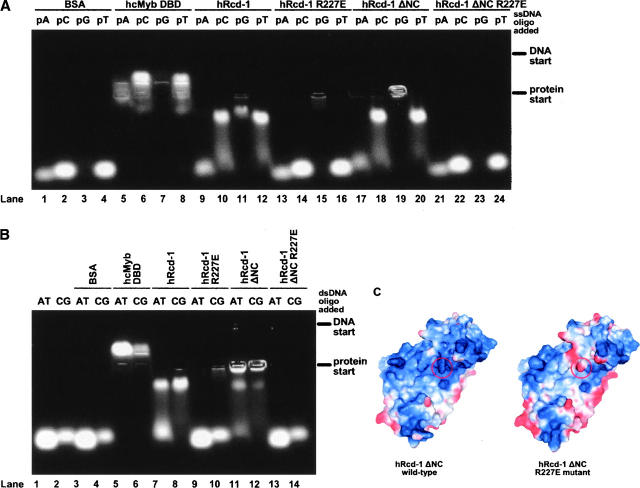
Intrigued by the presence of a cleft displaying multiple features of a potential nucleic acid binding site, we performed modified electrophoretic mobility shift assays (EMSAs) to test whether hRcd-1 or hRcd-1 ΔNC could associate with DNA in vitro: Protein and DNA samples were loaded in parallel sets of wells; interactions seen between protein samples and labeled DNA occurred within the gel matrix. Gels were not stained with ethidium bromide, thus all fluorescent signals seen are from the presence of fluorescein. The DNA-binding domain of human c-Myb (hcMyb DBD) was used as a positive control, while bovine serum albumin (BSA) served as a negative control (Fig. 4). 5′-Fluorescein-labeled DNA homopolymers were used to test the differences of affinity between wild-type, mutant, and truncated hRcd-1 proteins (Fig. 4A,B).
Figure 4.
Site-directed mutation studies on Rcd-1 and hRcd-1 ΔNC. Starting positions for DNA and protein samples are noted at right of gels. (A) In-gel ssDNA interaction assays on 5′-fluorescein labeled poly(A) (pA), poly(C) (pC), poly(G) (pG), or poly(T) (pT) DNA homopolymers. Approximately equivalent amounts of protein were used for the binding experiments. The proteins loaded are shown at the top of each lane along with type of homopolymer used. All lanes are numbered at the bottom of the gel. (B) In-gel dsDNA interaction assays. The type of 5′-fluorescein-labeled annealed oligomers loaded is noted at the top of each lane: (AT) poly(A)/poly(T) pair; (CG) poly(C)/poly(G) pair. The proteins loaded are shown at the top of each lane. All lanes are numbered at the bottom of the gel. (C) Electrostatic surface potential contourings of hRcd-1 ΔNC monomers (wild-type and R227E mutant) using PyMOL. The location of residue 227 is circled in red. Positively-charged regions are shown in increasing blue, while negatively-charged regions are shown in increasing red.
A point mutation was generated that was computed to have a dramatic effect on the electrostatic potential of the cleft region of hRcd-1 (Fig. 4C). Arginine 227 was changed into a glutamate for full-length hRcd-1 and hRcd-1 ΔNC. Based on the surface location of this arginine residue, changing it into another hydrophilic amino acid should have negligible effects on the overall structure. Expression, purification, and gel filtration results confirmed this conclusion; both point mutants behaved analogously to their wild-type counterparts (data not shown). Circular dichroism spectra were also the same for native and mutant proteins (data not shown). However, the point mutants’ ability to bind to homopolymer nucleic acids was abolished (Fig. 4A).
The migration distances of labeled DNA oligonucleotides in the presence or absence of BSA were identical (see Supplemental Fig. 2). hcMyb DBD bound but did not show any particular preference between the DNA homopolymers without any of the sequence specificity previously reported (Fig. 4A, lanes 5–8; Howe and Watson 1991; Tanikawa et al. 1993). Although equal amounts were loaded, the poly(G) oligomer had the poorest signal likely because of lower fluorescein labeling efficiency during synthesis.
Both hRcd-1 and hRcd-1 ΔNC appeared to interact with poly(G), poly(C), and poly(T) homopolymers and showed no detectable affinity for poly(A) sequences (Fig. 4A, lanes 9–12, 17–20). The R227E mutant of full-length hRcd-1 abolished all homopolymer interactions except for poly(G), which retained some weak binding (Fig. 4A, lanes 13–16). The hRcd-1 ΔNC R227E mutant shows an analogous loss, but with no detectable oligonucleotide affinity remaining (Fig. 4A, lanes 21–24). The notable difference in affinity for poly(G) sequences between the R227E mutants again suggests that the N and C termini play a role, possibly regulatory, in oligonucleotide interaction.
