Abstract
Myosins play essential roles in migration, cytokinesis, endocytosis, and adhesion. They are composed of a large N-terminal motor domain with ATPase and actin binding sites and C-terminal neck and tail regions, whose functional roles and structural context in the protein are less well characterized. The tail regions of myosins I, IV, VII, XII, and XV each contain a putative SH3 domain that may be involved in protein–protein interactions. SH3 domains are reported to bind proline-rich motifs, especially “PxxP” sequences, and such interactions serve regulatory functions. The activity of Src, PI3, and Itk kinases, for example, is regulated by intramolecular interactions between their SH3 domain and internal proline-rich sequences. Here, we use NMR spectroscopy to reveal the structure of a protein construct from Dictyostelium myosin VII (DdM7) spanning A1620–T1706, which contains its SH3 domain and adjacent proline-rich region. The SH3 domain forms the signature β-barrel architecture found in other SH3 domains, with conserved tryptophan and tyrosine residues forming a hydrophobic pocket known to bind “PxxP” motifs. In addition, acidic residues in the RT or n–Src loops are available to interact with the basic anchoring residues that are typically found in ligands or proteins that bind SH3 domains. The DdM7 SH3 differs in the hydrophobicity of the second pocket formed by the 310 helix and following β-strand, which contains polar rather than hydrophobic side chains. Most unusual, however, is that this domain binds its adjacent proline-rich region at a surface remote from the region previously identified to bind “PxxP” motifs. The interaction may affect the orientation of the tail without sacrificing the availability of the canonical “PxxP”-binding surface.
Keywords: myosin, SH3 domain, PxxP motif, NMR spectroscopy
Myosins are a superfamily of over 20 distinct actin-based motor proteins (each given a numerical designation) that participate in a range of cellular functions such as migration, cytokinesis, endocytosis, and adhesion (Kieke and Titus 2003; Krendel and Mooseker 2005). They are typically comprised of an N-terminal motor domain that harbors both ATPase and actin binding sites and a C-terminal tail region that specifies their intracellular localization and cargo for transport. Much progress has been made in characterizing the structural and enzymatic properties of several myosin motor domains and analysis of myosin tail sequences has revealed the presence of several well-characterized domains such as SH3 (M1, M4, M7, M15) and FERM (M4, M7, M10, M15) domains (Fig. 1A). However, a clear understanding of the functional and structural features of myosin tail regions is lacking.
Figure 1.
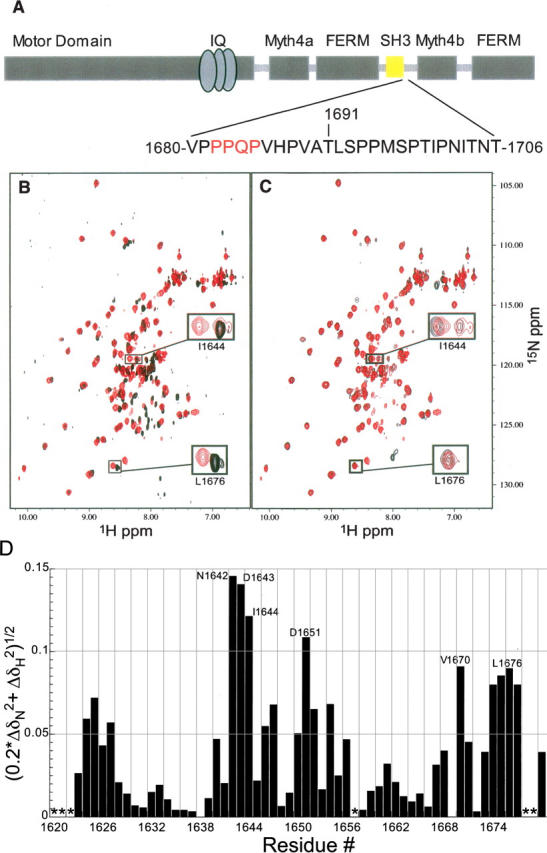
The DdM7 SH3 domain interacts with an adjacent proline-rich region. (A) Architecture of DdM7 with the SH3 domain highlighted in yellow. The sequence immediately following the SH3 domain spanning residues V1680–T1706 is also displayed with the interacting residues in red. (B) Superimposed [1H,15N] HSQC spectrum of the construct containing only the SH3 domain (A1620–V1680) (black) with that of a longer construct containing the adjacent proline-rich region (A1620–T1706) (red). This comparison reveals that certain resonance cross-peaks in the SH3 domain shift due to the presence of the proline-rich extension. (C) Superimposed [1H,15N] HSQC spectra of A1620–T1706 (red) and A1620–H1687 (black) reveal that P1681–H1687 contains the residues responsible for the chemical shift perturbations observed in the SH3 domain. (D) Amide chemical shift perturbation analysis reveals the residues of the SH3 domain most affected by the presence of the adjacent C-terminal region. For this analysis, Equation 1 was used to compare the amide chemical shift values of the SH3 in the shorter construct spanning A1620–V1680 with those in the construct spanning A1620–T1706. Asterisks represent residues lacking chemical shift assignments.
