Abstract
Collagen is the most abundant protein in animals. Every third residue in a collagen strand is a glycine with ϕ, ψ = −70°, 175°. A recent computational study suggested that replacing these glycine residues with d-alanine or d-serine would stabilize the collagen triple helix. This hypothesis is of substantial importance, as the glycine residues in collagen constitute nearly 10% of the amino acid residues in humans. To test this hypothesis, we synthesized a series of collagen mimic peptides that contain one or more d-alanine or d-serine residues replacing the canonical glycine residues. Circular dichroism spectroscopy and thermal denaturation experiments indicated clearly that the substitution of glycine with d-alanine or d-serine greatly disfavors the formation of a triple helix. Host–guest studies also revealed that replacing a single glycine residue with d-alanine is more destabilizing than is its replacement with l-alanine, a substitution that results from a common mutation in patients with collagen-related diseases. These data indicate that the glycine residues in collagen are not a surrogate for a d-amino acid and support the notion that the main-chain torsion angles of a glycine residue in the native structure (especially, ϕ > 0°) are critical determinants for its beneficial substitution with a d-amino acid in a protein.
Keywords: d-alanine, collagen, conformational stability, Ramachandran plot, d-serine
Most natural proteins are comprised of 19 l-amino acids and glycine, which is achiral. Although not usually found in natural proteins (Mitchell and Smith 2003), d-amino acid residues have conformational attributes that are useful for the imposition of conformational stability and as structural probes (Fairman et al. 1992; Krause et al. 2000). d-Proline, in particular, has been a demonstrably valuable component of artificial reverse turns (Imperiali et al. 1992; Struthers et al. 1996; Haque and Gellman 1997). Moreover, the nonnatural stereochemistry of d-amino acids endows resistance to proteolytic degradation (Fischer 2003), an attribute important in chemotherapeutic applications. Indeed, many compounds with antimicrobial and antitumor activity contain d-amino acids (Pohl et al. 1994; Frau and Price 1996).
The inversion of stereochemistry at Ci α makes the Ramachandran plot of d-amino acid residues differ by a 180° rotation from that of l-amino acid residues (Fig. 1). The absence of a stereocenter in glycine endows its Ramachandran plot with internal C2 symmetry, and the absence of a side chain provides access to most ϕ (Ci − 1′–Ni–Ci α–Ci′) and ψ (Ni–Ci α–Ci′–Ni + 1) torsion angles. Many of the angles accessible to glycine residues are accessible to d-amino acids but not l-amino acids. Accordingly, glycine can be a suitable target for substitution with a d-amino acid. Raleigh and coworkers, in particular, have replaced glycine residues with d-amino acids in several globular proteins and found that the incorporation of d-amino acids can greatly enhance conformational stability (Anil et al. 2004, 2006). In addition, d-amino acids can be better than l-amino acids as glycine surrogates, retaining protein function without perturbing protein structure (Valiyaveetil et al. 2004; Xie et al. 2005; Bang et al. 2006). These advantageous d-amino acid substitutions have a common feature—the target glycine residue always has ϕ > 0°.
Figure 1.
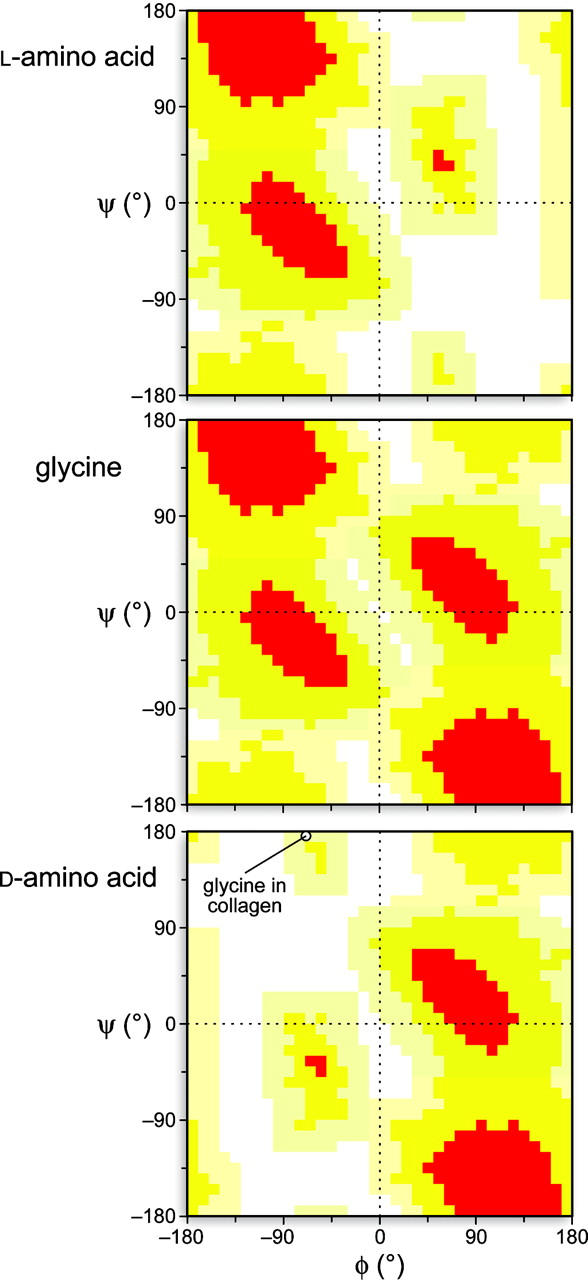
Ramachandran plots for l-amino acids (top), glycine (middle), and d-amino acids (bottom). The circle in the bottom panel indicates the values of ϕ = −70° and ψ = 175° for the glycine residues in an idealized collagen triple helix (Schumacher et al. 2005).