Complementary 5′-fluorescein-labeled DNA homopolymers were annealed, and in-gel interaction assays were performed with the resultant dsDNA. Again, BSA showed no interaction while cMyb DBD bound the AT and GC paired dsDNA equivalently well (Fig. 4B, lanes 1–6). Both hRcd-1 and hRcd-1 ΔNC were found to interact with both AT and GC paired dsDNA, but showed lower affinity for the AT pairing (Fig. 4B, lanes 7,8,11,12). The R227E point mutants on both the full-length and truncated forms of hRcd-1 lose their ability to interact with the hybridized DNA homopolymers, although the full-length mutant retains some very weak residual binding affinity for the GC dsDNA (Fig. 4B, lanes 7–14). It is, however, possible that the remaining bound signal is due to the presence of a small amount of unhybridized ssDNA in the DNA solution.
Discussion
Cellular differentiation is a required trait for all multicellular organisms. It also offers benefits to unicellular organisms faced with hostile environments. However, the actual molecular players that allow cells to differentiate are still the subject of intense investigation. Studies suggest the mechanisms and factors involved in differentiation are neither very cell type specific nor organism specific (Yamamoto et al. 1999; Hiroi et al. 2002). Thus, it is not very surprising that a gene like rcd1 in S. pombe, identified as crucial for yeast cellular differentiation, is conserved throughout eukaryotic genomes (Okazaki et al. 1998) (Fig. 1A).
According to the Pfam Protein Families Database (Bateman et al. 2004), Rcd-1 contains a single domain (pfam accession no. 04078). Although Rcd-1 shares both sequence and functional similarities across species, no specific function or fold had been assigned to this conserved protein. Our crystal structure analysis reveals that this domain possesses an armadillo-repeat-like fold. Using a Cα trace from the refined human Rcd-1 structure, the DALI server returned as the most structurally similar protein karyopherin α, consisting of 10 arm motifs. Arm motifs have been found in many functionally unrelated proteins from β-catenin to karyopherin α (Peifer et al. 1994; Conti et al. 1998). Primary sequence similarity between arm motifs within the same or between different proteins is low (Fig. 1B). The hallmark of this structural motif is its recurring three-α-helices pattern packed in a clockwise spiraling fashion (Figs. 1B, 2A). This arrangement of helices gives rise to a cleft and curving C-shaped appearance to the overall arm-repeat region. Arm repeats classically fold to create a surface for protein–protein interactions (Huber et al. 1997; Conti et al. 1998; Conti and Kuriyan 2000; Daniels et al. 2001). Presently, the PDB holds entries for the following arm-like-repeat proteins: β-catenin, importin/karyopherin α, plakophilin, mouse protein 25, HspBP1, and diaphanous homology domain as well as the IP3-receptor and epsin domain. The latter two only contain a small number of arm repeats.
While Rcd-1 shares close tertiary structure similarity with karyopherin α, their quaternary structures are rather different. In the case of karyopherin α, the two monomers face each other, both C termini partially cradled in the groove formed by the arm motifs. On the other hand, the structure of human Rcd-1 shows the interaction between monomers occurring on the face opposite to the cleft formed by the arm motifs (Fig. 2A). The dimer interface consists of hydrophobic interactions exclusively with no discernable hydrogen-bonding patterns. Moreover, the exposed sides on the dimer reveal that the cleft formed by the arm repeats is lined with numerous positively-charged residues. An electrostatic map (calculated by PyMOL; DeLano 2002) of the exposed face shows the extremely strong positive charge of the roughly 20–21 Å cleft (Fig. 3).
Although the charge and shape of this cleft are suggestive of nucleic acid binding, a search of the RSCB database for protein structures containing arm motifs yielded no DNA- or RNA-binding proteins. Furthermore, murine Rcd-1 was recently reported to not interact with a DNA-Sepharose column (Haas et al. 2004). Nevertheless, we undertook electrophoretic mobility shift assays using single- and double-stranded DNA. The DNA-binding domain of human c-Myb provided a positive control (Tahirov et al. 2002).
Using an in-gel protein–DNA interaction assay, clear binding and base preference was seen in in vitro experiments with four 5′-fluorescein-labeled DNA homopolymers (Fig. 4A,B); hRcd-1 and hRcd-1 ΔNC preferred poly(G), poly(C), and poly(T) ssDNA with notable affinity (Fig. 4A, lanes 9–12, 17–20). Surprisingly, poly(A) oligomers did not interact significantly with any hRcd-1 construct, though all four homopolymers were bound tightly to hcMyb DBD (Fig. 4A, lanes 5–12, 17–20). These results strongly suggest that interaction between hRcd-1 and oligomers is base-selective, discriminating against poly(A) sequences, ruling out mere nonspecific interactions that would have to occur through the backbone phosphates, and could not explain the strong differences in affinities.