The SH3 domains of amoeboid M1s have recently been demonstrated to play a role in recruiting the Arp2/3 actin polymerization machinery to the plasma membrane in yeasts and Dictyostelium (Evangelista et al. 2000; Lechler et al. 2000; Lee et al. 2000; Jung et al. 2001). Cryoelectron microscopy and sedimentation analysis suggests that the M1 SH3 domain of Acanthamoeba M1B or M1C interacts with adjacent proline-rich regions of the GPA domain, as the tail forms a compact structure (Lee et al. 1999; Ishikawa et al. 2004). Binding of the SH3 domain to adjacent sites on the tail could provide a mechanism for promoting or regulating either the structure of the M1 tail or its interaction with binding partners.
The tail region of DdM7 is comprised of a tandem repeat of two MyTH/FERM domains that are preceded at the N terminus by a proline-rich region and separated by an SH3 domain and a second proline-rich region. M7 is important for cell adhesion, an integral step in phagocytosis and cell motility in Dictyostelium (Maniak 2001; Tuxworth et al. 2001). By using NMR spectroscopy, we have determined the structure of the DdM7 SH3 domain and found that it interacts with its C-terminal proline-rich region. Interestingly, the binding does not involve the hydrophobic surface previously identified to bind proline-rich regions.
Results
Residues in the M7 SH3 domain interact with its C-terminal proline-rich region
With the exception of S1632, we were able to obtain complete chemical shift assignments for residues A1620–T1691, which encompasses the SH3 domain and first 15 residues of the adjacent proline-rich region. We expect that this is the only region within this fragment with higher order structure, as all of the spin systems that exhibited NOE cross-peaks in 15N- or 13C-dispersed NOESY spectra were assigned to these residues. We were unable to confidently identify any resonances from residues C-terminal to T1691. This inability was due to their absence in triple resonance experiments and lack of NOE cross-peaks in NOESY spectra.
Following the SH3 domain of DdM7 are sequences that contain “PxxP” motifs (Fig. 1A), to which other SH3 domains are reported to bind (Feng et al. 1994; Lim et al. 1994). Therefore, we hypothesized that intramolecular interactions exist between these regions. To test this, we produced protein fragments that contain the SH3 domain alone (A1620–V1680) and with varying lengths of the adjacent proline-rich sequence (A1620–H1687 and A1620–T1706). We compared the [1H, 15N] HSQC spectrum of each of these constructs to determine how the presence of the proline-rich region impacts resonances of the SH3 domain (Fig. 1B,C). The comparison is useful because the chemical shift value at which an atom resonates is sensitive to its chemical environment (Wüthrich 1986), making this analysis a powerful method for identifying residues at contact surfaces (Walters et al. 2001; Kang et al. 2006) or for identifying structural changes (Walters et al. 2003; Wang et al. 2003; Liu et al. 2006a). Strikingly, we observed distinct amide chemical shift perturbations in the SH3 domain of the two longer constructs compared to that of the SH3 domain alone. The data were quantified according to Equation 1
where ΔδN and ΔδH represent the chemical shift perturbation value of the amide nitrogen and proton, respectively (Fig. 1D). Significant perturbations (>0.08 ppm) were identified for residues N1642–I1644, D1651, V1670, I1675, and L1676. This result was unexpected as these residues are not predicted to be part of the SH3 domain surface previously identified to bind “PxxP” motifs (Feng et al. 1994; Lim et al. 1994).
The region spanning P1682–P1685 forms close contacts with the SH3 domain.