Collagen consists of three individual peptide strands folded into a right-handed triple helix. Each strand is a left-handed polyproline II helix with repeats of the sequence: Xaa–Yaa–Gly, in which Xaa is often 2S-proline (Pro) and Yaa is often (2S,4R)-4-hydroxyproline (Hyp). Of these residues, the glycine is perhaps the most important, as the structure of a collagen-like peptide containing a single glycine-to-alanine substitution suffers from substantial distortion and destabilization (Bella et al. 1994). Moreover, genetic mutations that lead to the replacement of a glycine residue greatly destabilize the triple helix and lead to human diseases (Beck et al. 2000).
A recent computational study by Dannenberg and coworkers suggested that replacing the glycine residues in collagen strands with d-alanine or d-serine residues would stabilize the triple helix (Tsai et al. 2005). Moreover, these workers suggested that d-serine would have a larger stabilizing effect than d-alanine because of the formation of a hydrogen bond between its side-chain hydroxyl group and a carbonyl group in another strand of the triple helix. The implications of these suggestions are enormous, as the glycines of collagen comprise ∼10% of the amino acid residues in humans (that is, one-third of the protein in humans times one-third of the residues in collagen). We were skeptical of this hypothesis, however, as the glycine residues in the collagen triple helix have ϕ < 0° (Fig. 1), which has not been conducive to favorable d-amino acid substitution in other proteins (Anil et al. 2004, 2006).
In this study, we report on the synthesis of collagen mimics containing d-alanine or d-serine in place of the canonical glycine residues. For comparison, we have also synthesized a collagen mimic with l-alanine substitution, as well as a mimic of “natural” collagen. Our experimental analyses of these peptides reveal whether the glycine residues are truly surrogates for a d-amino acid and provide insight on the use of d-amino acids in the design of stable peptides and proteins.
Results and Discussion
Conformational stability of (ProHyp-d-Ala)7 and (ProHyp-d-Ser)7 triple helices
We prepared six peptides by chemical synthesis (Table 1). All of these peptides are based on the sequence: (Pro-Hyp-Zaa)7, as Pro-Hyp-Gly is the most prevalent triplet in natural collagen (Ramshaw et al. 1998), and triple-helical (Pro-Hyp-Gly)7 [(POG)7] has a convenient T m = 36°C, where T m refers to the temperature at the midpoint of the thermal transition between the folded and unfolded states (Bretscher et al. 2001).
Table 1.
Peptides used in this study
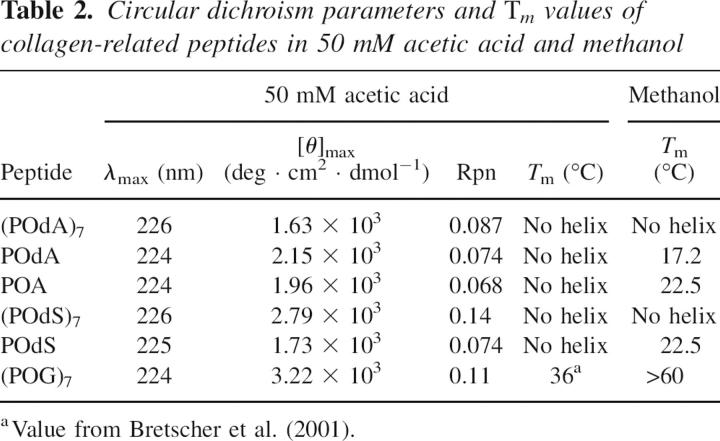
First, we synthesized (ProHyp-d-Ala)7 [(POdA)7] and (ProHyp-d-Ser)7 [(POdS)7] and examined their properties. (POdA)7 has a CD spectrum similar to that of (POG)7 at 4°C, but has a much smaller ellipticity maximum (1.63 × 103 vs. 3.22 × 103 deg cm2 dmol−1) (Fig. 2; Table 2). In addition, (POdA)7 has a very shallow ellipticity minimum, indicative of a very low propensity to form a triple helix. For (POdA)7, the Rpn value (which refers to the ratio of positive maximum to negative minimum and can be indicative of triple-helix formation) (Feng et al. 1996) is only 0.087, which is smaller than that of (POG)7 (Rpn = 0.11), and thereby provides additional evidence that monomeric strands are the dominant species in a solution of (POdA)7. Heating a solution of (POdA)7 results in only a linear decrease in ellipticity (Fig. 3A), rather than the cooperative transition characteristic of triple-helix denaturation. These results suggest that (POdA)7 does not form a triple helix.