To support our hypothesis that oligonucleotide binding is occurring within the positively-charged cleft seen in the model, both full-length and the hRcd-1 ΔNC constructs were mutated, changing residue Arg227 into Glu (numbering based on the full-length sequence). Gel filtration peaks and circular dichroism results analogous to their wild-type counterparts suggested that the fold of the mutants remained intact. However, the mutation abolished oligonucleotide binding ability of full-length and ΔNC truncated forms of hRcd-1 (Fig. 4A, lanes 9–24). These results reinforce our previous findings that hRcd-1 cannot only interact with DNA but does so using the highly positively-charged cleft.
In binding studies using annealed complementary 5′-fluorescein-labeled DNA homopolymers, hRcd-1 displayed a clear preference for the poly(C)/poly(G) pairing, although it still had affinity for the poly(A)/poly(T) pairing (Fig. 4B). For hRcd-1 and hRcd-1 ΔNC, an R227E point mutation abolished binding to the paired dsDNA homopolymers, although some weak signal remained from the poly(C)/poly(G) homopolymer with full-length hRcd-1 (Fig. 4B, lanes 7–14). This agrees with the ssDNA homopolymer interaction results with full-length Rcd-1, although it is also possible that the weaker, smearing signals are the result of some ssDNA remaining after the annealing protocol.
Considering that the homolog of Rcd-1, CAF40, was identified as a component of the CCR4–NOT transcription complex in S. cerevisiae (Chen et al. 2001), human Rcd-1 interaction with DNA would not be that surprising. Rcd-1 associates with the CCR4–NOT complex, which has mRNA manipulating properties, including deadenylation activity (Chen et al. 2001). This places Rcd-1 near nucleic acid and its discrimination against poly(A) seems conspicuous in the context of the complex's enzymatic activity. We postulate that the binding of Rcd-1 with other proteins in the complex could cause the N and C termini to shift, thus either allowing or disallowing easy access of oligonucleotides to the binding cleft. The truncated portions of Rcd-1 are rich in prolines and might be flexible (Fig. 1A). Although currently only limited in vitro evidence is available, it is tempting to speculate that Rcd-1 exerts some of its influence on transcription through a regulated nucleotide-binding activity.
While our studies identify a novel in vitro activity for hRcd-1, our results do not contradict other protein-complex formations involving hRcd-1 as has been previously shown or alluded to in other studies (Okazaki et al. 1998; Hiroi et al. 2002; Haas et al. 2004). Arm repeat-containing proteins are well known for their involvement in protein–protein interactions (Huber et al. 1997; Conti et al. 1998; Conti and Kuriyan 2000; Daniels et al. 2001). In numerous structural studies, the cleft formed by arm-containing proteins is the site involved in interaction with binding partners (Conti et al. 1998; Conti and Kuriyan 2000; Daniels and Weis 2002; Ha et al. 2004). β-Catenin also possesses a positively-charged groove, which has been shown to interact with sections of APC, cadherins, and Tcf family members (Huber et al. 1997; Daniels and Weis 2002; Ha et al. 2004), although the curvature of the cleft is significantly different from that found in Rcd-1. Protein–protein interactions undoubtedly play a role in hRcd-1’s involvement in cellular differentiation processes. Whether protein–protein interactions may serve to regulate hRcd-1’s nucleic acid binding ability is an area in need of further investigation. Additionally, the relatively small hydrophobic interface in the hRcd-1 dimer could become exposed in some larger protein complexes, supporting incorporation of hRcd-1 monomers into more complex structures.
The molecular structures of only a small number of arm-like proteins have been determined so far. All arm repeat-containing protein structures known today, from β-catenin to karyopherin α, contain a superhelix of repeating three (short-short-long) α-helical bundles (Huber et al. 1997; Conti et al. 1998; Conti and Kuriyan 2000; Daniels and Weis 2002; Ha et al. 2004); as noted earlier, the DALI server (Holm and Sander 1994) identified karyopherin α as the protein whose fold is most closely related to Rcd-1. The latter possesses the “short-short-long” arrangement of repeated three helices (Fig. 1A,B; α1 and α2 would be the short-long helices in the triad with α3-, α4-, α5-, and subsequent repeats afterward following the short-short-long arrangement); the tertiary fold prominently reveals the archetypical arm superhelix of helices seen in previously determined structures (Fig. 2A), which all contain the fingerprint sequence PTxxxHxxHHPxTxxHLxxLHPHLT(x)24PxPLxTxSHHSLTPLSx (Schultz et al. 1998). However, currently known arm-containing proteins share a sequence similarity of only 0.170 (calculated using Clustal W; Thompson et al. 1994). Rcd-1 is the first example that adopts an arm-repeat fold but cannot be identified by sequence information alone, sharing a sequence similarity of only 0.060 when compared to the structurally aligned region on karyopherin α. Given the very limited set of structurally characterized arm proteins, it might not be too surprising to find a new family member with no recognizable sequence conservation that deviates from what has previously been the only established function, namely, as a protein-binding partner. For a similar example, we point to the TIM-barrel family of proteins for which Nature seems to have found an enormous number of different roles (Nagano et al. 2002; Anantharaman et al. 2003).