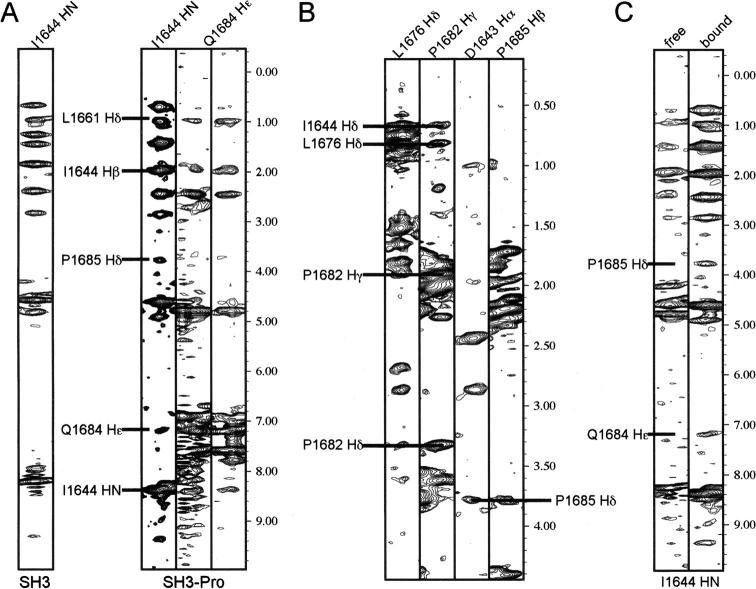
In addition to the chemical shift perturbation data, we identified 11 NOE interactions between the SH3 domain and residues P1682, Q1684, and P1685 of the proline-rich region (Fig. 2A,B). “PxxP” motifs that bind SH3 domains are generally flanked by an arginine or lysine basic residue, which is positioned in an acidic pocket proximal to the two hydrophobic ones (Feng et al. 1994; Lim et al. 1994). The proline-rich region resembles such a motif but with a smaller, less basic histidine residue substituted for arginine or lysine, 1682PPQPVH1687 (Fig. 1A). The assignment of these NOEs was confirmed by two approaches. First, the [1H, 15N] HSQC spectrum of the shorter construct demonstrated the same chemical shift perturbations as the longer one (Fig. 1B,C). This finding indicates that residues P1681–H1687 are sufficient for SH3 domain interaction and that no additional interactions occur with P1688–T1706. Second, a comparison of 15N-dispersed NOESY experiments recorded on the SH3 domain with and without the adjacent proline-rich region aided in distinguishing interactions within the SH3 domain from those involving the proline-rich sequence (V1680–T1706). Figure 2A illustrates this approach for I1644. This residue's amide proton interacts with Q1684 Hɛ and P1685 Hδ atoms, and the corresponding NOEs are absent in the spectrum recorded on the SH3 domain alone. These interactions are also verified by those observed in the 13C-dispersed NOESY spectrum (Fig. 2B). It is worth noting that it is challenging to assign the backbone of the C-terminal proline-rich sequence in A1620–T1706 by using the standard triple resonance experiments we employed, as prolines lack the amide protons necessary for such experiments. Although additional methods are available to assign proline-rich regions (Kanelis et al. 2000), we found it unnecessary to implement these for the present study because the shorter construct A1620–H1687, which contains only four prolines in the C-terminal tail, exhibited similar chemical shift perturbations compared to the longer one (Fig. 1B,C). In addition, the 13C dispersed NOESY aided the assignment of these prolines in the tail that interacts with the SH3 domain.
Figure 2.
NOE interactions confirm that the DdM7 SH3 domain interacts with the adjacent proline-rich region. (A) NOE interactions are displayed between the amide proton of I1644 and P1685 Hδ and Q1684 Hɛ atoms. As expected, these NOEs are absent in the shorter construct spanning A1620–V1680 (A) and supported by NOEs involving neighboring atoms in the 13C dispersed NOESY spectrum (B). (C) Two sets of resonances are observed for I1644, one of which lacks the NOE interactions with P1685 and Q1684. This set has similar amide chemical shift values compared to I1644 of the shorter construct (Fig. 1B).
It is worth noting that two amide resonance cross-peaks are observed in [1H, 15N] HSQC spectra for I1644 in the constructs that include the proline-rich region; the weaker of which overlaps with that of I1644 from the construct containing only the SH3 domain (Fig. 1B). We expect it to correspond to a minor population in the sample that does not interact with the proline-rich region. This hypothesis was confirmed in the 15N-dispered NOESY spectrum of A1620–T1706. In this spectrum, the set of resonances with the same amide chemical shift values of I1644 in shorter fragment lacks the NOE interactions with Q1684 and P1685 (Fig. 2C). This finding suggests that the sample contains protein in which the SH3 domain is bound to its C-terminal extension as well as protein that lacks this interaction. Based on the difference in their resonance volumes, the bound state is more populated.