Figure 2.
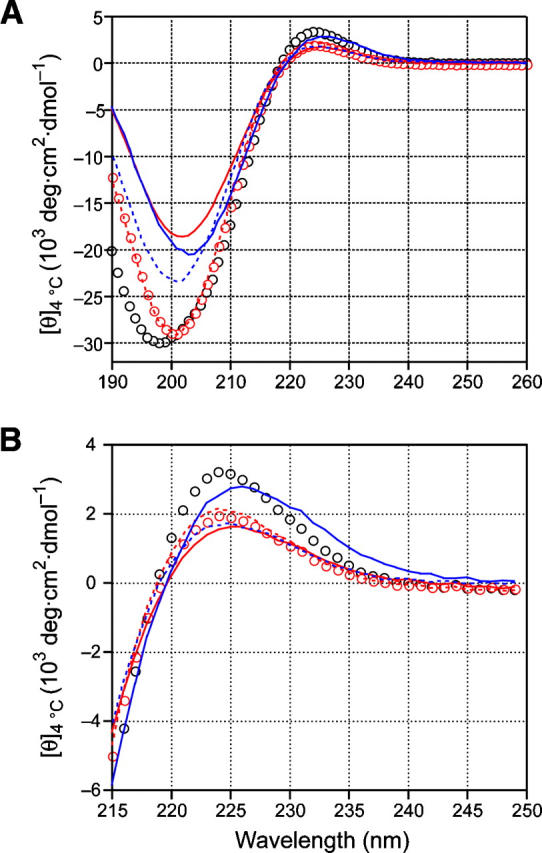
CD spectra of (POG)7 (open black circles), (POdA)7 (red line), POdA (red dashes), POA (open red circles), (POdS)7 (blue line), and POdS (blue dashes) at 4°C in 50 mM acetic acid at pH 2.9. The peptide concentrations were 0.2 mg/mL (0.1 mM). (A) Far-UV region. (B) Ellipticity maxima.
Table 2.
Circular dichroism parameters and Tm values of collagen-related peptides in 50 mM acetic acid and methanol
Figure 3.
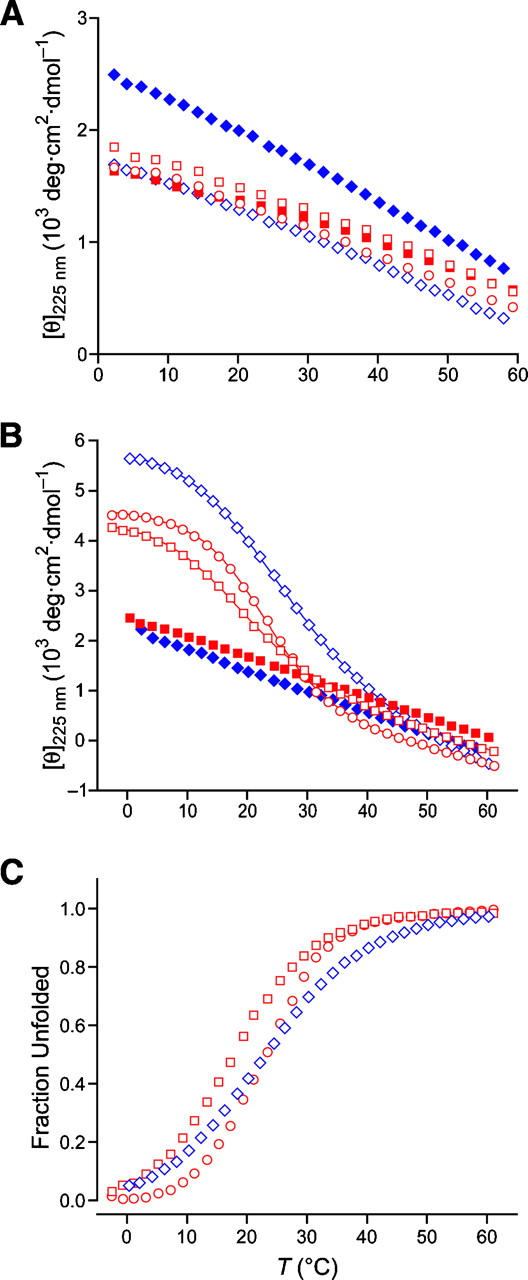
Thermal unfolding transition curves for (POdA)7 (red squares), POdA (open red squares), POA (open red circles), (POdS)7 (blue diamonds), and POdS (open blue diamonds). (A) In 50 mM acetic acid at pH 2.9; peptide concentrations, 0.4–0.8 mg/mL (0.2–0.4 mM). (B,C) In methanol; peptide concentrations, 0.6 mg/mL (0.3 mM). Lines depict the best fit of the data to a two-state model.