In conclusion, we have determined the crystal structure of the conserved region of human Rcd-1. The atomic model revealed its structural similarity to arm repeat-containing proteins, while the shape and electrostatic potential of its surface suggested nucleic acid binding properties. Indeed, in vitro in-gel interaction assays confirm discriminative nucleotide binding. To the best of our knowledge, Rcd-1 is the first arm repeat protein exhibiting this property, adding to the protein family's characterized potential to interact with other proteins or small ligands (Huber et al. 1997; Conti et al. 1998; Conti and Kuriyan 2000; Daniels and Weis 2002; Ha et al. 2004). Rcd-1’s nucleic acid binding suggests an additional mechanistic feature by which it could exert its role in cellular differentiation (Okazaki et al. 1998; Gregory et al. 2000; Chen et al. 2001; Friedman 2002; Hiroi et al. 2002). However, understanding the process of cellular differentiation lies not in any single protein but, rather, in the way larger protein complexes and signaling pathways orchestrate the changes in transcription and translation, thereby bringing about changes to the cell as a whole; whether Rcd-1’s nucleotide interactions occur in vivo, regulated by other proteins, represents an attractive hypothesis but remains to be investigated further. Obviously, understanding the means through which Rcd-1 interacts with nucleic acids would be aided dramatically by a crystal structure of a complex between Rcd-1 and an oligonucleotide or, even better, of Rcd-1 together with members of the CCR4–NOT complex. Additionally, other protein-binding partners may yet be identified that account for downstream effects. Structure-based mutations can be explored to study their effects in various cellular differentiation assays. The structure we present offers a starting scaffold on which future experiments can build.
Materials and methods
Cloning and mutagenesis
Wild-type human and mouse Rcd-1 genes were cloned by polymerase chain reaction (PCR) from human and mouse cDNA libraries derived from cell lines. The full-length human Rcd-1 gene (hRcd-1) was amplified using primers 5′-GCTAACCATGGCACACAGCCTGGCGACGGCTGCGCCTGTG-3′ and 5′-GCTAACTCGAGTCACTGAGGGGGCAGGGGGATACCCCG-3′ (NcoI and XhoI sites are underlined).
The full-length mouse Rcd-1 gene (mRcd-1) was amplified using primers 5′-GCTAACCATGGCACACAGCCTGGCAACGGCAGCGGCTGTG-3′ and 5′-GCTAACTCGAGTCACTGAGGGGGCAGGGGAATCCCCCG-3′ (NcoI and XhoI sites are underlined). PCR products were amplified using the Expand kit (Roche Diagnostics), gel purified, digested with NcoI and XhoI (New England Biolabs), and ligated into the pET32 vector (Novagen). All clones were expressed as N-terminal 6-His-thioredoxin fusion proteins in BL21/codonplus Escherichia coli cells (Stratagene). Clones were verified by sequencing (University Health Network DNA Sequencing Facility).
Three human Rcd-1 truncations were constructed by PCR using modified primer pairs and cloned into pET32 (Novagen). The N-terminally truncated construct (hRcd-1 ΔN) used the 5′ primer (5′-GCTAACCATGGATAGAGAAAAGATCTATCAGTGGATC-3′) paired with the original 3′ primer. The resulting construct begins at residue 18 of the wild-type form. The C-terminally truncated construct (hRcd-1 ΔC) used the 3′ primer (5′-GCTAACTCGAGTCACTCTTGCAGGTTCTTCACCAGTTGTGC-3′) paired with the original 5′ primer. The resulting construct begins at residue 1 and ends at residue 285 of the wild-type form. The N- and C-terminally truncated construct (hRcd-1 ΔNC) was amplified using both truncation primers, yielding a fragment spanning codons 18–285 of the full-length protein. All human Rcd-1 truncated proteins were also expressed in BL21/codonplus E. coli cells.
Mutants of hRcd-1 and hRcd-1 ΔNC were generated using PCR-based site-directed mutagenesis. A point mutation changing residue Arg227 into a Glu was inserted into both the full-length and the N- and C-terminally truncated forms of hRcd-1. The primer pair containing the mutation consisted of primers 5′-CCAAAGAGCCTTCTGCCGAACTGCTGAAGCATGTAGTGAG-3′ and 5′-CTCACTACATGCTTCAGCAGTTCGGCAGAAGGCTCTTTGG-3′ (mutated codon is denoted in bold). Mutated Rcd-1 proteins were expressed and purified analogously to their native counterparts.