SH3 domain interaction with the C-terminal proline-rich region is intra- rather than intermolecular
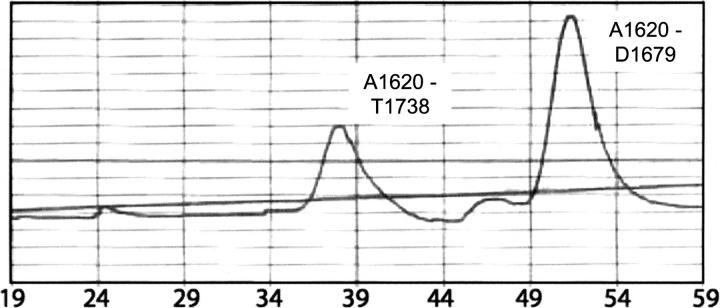
For the final stage of purification, the M7 protein fragments are subjected to size exclusion chromatography on an FPLC system. In all cases, the purified protein eluted in fractions expected for a monomer of the corresponding molecular weight. However, to provide further evidence that the observed interaction between the SH3 domain and its C-terminal proline-rich region is intra- rather than intermolecular, we mixed an M7 fragment containing the SH3 domain only (A1620–D1679) with one containing the C-terminal proline-rich region in addition (A1620–T1738) and subjected the mixture to FPLC size exclusion chromatography. Were the longer fragment forming dimers, a population would be expected in which these two constructs interact to elute in fractions between dimeric A1620–T1738 protein and monomeric A1620–D1679 protein. No such population was observed in either the UV spectrogram or by gel electrophoresis and Coomassie staining. The spectrogram revealed the presence of two peaks centered on fractions 37 and 50 (Fig. 3) and subsequent gel electrophoresis and Coomassie staining of protein in these fractions confirmed the first to contain only DdM7 A1620–T1738, which is 13.6 kDa, and the second to contain only DdM7 A1620–D1679, which is 6.9 kDa (data not shown). A small peak was observed in the spectrogram at fraction 24; however, no protein was revealed by gel electrophoresis and Coomassie staining in the fractions spanning 20–31 (Fig. 3). Although it is possible that the interactions between the SH3 domain and its proximal proline-rich region are intermolecular at high protein concentrations, we conclude that the DdM7 constructs containing the SH3 domain and adjacent proline-rich regions are largely monomeric at the concentrations used in this study.
Figure 3.
The DdM7 SH3 domain binds the C-terminal proline-rich regions in an intramolecular fashion. Equimolar quantities of constructs containing the SH3 domain alone (A1620–D1679) or with the adjacent proline-rich region (A1620–T1738) were incubated overnight and the resulting components separated according to their molecular weight by using an FPLC system equipped with a Superdex 75 column (Pharmacia). The UV spectrogram revealed two peaks centered on fractions 37 and 50, which is the expected position for molecular weights of 6.9 kDa (that of A1620–D1679) and 13.6 kDa (that of A1620–T1738), respectively. Gel electrophoresis and Coomassie staining confirmed this prediction, and although a small peak was recorded in the spectrogram at fraction 24, no protein was detected in fractions 20–31 by Coomassie staining.
M7 contains a canonical SH3 domain that binds its C-terminal proline-rich region in an unexpected manner
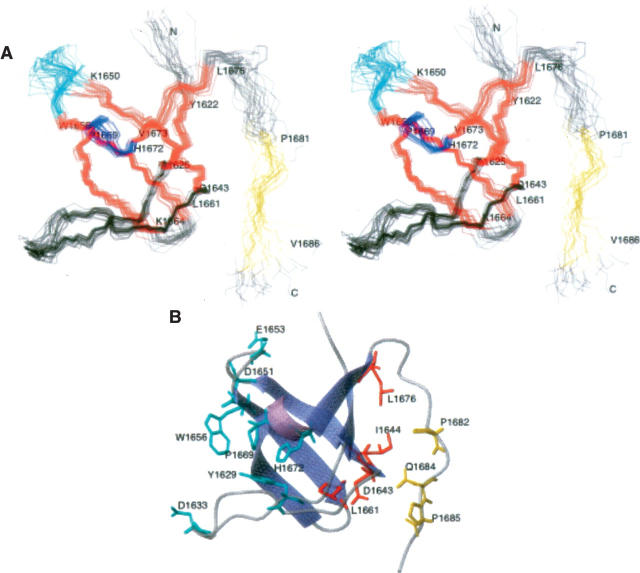
The structure of A1620–A1690 was determined by using the experimental parameters of Table 1. We determined the DdM7 SH3 domain contains five antiparallel β-strands spanning residues Y1622–A1625, D1643–K1650, W1656–L1661, K1664–P1669, and V1673–L1676 and a 310 helix spanning residues V1670–H1672 (Fig. 4A). Its fold resembles that of other SH3 domains reported in the literature (Musacchio et al. 1992; Yu et al. 1992; Noble et al. 1993), with a β-barrel architecture and two prominent loops, termed the n-Src and RT loops. W1656 and Y1629 form a conserved hydrophobic pocket common to SH3 domains that bind “PxxP” motifs. Anchoring basic residues at either end of the motif often engage in salt bridges with acidic residues of the n-Src or RT loops (Weng et al. 1995). D1651 and E1653, of the n-Src loop, or D1633, of the RT loop, are suitably positioned to serve such a role. A second hydrophobic pocket exists in other SH3 domains, and is formed by residues in the 310 helix and following β-strand. This region contains highly conserved P1669, but is less hydrophobic than other reported SH3 domains (Musacchio et al. 1992; Noble et al. 1993) due to the presence of D1671 and H1672 (Fig. 4B).