Like (POdA)7, (POdS)7 peptide also has a CD spectrum at 4°C similar to that of (POG)7 but with a smaller ellipticity maximum (2.79 × 103 vs. 3.22 × 103 deg cm2 dmol−1) (Fig. 2; Table 2). Compared to (POG)7, (POdS)7 has a shallow ellipticity minimum, which results in an Rpn value of 0.14. As with (POdA)7, heating a solution of (POdS)7 results in a linear decrease in ellipticity, indicating that no triple helix is formed in solutions of (POdS)7 (Fig. 3A).
Next, we attempted to use methanol to induce the formation of a triple helix by (POdA)7 and (POdS)7, as methanol is known to stabilize the collagen triple helix (Engel et al. 1977). According to CD measurements, no cooperative thermal denaturation curves were observed for (POdA)7 or (POdS)7 in methanol (Fig. 3B). The results indicate that in (POdA)7 and (POdS)7, the destabilization imposed by d-alanine and d-serine is severe.
Host–guest studies of (ProHypGly)3(ProHypZaa)(ProHypGly)3 (Zaa = d-Ala, l-Ala, d-Ser)
We used a host–guest strategy to measure the destabilization caused by a single d-Ala or d-Ser substitution in the middle of a triple helix. Specifically, we synthesized (ProHypGly)3(ProHyp-d-Ala)(ProHypGly)3 (POdA) and (ProHypGly)3(ProHyp-d-Ser)(ProHypGly)3 (POdS), and examined their properties. At 4°C, POdA has a CD spectrum similar to that of (POG)7 but with a smaller ellipticity maximum (2.15 × 103 vs. 3.22 × 103 deg cm2 dmol−1) (Fig. 2), indicating that POdA has a lower propensity to form a triple helix. The ellipticity maximum of POdA is slightly larger than that of (POdA)7, but the Rpn value of POdA (0.073) is smaller than that of (POG)7, indicating that monomers dominate in a solution of POdA. Heating the POdA solution results in a linear decrease in ellipticity at 225 nm rather than a cooperative transition.
Similar to POdA, POdS has a lower maximum molar ellipticity than does (POG)7 (1.73 × 103 vs. 3.22 × 103 deg cm2 dmol−1), and its Rpn value is 0.074 (Fig. 2; Table 2). The lack of cooperative thermal unfolding transition also indicates that POdS does not fold into a triple helix in aqueous solution. These results indicate that POdA and POdS do not form a triple helix under the normal condition. Thus, even a single d-Ala or d-Ser substitution for glycine can prevent the folding of collagen strands into a triple helix.
We also conducted CD measurements of POdA and POdS in methanol. Both peptides were found to form a triple helix. As shown in Figure 3B, thermal denaturation showed the cooperative transition characteristic of a triple helix. The T m determined from the thermal denaturation is 17.2°C for triple-helical POdA and 22.5°C for triple-helical POdS (Fig. 3C; Table 2). Thus, substitution of glycine with a d-serine residue is more favorable than is substitution with a d-alanine residue, as predicted by Dannenberg and coworkers (Tsai et al. 2005). The capability of POdA and POdS but not (POdA)7 and (POdS)7 to form a triple helix in methanol suggests that the destabilization imposed by d-amino acid substitution is additive.
Besides POdA, we also synthesized (ProHypGly)3(ProHyp-l-Ala)(ProHypGly)3 (POA) to compare the effect of d-Ala and l-Ala substitution. As expected, POA does not form a triple helix in 50 mM acetic acid and demonstrated very similar properties to POdA according to CD measurements (Figs. 2, 3; Table 2). The Rpn value of POA solution (0.068) is similar to that of POdA. Likewise, POA forms a triple helix in methanol as confirmed by its having a cooperative thermal unfolding transition (Fig. 3B). From the thermal unfolding curve, triple-helical POA was found to have a T m value of 22.5°C (Fig. 3C).
A glycine-to-alanine substitution is known to destabilize the collagen triple helix because of deleterious steric effects imposed by its side chain (Beck et al. 2000). Yet, d-alanine appears to be even less competent than l-alanine as a glycine surrogate, as triple-helical POdA has a lower T m value in methanol (17.2°C vs. 22.5°C) (Fig. 3C; Table 2). Thus, despite the presumed lack of steric effects induced by the side chain of a d-alanine or d-serine residue (Tsai et al. 2005), the preferred ϕ > 0° of these residues (Fig. 1) makes them unsuitable for forming a triple helix.