The part of human c-Myb coding for its DNA-binding domain (hcMybDBD) was amplified from a cDNA clone (Open Biosystems; Clone ID 6069320) and used as a control for oligonucleotide-binding experiments. The region spanning residues 37–193 of hc-Myb was amplified using primers 5′-GGCAAACCATGGGCCACTTGGGGAAAACAAGGTGGACCCGG-3′ and 5′-GGTCTCGAGTCAGACCTTCCGACGCATTGTAGAATTCCAGTG-3′ and cloned into a pET32 vector (Novagen). hcMybDBD was also expressed in BL21/codonplus E. coli cells.
Protein expression and purification
Rcd-1 expressing BL21/codonplus cells were grown at 37°C in Luria-Bertani broth supplemented with 50 μM carbenicillin in a shaker to an OD600 of 0.8. Cells were induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG), grown for 4 h at room temperature, then placed overnight at 4°C. Cells were pelleted at 4000 revolutions per minute (rpm), resuspended in buffer (50 mM Tris at pH 7.5, 500 mM NaCl, 5% glycerol, 1 mM PMSF, 5 mM β-mercaptoethanol), and sonicated using five 1-min pulses on a Branson Sonifier. The lysed cell solution was centrifuged for 1 h at 17,000 rpm, then the supernatant was mixed with 1 mL of Ni-NTA resin (QIAGEN) per liter of lysed cells and gently rocked overnight at 4°C. The Ni-NTA resin was batch washed three times using 50 mL of 50 mM Tris (pH 7.5), 500 mM NaCl, 30 mM imidazole, 5% glycerol, and 5 mM β-mercaptoethanol for every 2 mL of Ni-NTA resin. Protein was batch eluted using 10 mL of 50 mM Tris (pH 7.5), 500 mM NaCl, 250 mM imidazole, 5% glycerol, and 5 mM β-mercaptoethanol. The presence and quality of protein were verified by running samples of the elute on 15% SDS-PAGE gels and visualizing them by Coomassie blue staining. CaCl2 was added to the combined fractions containing Rcd-1 to a final concentration of 2.5 mM followed by 0.5 units of thrombin (Calbiochem). The enzymatic digest proceeded overnight at 4°C in 3500 MWCO SnakeSkin Pleated Dialysis Tubing (Pierce) suspended in 4 L of dialysis buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 2.5 mM CaCl2, 5 mM β-mercaptoethanol).
Then, the sample was batch-bound onto pre-equilibrated Ni-NTA resin for 1 h at 4°C with gentle shaking followed by transfer into a column. Flow-through fractions had TCEP added before being concentrated to 1 mL. This was followed by buffer exchange on a Superdex 200 column (Amersham-Pharmacia) pre-equilibrated with 20 mM Tris (pH 7.5), 50 mM NaCl. One-milliliter fractions were collected and their Rcd-1 content and purity verified on a gel, then fractions were pooled and concentrated.
Identical purification protocols were used for the full-length human and mouse Rcd-1 constructs as well as the N-, the C-, and the N- plus C-terminally truncated human constructs including both point-mutated protein constructs.
For the expression of the seleno-methionine derivatives (Se-Met), vectors containing the genes hRcd-1 and hRcd-1 ΔNC were transformed into B834 (DE3) cells (Novagen). Proteins were expressed in M9 media enriched with seleno-methionine and the other 19 amino acids (Ramakrishnan et al. 1993). Purification was the same as for the native protein except that subsequent buffer-exchange steps were performed in the presence of 0.5 mM TCEP (BioShop).
Size exclusion column chromatography
All protein chromatographic steps were performed using an ÄKTA FPLC system (Amersham-Pharmacia). Protein samples (1 mL) were prepared as above and size-fractionated by gel filtration on a Superdex 200 (Amersham-Pharmacia) column at 4°C. Protein standards used for calibrating the column were ribonuclease A, chymotrypsin, ovalbumin, albumin, and aldolase (Amersham). Peak samples were loaded onto SDS-PAGE gels and visualized using Coomassie blue staining. For each protein preparation, at least three independent experiments were performed with consistent results.
Circular dichroism
All circular dichroism experiments were done using an Aviv Circular Dichroism Spectrometer (Model G2A DS). Scans were performed at 25°C from 200 nm to 260 nm. Data were analyzed using KaleidaGraph, version 3.09 (Synergy Software).