Table 1.
Structural statistics for the NMR structure of DdM7 A1620–A1690, which includes its SH3 domain
Figure 4.
The DdM7 SH3 domain interacts with an adjacent “PxxP” motif through an unexpected surface. (A) A stereoview of the 20 structures with no NOE violations above 0.5 Å are displayed with β-strands in red and the helix in blue. The region that has NOE interactions with the SH3 domain is highlighted in yellow, whereas the RT and n-Src loops are displayed in black and cyan, respectively. (B) Representative ribbon diagram of the residues A1620–H1687. The side chain atoms of residues in the region previously identified for binding “PxxP” motifs are included in cyan. Interacting residues of the SH3 domain or proline-rich region are displayed in red or yellow, respectively.
Most unique to the DdM7 SH3 domain is its interaction with the adjacent C-terminal proline-rich region (Fig. 4B). The surface used to interact with this region is opposite the hydrophobic surface previously identified to bind “PxxP” motifs. Interestingly, no interaction was observed between the SH3 domain and the PxxP sequence spanning P1695–P1698. In addition, we acquired a [1H, 15N] HSQC spectrum on a longer construct containing many more prolines, namely that spanning A1620–T1738. The SH3 domain resonances of this HSQC spectrum overlap with those of the construct spanning A1620–T1706 (see Supplemental Fig. 1). An 15N-dispersed NOESY spectrum recorded on this construct retains the interactions between the SH3 domain and P1682–P1685, and no new NOE interactions are observed for residues in the SH3 domain (data not shown).
Discussion
The DdM7 SH3 domain is structurally similar to other SH3 domains. The protein's fold consists of perpendicular β-sheets and a hydrophobic surface contains the conserved residues of Y1629, W1656, and P1669. Analogous residues in Src, Fyn, and Grb2 are positioned in a similar orientation and interact with “PxxP” motifs (Lim et al. 1994; Agrawal and Kishan 2002). We expect that these residues in DdM7 form similar interactions with “PxxP” motifs in other proteins. To our surprise, however, residues at the opposite side of the SH3 domain interact with an adjacent “PxxP” motif. In particular, the sequence C-terminal to the SH3 domain bends to enable I1644, D1643, L1661, and L1676 of the SH3 domain to interact with P1682, Q1684, and P1685.
Intramolecular interactions involving SH3 domains occur in Src, PI3, and Itk kinases (Koch et al. 1991; Kapeller et al. 1994; Andreotti et al. 1997). In these proteins, however, the interactions perform an autoregulatory role as polyproline motifs bind to residues in the hydrophobic binding pockets to inactivate these kinases. We demonstrate here that the binding of the proline-rich sequence immediately following the SH3 domain of DdM7 does not preclude binding to the surface typically used to bind “PxxP” motifs. The interaction we observe may perform a structural role in orienting the conformation of the region following the SH3 domain. There is precedent for myosins assuming a folded conformation. For example, the tail of M5 has recently been found to be capable of folding over and interacting with the motor domain, an interaction that is thought to regulate the activity of this motor protein (Liu et al. 2006b; Thirumurugan et al. 2006). It is tempting to speculate that intramolecular interactions in the DdM7 tail, perhaps promoted by binding of the SH3 domain with its nearby proline-rich region either plays a similar role in regulating the motor activity of this myosin or is required for interaction of DdM7 with its binding partners. Ongoing efforts to identify DdM7 SH3 domain binding partners through a combination of affinity and yeast two hybrid approaches, and testing of the requirement for the SH3 domain and adjacent Pro-rich sequence for DdM7 function using a complementation approach should provide insight into the functional contribution of the SH3 and proline domains.