Conclusions
We have used experimental approaches to demonstrate that glycine is not a surrogate for a d-amino acid in the collagen triple helix. Although no deleterious steric consequences are presumed to arise from the side chains of d-alanine and d-serine upon forming a collagen triple helix (Tsai et al. 2005), the torsion angles in their main chain appear to disfavor triple-helix formation (Fig. 1). Rather than adopting the requisite polyproline II helix, a Hyp-d-Ala sequence could, for example, prefer to adopt a “bent” conformation like that observed in crystalline Pro-d-AlaOH (Ananthanarayanan and Cameron 1988). Some previous studies in which glycine residues were replaced with d-amino acids did lead to enhanced conformational stability, purportedly because of a decrease in the entropy of the unfolded state (Matthews et al. 1987; Ganter and Plückthun 1990; Stites and Pranata 1995). In each instance, however, the glycine residue had ϕ > 0°, which appears to be a prerequisite for a d-amino acid substitution to yield a more stable protein (Anil et al. 2004, 2006).
Materials and methods
General
Chemical reagents were obtained from Aldrich Chemical or Fisher Scientific and used without further purification. Amino acids and their derivatives were obtained from Novabiochem or Chem-Impex International. DMF and CH2Cl2 were drawn from a Baker CYCLE-TAINER. Flash chromatography was performed with columns of silica gel 60, 230–400 mesh (Silicycle). FmocProHypGlyOH was synthesized in solution as described previously (Bretscher et al. 2001), and FmocProHyp-d-AlaOH and FmocProHyp-d-Ser(tBu)OH were synthesized as shown in Schemes 1 and 2, respectively.
Scheme 1.
Scheme 2.
The term “concentrated under reduced pressure” refers to the removal of solvents and other volatile materials using a rotary evaporator at water aspirator pressure (<20 torr) while maintaining the water-bath temperature below 40°C. Residual solvent was removed from samples at high vacuum (<0.1 torr). The term “high vacuum” refers to vacuum achieved by a mechanical belt-drive oil pump.
NMR spectra were recorded with Bruker DMX-400 and DMX-500 spectrometers in the National Magnetic Resonance Facility at Madison (NMRFAM). CD experiments were performed with an Aviv Model 202SF circular dichroism spectrometer in the Biophysics Instrumentation Facility. Mass spectrometry was performed with a Micromass LCT instrument (electrospray ionization, ESI) in the Department of Chemistry or with an Applied Biosystems Voyager DE-Pro (matrix-assisted laser desorption ionization, MALDI–TOF) instrument in the Biophysics Instrumentation Facility.
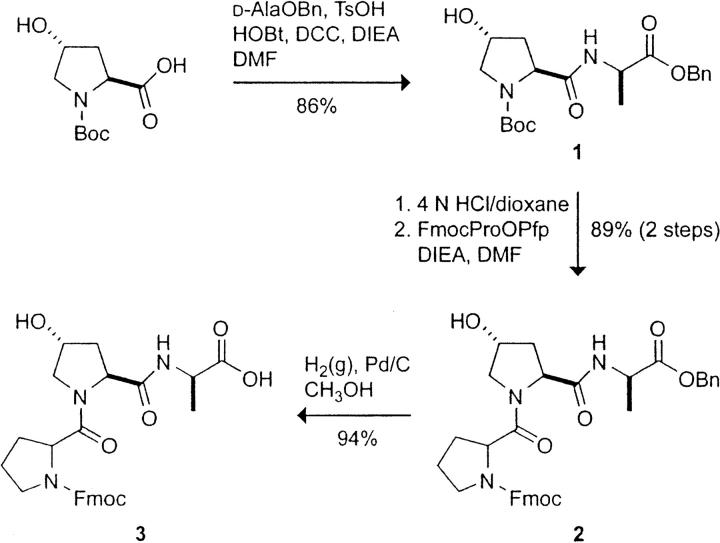
Synthesis of BocHyp-d-AlaOBn (1)
To a solution of BocHypOH (1.97 g, 8.52 mmol) in DMF (100 mL) was added d-AlaOBn · TsOH (3.0 g, 8.53 mmol), DCC (1.76 g, 8.53 mmol), HOBt (1.16 g, 8.58 mmol), and DIEA (4.5 mL, 25.8 mmol). The addition of DIEA caused the suspension to dissolve and white precipitate to appear after stirring for ∼30 min. The mixture was stirred under Ar(g) at room temperature overnight. The reaction mixture was filtered to remove all white precipitate and then concentrated under reduced pressure to an oily residue. The residue was dissolved in 100 mL of ethyl acetate, and the resulting solution was washed with NaHCO3(aq) [5% (w/v); twice with 50 mL], KHSO4(aq) [5% (w/v); twice with 50 mL], and saturated NaCl(aq) (50 mL). The organic layer was dried over anhydrous MgSO4(s) and concentrated under reduced pressure. The crude product was purified by flash chromatography, eluting with ethyl acetate, to yield peptide 1 as a white solid (2.90 g, 86%).