Analytical ultracentrifugation
Sedimentation equilibrium experiments were performed at 4°C on an Optima XL-A/XL-I Analytical Ultracentrifuge (Beckman Instruments) using an AN50-Ti rotor, quartz windows, and standard six-sector charcoal-filled Epon centerpieces. Samples were centrifuged at 10,000 rpm, 15,000 rpm, 20,000 rpm, and 25,000 rpm for 26 h at each speed to ensure equilibrium was reached before absorbance measurements were taken. Global analysis of the data was performed using XL-A/XL-I data analysis software (Origin version 4.1) from Beckman Instruments. Global analysis of the data was fitted to a single ideal species.
Crystallization, data collection, and processing
Initial crystallization conditions for hRcd-1 and its truncated forms were determined using Crystal Screens I and II as well as Index Screen (Hampton Research). hRcd-1 ΔNC was crystallized by vapor diffusion against 100 mM MES (pH 5.7), 115 mM MnSO4, 16%–18% PEG 3350, and 1% glucose. The crystals were cryoprotected using a stepwise soak of increasing PEG 3350 concentrations to a final concentration of 28%–30% PEG 3350. Optimal crystallization conditions for the native and Se-Met derivatives of hRcd-1 ΔNC were identical.
All data sets were collected at the Advanced Photon Source beamline 14B (BioCARS) using a Q4 area detector. A native data set was obtained from an hRcd-1 ΔNC crystal at λ = 0.9407 Å, MAD data sets (λremote = 0.9568 Å, λpeak = 0.9795 Å, λedge = 0.9797 Å) from an hRcd-1 ΔNC Se-methionine crystal. Data were reduced using DENZO and scaled using SCALEPACK (Otwinowski and Minor 1997).
Phase determination
The structure for hRcd-1 ΔNC was determined using the MAD method of phasing. Initial positions of the Se atoms were determined using the program BnP version 1.01 (Weeks et al. 2002). Fourteen high occupancy sites were found, three or four for each monomer of the dimer with two dimers occupying the unit cell, and the overall figure of merit was 0.57 for data to 2.5 Å resolution. The density-modified map was traced manually using the program O (Jones et al. 1991). Subsequent refinements were done against the native data set using the program CNS, version 1.1 (Brünger et al. 1998). Refinement statistics are given in Table 1. Figures were prepared using the programs SPOCK (Christopher 1998), MOLSCRIPT (Kraulis 1991), and RASTER3D (Merritt and Murphy 1994). Atomic coordinates and structure factors of hRcd-1 ΔNC have been deposited in the RCSB Protein Databank under the accession code 2FV2 (RCSB ID code RCSB036346).
In-gel DNA interaction assays
hRcd-1, hRcd-1 ΔNC, and point-mutated proteins were purified as described above. Each binding assay was performed in a 1% TAE-agarose gel containing two rows of wells 1 cm apart. In wells closer to the anode, 100 pmol of 5′-fluorescein-labeled ssDNA or annealed 5′-fluorescein-labeled dsDNA oligonucleotides were loaded. Approximately 100 pmol of purified proteins were loaded in the wells closer to the cathode. Gels were run for 1 h such that the bromophenol blue loading dye marker from the upper, DNA-containing lanes migrated well beyond the xylene cyanol loading dye marker from the lower protein lanes. (This protocol is based on personal communications with Jeroen Mesters, University of Lübeck; it is a modified version of zone-interference gel eletrophoresis [Abrahams et al. 1988].) Positions of fluorescein-labeled bands were visualized on a UV-light table.
The following oligonucleotides were used to test for selective binding (f stands for fluorescein):
f-poly(A) DNA (20): 5′-f-AAAAAAAAAAAAAAAAAAAA-3′
f-poly(C) DNA (20): 5′-f-CCCCCCCCCCCCCCCCCCCC-3′
f-poly(G) DNA (20): 5′-f-GGGGGGGGGGGGGGGGGGGG-3′
f-poly(T) DNA (20): 5′-f-TTTTTTTTTTTTTTTTTTTT-3′
Duplex DNA pairs were generated by mixing equal amounts of complementary oligonucleotides, heating them for 10 min at 95°C, then allowing the samples to slowly cool down to room temperature in the heating block with power switched off.
Acknowledgments
We thank the staff at BioCARS sector 14 beamlines at the Advanced Photon Source, Argonne National Laboratories, for their generous time commitments and support. R.G.G. thanks Ning Wu for her support and advice. We thank A. Holdorf and A. Pawson for the gift of human and mouse cDNA libraries. This work was supported by the Ontario Cancer Institute/Princess Margaret Hospital Foundation, Toronto (R.G.G.), the Canada Research Chairs Program, and the National Sciences and Engineering Research Council, as well as the Ontario Research and Development Challenge Fund (E.F.P.). Use of the Advanced Photon Source was supported by the Basic Energy Sciences, Office of Science, United States Department of Energy, under Contract W-31-109-Eng-38. Use of the BioCARS sector 14 was supported by the National Center for Research Resources, National Institutes of Health, under Grant RR07707.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Emil F. Pai, Department of Biochemistry, University of Toronto, 1 King's College Circle, Toronto, ON, M5S 1A8, Canada; e-mail: pai@hera.med.utoronto.ca; fax: (416) 581-7545.