Materials and methods
Cloning and production of M7 SH3 domains
A standard PCR-based approach was used to generate a fusion protein between GST and the DdM7 SH3 domain plus a segment of the adjacent proline-rich region. The regions of the gene encoding amino acids A1620–T1738, A1620–T1706, A1620–H1687, A1620–V1680, and A1620–D1679 of the DdM7 tail region were amplified using primers with restriction enzyme sites for directional cloning. The products were digested and ligated to the pGEX-6P-1 or pGEX-2TK (Amersham Biosciences) vector. Clones were identified by PCR and verified by automated DNA sequencing. The expression plasmid of interest was transformed into BL21(DE3) cells (Invitrogen). A 5-mL overnight culture was used to inoculate 1 L of LB growth media or M9 minimal media containing 15NH4Cl and/or 13C-labeled glucose to isotopically label samples. Protein expression was induced with 0.5 mM IPTG at 37°C for 4 h. Cells were harvested by centrifugation (5000g × 20 min.), frozen overnight at –80°C, lysed by sonication in PBS buffer (pH 7.5), and then centrifuged (15,000 × g for 30 min.). Cells were sonicated and centrifuged a second time for maximum protein yield in the case of 13C/15N single- and double-labeled samples. Glutathione-Sepharose resin (Amersham Biosciences) was added to the supernatant and the mixture incubated with gentle agitation at room temperature for 45 min in the presence of Complete Protease Inhibitor (Roche). The resin was then washed five times with PBS at 4°C and the fusion protein was cleaved by incubation with PreScission protease (Amersham Biosciences) at 4°C for 15 h. Cleaved products were purified on a Superdex 75 column (Pharmacia) equilibrated in 20 mM NaPO4, 100 mM NaCl (pH 6.4), and concentrated for NMR studies. 2D and 13C edited NOESY experiments as well as 2D TOCSY and DQ-COSY experiments required lyophilizing the sample to remove water followed by dissolving in D2O.
FPLC experiments to test for the presence of intermolecular interactions between the SH3 domain and its C-terminal proline-rich region
DdM7 A1620–D1679 and DdM7 A1620–T1738 were incubated together overnight at 0.3 mM protein concentration and 25°C in 20 mM NaPO4 and 100 mM NaCl, pH 6.4. The protein components of the mixture were then separated according to their molecular weight by using an FPLC system equipped with a Superdex 75 column (Pharmacia) in a buffer containing 20 mM NaPO4 and 100 mM NaCl (pH 6.4). Fractions were collected every 2 mL and analyzed for the presence of protein using gel electrophoresis and FPLC UV detection.
NMR Spectroscopy
All experiments were performed on Varian INOVA 600 or 800 MHz spectrometers equipped with HCN triple resonance probes and at 25°C with samples dissolved in 20 mM NaPO4, 100 mM NaCl at pH 6.4. Experimental data were processed by NMRPIPE (Delaglio et al. 1995) and the resulting spectra visualized in XEASY (Bartels et al. 1995) on Octane2 Silicon Graphics workstations. To assign chemical shift values for the backbone atoms of the A1620–T1706 fragment, a series of three heteronuclear NMR experiments were performed, including [1H,15N,13C] HNCA, [1H,15N,13C] HN(CO)CA, [1H,15N,13C] HNCO experiments. An 15N-edited TOCSY was used for side chain assignments. Two-dimensional homonuclear as well as 15N- and 13C-dispersed NOESY experiments were recorded on the fragment spanning A1620–T1706. The 15N- and 13C-dispersed NOESY experiments were recorded with mixing times of 120 and 80 msec, respectively. In addition, 15N-dispersed NOESY spectra were recorded on the fragments spanning A1620–D1679 and A1620–T1738.