1H NMR (400 MHz, CDCl3): δ 7.43–7.32 (m, 5H), 5.20–5.11 (m, 2H), 4.63 (s, 1H), 4.40 (s, 2H), 3.64–3.39 (m, 2H), 2.85–1.90 (m, 3H), 1.44–1.41 (m, 12H).
HRMS (ESI): m/z calculated for C20H28N2O6Na ([M + Na]+) 415.1845, found 415.1838.
Synthesis of FmocProHyp-d-AlaOBn (2)
To a solution of peptide 1 (2.90 g, 7.39 mmol) in anhydrous dioxane (30 mL) was added 4 N HCl/dioxane (19 mL, 76 mmol). An additional 5 mL of 4 N HCl/dioxane was added during the stirring. The mixture was stirred under Ar(g) at room temperature for 1.5 h and then concentrated under reduced pressure to a white solid. The white solid was dried at reduced pressure for 6 h. To the solid was added DMF (80 mL), FmocProOPfp (3.72 g, 7.39 mmol), and DIEA (2.6 mL, 14.9 mmol). The mixture became a clear, light yellow solution upon DIEA addition. The reaction mixture was stirred under Ar(g) at room temperature overnight and then concentrated under reduced pressure to a brown, oily residue. The crude product was purified by flash chromatography, eluting with CH3OH [2% (v/v)] in ethyl acetate, to yield peptide 2 as a white powder (4.0 g, 89%).
1H NMR (400 MHz, CDCl3, 2 rotamers): δ 8.00–7.28 (m, 13H), 5.17–5.06 (m, 2H), 4.81 (t, J = 7.2 Hz, 0.8H), 4.69–4.66 (m, 0.2H), 4.60–4.57 (m, 6H), 3.89–3.42 (m, 4H), 2.78–1.80 (m, 7H), 1.42–1.36 (m, 3H).
HRMS (ESI): m/z calculated for C35H37N3O7Na ([M + Na]+) 634.2529, found 634.2451.
Synthesis of FmocProHyp-d-AlaOH (3)
A small amount of CH3OH was added to peptide 2 (4.00 g, 6.54 mmol), and the mixture was flashed with Ar(g) before the addition of Pd/C (0.47 g) and more CH3OH (70 mL). The mixture was stirred under H2(g) at room temperature for 1 h, filtered through Celite, and concentrated under reduced pressure. The crude product was purified by flash chromatography, eluting with CH2Cl2/CH3OH/HCOOH (90:10:0.5), to yield peptide 3 as a white powder (3.23 g, 94%).
1H NMR (400 MHz, CDCl3, 2 rotamers): δ 7.76–7.36 (m, 8H), 4.71 (t, J = 7.4 Hz, 1H), 4.51–4.34 (m, 4H), 4.23–4.17 (m, 2H), 3.68–3.48 (m, 4H), 2.25–1.85 (m, 7H), 1.10 (d, J = 6.8 Hz, 3H).
13C NMR (100 MHz, CDCl3, 2 rotamers): δ 174.8, 171.7, 171.1, 155.6, 144.1, 143.8, 141.5, 141.4, 128.0, 127.3, 125.5, 125.4, 125.2, 120.1, 70.3, 68.2, 59.3, 58.9, 55.0, 48.4, 47.2, 47.1, 37.4, 29.3, 24.4, 18.0.
HRMS (ESI): m/z calculated for C28H31N3O7Na ([M + Na]+) 544.2060, found 544.2070.
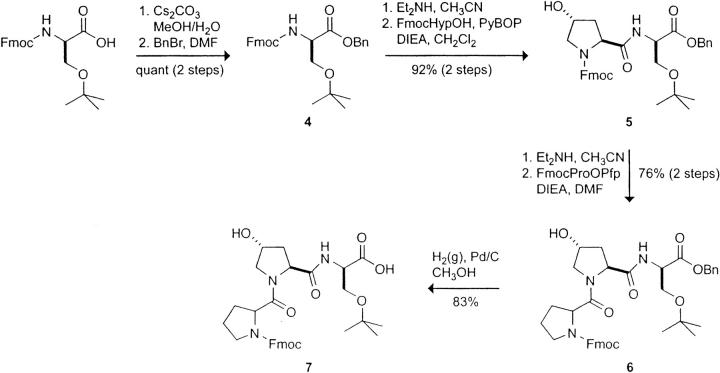
Synthesis of Fmoc-d-Ser(tBu)OBn (4)
A solution of Cs2CO3 (0.64 g, 1.96 mmol) in H2O (13 mL) at 0°C was added to a solution of Fmoc-d-Ser(tBu)OH (1.50 g, 3.91 mmol) in MeOH (20 mL) at 0°C. The mixture was stirred for 5 min and then concentrated under reduced pressure. Residual solvent was removed as an azeotrope with MeOH (2×) and by high vacuum to give a white solid [Fmoc-d-Ser(tBu)O−Cs+]. The solid was dissolved in DMF (30 mL), and the resulting solution was cooled to 0°C under Ar(g). Benzyl bromide (0.47 mL, 3.91 mmol) was added, and the resulting mixture was allowed to warm to room temperature and then stirred under Ar(g) for 22 h. The mixture was filtered to remove insoluble CsBr and evaporated to a white solid. The solid was dissolved in EtOAc (80 mL), and the organic layer was washed with H2O (twice with 40 mL), dried over MgSO4(s), and concentrated under reduced pressure. The crude product was purified by flash chromatography, eluting with EtOAc/hexanes (1:1), to yield Fmoc-d-Ser(tBu)OBn (4) as a white solid (1.85 g, quantitative).