Abbreviations: Rcd-1, required for cell differentiation.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062600507.
References
- Abrahams, J.P., Kraal, B., and Bosch, L. 1988. Zone-interference gel electrophoresis: A new method for studying weak protein–nucleic acid complexes under native equilibrium conditions. Nucleic Acids Res. 16: 10099–10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman, V., Aravind, L., and Koonin, E.V. 2003. Emergence of diverse biochemical activities in evolutionarily conserved structural scaffolds of proteins. Curr. Opin. Chem. Biol. 7: 12–20. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., and Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach, D., Rodgers, L., and Gould, J. 1985. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr. Genet. 10: 297–311. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., and Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Chen, J., Rappsilber, J., Chiang, Y.C., Russell, P., Mann, M., and Denis, C.L. 2001. Purification and characterization of the 1.0 MDa CCR4–NOT complex identifies two novel components of the complex. J. Mol. Biol. 314: 683–694. [DOI] [PubMed] [Google Scholar]
- Chen, J., Chiang, Y.C., and Denis, C.L. 2002. CCR4, a 3′–5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21: 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher, J.A. 1998. SPOCK. Texas A&M University, The Center for Macromolecular Design. College Station, TX.
- Coates, J.C. 2003. Armadillo repeat proteins: Beyond the animal kingdom. Trends Cell Biol. 13: 463–471. [DOI] [PubMed] [Google Scholar]
- Collart, M.A. and Struhl, K. 1993. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 12: 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti, E. and Kuriyan, J. 2000. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin α. Structure 8: 329–338. [DOI] [PubMed] [Google Scholar]
- Conti, E., Uy, M., Leighton, L., Blobel, G., and Kuriyan, J. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94: 193–204. [DOI] [PubMed] [Google Scholar]
- Daniels, D.L. and Weis, W.I. 2002. ICAT inhibits β-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol. Cell 10: 573–584. [DOI] [PubMed] [Google Scholar]
- Daniels, D.L., Eklof Spink, K., and Weis, W.I. 2001. β-Catenin: Molecular plasticity and drug design. Trends Biochem. Sci. 26: 672–678. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- Denis, C.L. and Malvar, T. 1990. The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics 124: 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis, C.L., Chiang, Y.C., Cui, Y., and Chen, J. 2001. Genetic evidence supports a role for the yeast CCR4–NOT complex in transcriptional elongation. Genetics 158: 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, M.K. and McBurney, M.W. 1983. The concentration of retinoic acid determines the differentiated cell types formed by a teratocarcinoma cell line. Dev. Biol. 98: 187–191. [DOI] [PubMed] [Google Scholar]
- Friedman, A.D. 2002. Transcriptional regulation of granulocyte and monocyte development. Oncogene 21: 3377–3390. [DOI] [PubMed] [Google Scholar]
- Gregory, R.C., Lord, K.A., Panek, L.B., Gaines, P., Dillon, S.B., and Wojchowski, D.M. 2000. Subtraction cloning and initial characterization of novel epo-immediate response genes. Cytokine 12: 845–857. [DOI] [PubMed] [Google Scholar]
- Guex, N. and Peitsch, M.C. 1997. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Ha, N.C., Tonozuka, T., Stamos, J.L., Choi, H.J., and Weis, W.I. 2004. Mechanism of phosphorylation-dependent binding of APC to β-catenin and its role in β-catenin degradation. Mol. Cell 15: 511–521. [DOI] [PubMed] [Google Scholar]
- Haas, M., Siegert, M., Schurmann, A., Sodeik, B., and Wolfes, H. 2004. c-Myb protein interacts with Rcd-1, a component of the CCR4 transcription mediator complex. Biochemistry 43: 8152–8159. [DOI] [PubMed] [Google Scholar]
- Hiroi, N., Ito, T., Yamamoto, H., Ochiya, T., Jinno, S., and Okayama, H. 2002. Mammalian Rcd1 is a novel transcriptional cofactor that mediates retinoic acid-induced cell differentiation. EMBO J. 21: 5235–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, B.L., Taylor, A., and Adamson, E. 1981. Cell interactions modulate embryonal carcinoma cell differentiation into parietal or visceral endoderm. Nature 291: 235–237. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1994. Searching protein structure databases has come of age. Proteins 19: 165–173. [DOI] [PubMed] [Google Scholar]
- Howe, K.M. and Watson, R.J. 1991. Nucleotide preferences in sequence-specific recognition of DNA by c-myb protein. Nucleic Acids Res. 19: 3913–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, A.H., Nelson, W.J., and Weis, W.I. 1997. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell 90: 871–882. [DOI] [PubMed] [Google Scholar]