Structure calculations
NOESY experiments were used to generate distance constraints. NOE restraints were grouped into four distance ranges (2.5, 3.5, 4.5, and 6.0 Å) based on peak intensity. The 15N-dispersed NOESY spectrum was calibrated by setting the average integrated value for NOE interactions V1673 Hα to E1674 HN and I1675 Hα to L1676 HN to 2.5 Å, whereas the intraresidue Hα to Hβ cross-peaks for L1661 and L1637 was set to 2.5 Å to calibrate the 13C-dispersed NOESY spectrum. Hydrogen bonds were identified by 1HN–1HN and 1Hα–1Hα NOEs in regions of predicted antiparallel β-strands. In addition, the program TALOS was used to predict the ϕ and ψ dihedral angle constraints (Cornilescu et al. 1999). The NOE-derived distance constraints, hydrogen bonds, and dihedral angle constraints (Table 1) were used in XPLOR version 3.851 (Brünger 1993) to determine the structure of a DdM7 fragment spanning A1620–A1690, which includes the SH3 domain. Of the 21 structures calculated, only one had NOE or dihedral angle violations >0.5 Å or 5°, respectively. The 20 structures with no violations are available through the Protein Data Bank (PDB ID code 2I0N).
Acknowledgments
Many thanks for the generous support of the United States Army in funding M.A.D.'s Advanced Civil Schooling. C.L. is grateful for funding from the Richard C. Nelson Scholarship from the College of Biological Sciences and Minnesota Supercomputing Institute Intern Program. NMR data were acquired in the NMR facility of the UMN, and we thank Dr. David Live and Dr. Beverly Ostrowsky for their technical assistance. Data processing and visualization occurred in the Minnesota Supercomputing Institute Basic Sciences Computing Lab. This work was funded by grants from the National Institutes of Health CA097004 (to K.J.W.) and GM046486 (to M.A.T.), Minnesota Medical Foundation (NMR facility), and NSF BIR-961477 (NMR facility).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Margaret A. Titus, Department of Genetics, Cell Biology, and Development, University of Minnesota, Minneapolis, MN 55455, USA; e-mail: titus004@umn.edu; fax: (612) 626-6140; or Kylie J. Walters, Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis, MN 55455, USA; e-mail: walte048@umn.edu; fax: (612) 625-2163.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062496807.
Abbreviations: HSQC, heteronuclear single quantum coherence; M, myosin; IPTG, isopropyl-β-D-thiogalactopyranoside; LB, Luria-Bertani; NMR, nuclear magnetic resonance; NOE, nuclear Overhauser enhancement; ppm, parts per million; SH3, Src homology domain 3.
References
- Agrawal, V. and Kishan, K.V. 2002. Promiscuous binding nature of SH3 domains to their target proteins. Protein Pept. Lett. 9: 185–193. [DOI] [PubMed] [Google Scholar]
- Andreotti, A.H., Bunnell, S.C., Feng, S., Berg, L.J., and Schreiber, S.L. 1997. Regulatory intramolecular association in a tyrosine kinase of the Tec family. Nature 385: 93–97. [DOI] [PubMed] [Google Scholar]
- Bartels, C., Xia, T.-H., Billeter, M., Güntert, P., and Wüthrich, K. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6: 1–10. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T. 1993. XPLOR version 3.1: A system for X-ray crystallography and NMR. Yale University Press, New Haven, CT.
- Cornilescu, G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13: 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Klebl, B.M., Tong, A.H., Webb, B.A., Leeuw, T., Leberer, E., Whiteway, M., Thomas, D.Y., and Boone, C. 2000. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 148: 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., Chen, J.K., Yu, H., Simon, J.A., and Schreiber, S.L. 1994. Two binding orientations for peptides to the Src SH3 domain: Development of a general model for SH3-ligand interactions. Science 266: 1241–1247. [DOI] [PubMed] [Google Scholar]
- Ishikawa, T., Cheng, N., Liu, X., Korn, E.D., and Steven, A.C. 2004. Subdomain organization of the Acanthamoeba myosin IC tail from cryo-electron microscopy. Proc. Natl. Acad. Sci. 101: 12189–12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G., Remmert, K., Wu, X., Volosky, J.M., and Hammer 3rd, J.A. 2001. The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol. 153: 1479–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanelis, V., Donaldson, L., Muhandiram, D., Rotin, D., Forman-Kay, J., and Kay, L.E. 2000. Sequential assignment of proline-rich regions in proteins: Application to modular binding domain complexes. J. Biomol. NMR 16: 253–259. [DOI] [PubMed] [Google Scholar]
- Kang, Y., Vossler, R.A., Diaz-Martinez, L.A., Winter, N.S., Clarke, D.J., and Walters, K.J. 2006. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J. Mol. Biol. 356: 1027–1035. [DOI] [PubMed] [Google Scholar]
- Kapeller, R., Prasad, K.V., Janssen, O., Hou, W., Schaffhausen, B.S., Rudd, C.E., and Cantley, L.C. 1994. Identification of two SH3-binding motifs in the regulatory subunit of phosphatidylinositol 3-kinase. J. Biol. Chem. 269: 1927–1933. [PubMed] [Google Scholar]
- Kieke, M.C. and Titus, M.A. 2003. The myosin superfamily—An overview. In Molecular motors (ed. M. Schliwa), pp. 3–44. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany.