1H NMR (500 MHz, CDCl3, >85% major rotamer): δ 7.78 (d, J = 7.6 Hz, 2H), 7.63 (t, J = 6.6 Hz, 2H), 7.41 (t, J = 7.5 Hz, 2H), 7.39–7.29 (m, 8H), 5.74 (d, J = 9.0 Hz, 1H), 5.26 (d, J = 12.3 Hz, 1H), 5.18 (d, J = 12.3 Hz, 1H), 4.56 (app d, J = 8.9 Hz, 1H), 4.42 (dd, J = 10.6, 7.2 Hz, 1H), 4.35 (dd, J = 10.5, 7.5 Hz, 1H), 3.87 (dd, J = 8.9, 2.0 Hz, 1H), 3.61 (dd, J = 8.9, 2.6 Hz, 1H), 1.11 (s, 9 H).
HRMS (ESI): m/z calculated for C29H31NO5Na ([M + Na]+) 496.2100, found 496.2113.
Synthesis of FmocHyp-d-Ser(tBu)OBn (5)
To a solution of Fmoc-d-Ser(tBu)OBn (4) (1.85 g, 3.91 mmol) in CH3CN (80 mL) was added HNEt2 (16.2 mL, 156.4 mmol). The solution was stirred at room temperature for 4 h and concentrated under reduced pressure to a yellowish residue [d-Ser(tBu)OBn]. The residue was dissolved in CH2Cl2 (80 mL), FmocHypOH (1.38 g, 3.91 mmol) was added, and the suspension was cooled to 0°C under Ar(g). PyBOP (2.04 g, 3.91 mmol) and DIEA (2.1 mL, 11.73 mmol) were added to the mixture, giving a clear solution. The resulting mixture was allowed to warm to room temperature and stirred under Ar(g) for 20 h. The mixture was then concentrated under reduced pressure, and the remaining residue was dissolved in EtOAc (50 mL). The organic layer was washed with 0.1 N HCl(aq) (20 mL), saturated NaHCO3(aq) (20 mL), and H2O (20 mL), dried over MgSO4(s), and concentrated under reduced pressure. The crude product was purified by flash chromatography, eluting with EtOAc/hexanes (7:3), to yield FmocHyp-d-Ser(tBu)OBn (5) as a white solid (2.10 g, 92% over 2 steps).
1H NMR (500 MHz, CDCl3, ∼3:2 ratio of 2 rotamers) data were δ 7.80–7.72 (m, 2H), 7.61–7.50 (m, 2.6H), 7.44–7.27 (m, 10H), 6.84 (d, J = 8.6 Hz, 0.4H), 5.21 (d, J = 12.4 Hz, 0.6H), 5.15 (d, J = 12.1 Hz, 0.4H), 5.10–5.00 (m, 1H), 4.71 (bs, 1H), 4.60–4.47 (m, 1.6H), 4.46–4.38 (m, 1.4H), 4.34–4.28 (m, 0.6H), 4.27–4.17 (m, 1.4H), 3.86–3.72 (m, 1.6H), 3.67–3.51 (m, 2H), 3.29 (d, J = 8.3 Hz, 0.4H), 2.42–2.30 (m, 1H), 1.04 (s, 5.4H), 0.95 (s, 3.6H).
HRMS (ESI): m/z calculated for C34H38N2O7Na ([M + Na]+) 609.2577, found 609.2565.