- Hughes, D.A., Fukui, Y., and Yamamoto, M. 1990. Homologous activators of ras in fission and budding yeast. Nature 344: 355–357. [DOI] [PubMed] [Google Scholar]
- Iino, Y. and Yamamoto, H. 1985a. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol. Gen. Genet. 198: 416–421. [DOI] [PubMed] [Google Scholar]
- Iino, Y. and Yamamoto, M. 1985b. Negative control for the initiation of meiosis in Schizosaccharomyces pombe . Proc. Natl. Acad. Sci. 82: 2447–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kelly, M., Burke, J., Smith, M., Klar, A., and Beach, D. 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 7: 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT. J. Appl. Crystallogr. 24: 946–950. [Google Scholar]
- Maillet, L., Tu, C., Hong, Y.K., Shuster, E.O., and Collart, M.A. 2000. The essential function of Not1 lies within the Ccr4–Not complex. J. Mol. Biol. 303: 131–143. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A. and Murphy, M.E.P. 1994. Raster3D. Acta Crystallogr D Biol. Crystallogr. 50: 869–873. [DOI] [PubMed] [Google Scholar]
- Miyamoto, M., Tanaka, K., and Okayama, H. 1994. res2+, a new member of the cdc10+/SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J. 13: 1873–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano, N., Orengo, C.A., and Thornton, J.M. 2002. One fold with many functions: The evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 321: 741–765. [DOI] [PubMed] [Google Scholar]
- Okazaki, N., Okazaki, K., Watanabe, Y., Kato-Hayashi, M., Yamamoto, M., and Okayama, H. 1998. Novel factor highly conserved among eukaryotes controls sexual development in fission yeast. Mol. Cell. Biol. 18: 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Peifer, M., Berg, S., and Reynolds, A.B. 1994. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 76: 789–791. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V., Finch, J.T., Graziano, V., Lee, P.L., and Sweet, R.M. 1993. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362: 219–223. [DOI] [PubMed] [Google Scholar]
- Sakai, A., Chibazakura, T., Shimizu, Y., and Hishinuma, F. 1992. Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae . Nucleic Acids Res. 20: 6227–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. 1998. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. 95: 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, A., Iino, Y., Maeda, T., Watanabe, Y., and Yamamoto, M. 1991. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes & Dev. 5: 1990–1999. [DOI] [PubMed] [Google Scholar]
- Sugiyama, A., Tanaka, K., Okazaki, K., Nojima, H., and Okayama, H. 1994. A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J. 13: 1881–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov, T.H., Sato, K., Ichikawa-Iwata, E., Sasaki, M., Inoue-Bungo, T., Shiina, M., Kimura, K., Takata, S., Fujikawa, A., and Morii, H., et al. 2002. Mechanism of c-Myb-C/EBP β cooperation from separated sites on a promoter. Cell 108: 57–70. [DOI] [PubMed] [Google Scholar]
- Tanikawa, J., Yasukawa, T., Enari, M., Ogata, K., Nishimura, Y., Ishii, S., and Sarai, A. 1993. Recognition of specific DNA sequences by the c-myb protooncogene product: Role of three repeat units in the DNA-binding domain. Proc. Natl. Acad. Sci. 90: 9320–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M., Valencia-Sanchez, M.A., Staples, R.R., Chen, J., Denis, C.L., and Parker, R. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae . Cell 104: 377–386. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., Lino, Y., Furuhata, K., Shimoda, C., and Yamamoto, M. 1988. The S. pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 7: 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., Shinozaki-Yabana, S., Chikashige, Y., Hiraoka, Y., and Yamamoto, M. 1997. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 386: 187–190. [DOI] [PubMed] [Google Scholar]
- Weeks, C.M., Blessing, R.H., Miller, R., Mungee, R., Potter, S.A., Rappleye, J., Smith, G.D., Xu, H., and Furey, W. 2002. Towards automated protein structure determination: BnP, the SnB-Phases interface. Z. Kristallogr. 217: 686–693. [Google Scholar]
- Willer, M., Hoffmann, L., Styrkarsdottir, U., Egel, R., Davey, J., and Nielsen, O. 1995. Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe . Mol. Cell. Biol. 15: 4964–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, H., Tsukahara, K., Kanaoka, Y., Jinno, S., and Okayama, H. 1999. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol. Cell. Biol. 19: 3829–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]