- Koch, C.A., Anderson, D., Moran, M.F., Ellis, C., and Pawson, T. 1991. SH2 and SH3 domains: Elements that control interactions of cytoplasmic signaling proteins. Science 252: 668–674. [DOI] [PubMed] [Google Scholar]
- Krendel, M. and Mooseker, M.S. 2005. Myosins: Tails (and heads) of functional diversity. Physiology 20: 239–251. [DOI] [PubMed] [Google Scholar]
- Lechler, T., Shevchenko, A., and Li, R. 2000. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 148: 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.L., Ostap, E.M., Zot, H.G., and Pollard, T.D. 1999. Organization and ligand binding properties of the tail of Acanthamoeba myosin-IA. Identification of an actin-binding site in the basic (tail homology-1) domain. J. Biol. Chem. 274: 35159–35171. [DOI] [PubMed] [Google Scholar]
- Lee, W.L., Bezanilla, M., and Pollard, T.D. 2000. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J. Cell Biol. 151: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, W.A., Richards, F.M., and Fox, R.O. 1994. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature 372: 375–379. [DOI] [PubMed] [Google Scholar]
- Liu, F., Zhang, N., Zhou, X., Hanna, P.E., Wagner, C.R., Koepp, D.M., and Walters, K.J. 2006a. Protein aggregation and constitutive ubiquitylation of arylamine N-acetyltransferase. J. Mol. Biol. 361: 482–492. [DOI] [PubMed] [Google Scholar]
- Liu, J., Taylor, D.W., Krementsova, E.B., Trybus, K.M., and Taylor, K.A. 2006b. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature 442: 208–211. [DOI] [PubMed] [Google Scholar]
- Maniak, M. 2001. Cell adhesion: Ushering in a new understanding of myosin VII. Curr. Biol. 11: R315–R317. [DOI] [PubMed] [Google Scholar]
- Musacchio, A., Noble, M., Pauptit, R., Wierenga, R., and Saraste, M. 1992. Crystal structure of a Src-homology 3 (SH3) domain. Nature 359: 851–855. [DOI] [PubMed] [Google Scholar]
- Noble, M.E., Musacchio, A., Saraste, M., Courtneidge, S.A., and Wierenga, R.K. 1993. Crystal structure of the SH3 domain in human Fyn; Comparison of the three-dimensional structures of SH3 domains in tyrosine kinases and spectrin. EMBO J. 12: 2617–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumurugan, K., Sakamoto, T., Hammer III, J.A., Sellers, J.R., and Knight, P.J. 2006. The cargo-binding domain regulates structure and activity of myosin 5. Nature 442: 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxworth, R.I., Weber, I., Wessels, D., Addicks, G.C., Soll, D.R., Gerisch, G., and Titus, M.A. 2001. A role for myosin VII in dynamic cell adhesion. Curr. Biol. 11: 318–329. [DOI] [PubMed] [Google Scholar]
- Walters, K.J., Ferentz, A.E., Hare, B.J., Hidalgo, P., Jasanoff, A., Matsuo, H., and Wagner, G. 2001. Characterizing protein–protein complexes and oligomers by nuclear magnetic resonance spectroscopy. Methods Enzymol. 339: 238–258. [DOI] [PubMed] [Google Scholar]
- Walters, K.J., Lech, P.J., Goh, A.M., Wang, Q., and Howley, P.M. 2003. DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc. Natl. Acad. Sci. 100: 12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Goh, A.M., Howley, P.M., and Walters, K.J. 2003. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry 42: 13529–13535. [DOI] [PubMed] [Google Scholar]
- Weng, Z., Rickles, R.J., Feng, S., Richard, S., Shaw, A.S., Schreiber, S.L., and Brugge, J.S. 1995. Structure–function analysis of SH3 domains: SH3 binding specificity altered by single amino acid substitutions. Mol. Cell. Biol. 15: 5627–5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich, K. 1986. NMR of proteins and nucleic acids. Wiley, New York.
- Yu, H., Rosen, M.K., Shin, T.B., Seidel-Dugan, C., Brugge, J.S., and Schreiber, S.L. 1992. Solution structure of the SH3 domain of Src and identification of its ligand-binding site. Science 258: 1665–1668. [DOI] [PubMed] [Google Scholar]