Synthesis of FmocProHyp-d-Ser(tBu)OBn (6)
To a solution of peptide 5 (0.80 g, 1.36 mmol) in CH3CN (50 mL) was added HNEt2 (5.6 mL, 54.4 mmol). The resulting solution was stirred at room temperature for 4 h and evaporated to a clear residue [Hyp-d-Ser(tBu)OBn]. DMF (40 mL) was added to the residue, followed by FmocProOPfp (0.68 g, 1.36 mmol) and DIEA (0.48 mL, 2.72 mmol), and the resulting milky suspension was stirred at room temperature under Ar(g) for 24 h. The mixture was concentrated under reduced pressure, and the remaining residue was dissolved in EtOAc (100 mL). The organic layer was washed with 0.1 N HCl(aq) (50 mL), saturated NaHCO3(aq) (50 mL), and H2O (50 mL), dried over MgSO4(s), and concentrated under reduced pressure. The crude product was purified by flash chromatography, eluting with a linear gradient of EtOAc in hexanes [80%–100% (v/v) EtOAc in hexanes], to yield peptide 6 as a colorless oily solid (0.71 g, 76% over 2 steps).
1H NMR (500 MHz, CDCl3, ∼4:1 ratio of 2 rotamers): δ 7.79–7.69 (m, 3H), 7.63–7.52 (m, 2H), 7.30–7.27 (m, 10H), 5.25–5.20 (m, 1H), 5.08 (d, J = 12.3 Hz, 0.8H), 5.03 (d, J = 12.6 Hz, 0.2H), 4.91 (app t, J = 7.2 Hz, 0.8H), 4.80–4.74 (m, 0.2H), 4.66 (d, J = 7.9 Hz, 0.8H), 4.60 (d, J = 7.5 Hz, 0.2H), 4.53–4.42 (m, 2.2H), 4.40–4.27 (m, 2H), 4.26–4.19 (m, 1H), 4.05 (d, J = 11.1 Hz, 0.8H), 3.87–3.80 (m, 1H), 3.72–3.65 (m, 0.8H), 3.65–3.59 (m, 0.2H), 3.59–3.43 (m, 3H), 3.36 (s, 0.8H), 3.25–3.19 (m, 0.2H), 2.61–2.50 (m, 1H), 2.29–2.18 (m, 1H), 2.14–1.68 (m, 5H), 1.12 (s, 7.2H), 1.10 (s, 1.8H).
HRMS (ESI): m/z calculated for C39H45N3O8Na ([M + Na]+) 706.3104, found 706.3124.
Synthesis of FmocProHyp-d-Ser(tBu)OH (7)
A suspension of peptide 6 (0.70 g, 1.02 mmol) and Pd/C [0.07 g, 10% (w/w)] in MeOH (40 mL) was stirred under an atmosphere of H2(g) for 4 h. The mixture was filtered through a pad of Celite and concentrated under reduced pressure. The crude product was purified by flash chromatography, eluting with a linear gradient of MeOH in CH2Cl2 (MeOH/CH2Cl2, 5:95–10:90), to yield peptide 7 as a white solid (0.50 g, 83%).
1H NMR (400 MHz, DMSO-d 6, 2 rotamers): δ 7.92–7.84 (m, 2H), 7.68–7.50 (m, 3H), 7.45–7.29 (m, 4H), 5.16 (bs, 1H), 4.59–4.52 (m, 1H), 4.50–4.42 (m, 1H), 4.38–4.28 (m, 1H), 4.28–4.07 (m, 4H), 3.68–3.32 (m, 6H), 2.30–2.07 (m, 1H), 2.03–1.73 (m, 5H), 1.06 (app d, J = 1.3 Hz, 9H).
13C NMR (100 MHz, DMSO-d 6, 2 rotamers): 170.5, 170.4, 170.3, 170.2, 153.7, 143.9, 143.8, 140.7, 140.6, 127.7, 127.3, 127.1, 120.1, 120.0, 72.2, 68.7, 68.5, 66.8, 66.5, 61.9, 58.5, 58.4, 58.0, 57.6, 54.2, 53.9, 53.8, 47.0, 46.7, 46.6, 46.2, 37.5, 29.6, 28.5, 27.3, 23.6, 22.6.
HRMS (ESI): m/z calculated for C32H38N3O8 ([M−H]−) 592.2659, found 592.2647.
Attachment of FmocProHyp-d-AlaOH (3) to 2-chlorotrityl resin
Under Ar(g), 120 mg (0.19 mmol) of 2-chlorotrityl chloride resin (loading 1.6 mmol/g) was swollen in dry CH2Cl2 (3 mL). A solution of peptide 3 (100 mg, 0.19 mmol) and DIEA (0.1 mL, 0.57 mmol) in dry CH2Cl2 (1.5 mL) was added by syringe. An additional 2.0 mL of dry CH2Cl2 was used to ensure complete transfer. After 2 h, 2.5 mL of anhydrous CH3OH was added to the mixture to cap any remaining active sites on the resin. The resin-bound peptide was isolated by gravity filtration, washed with several portions of dry CH2Cl2 (∼30 mL), and dried under high vacuum. The mass of the resin-bound peptide was 194 mg. Loading was measured by UV spectroscopy using a reported protocol to be 0.64 mmol/g (Applied Biosystems Technical Note 123485, Rev 2).
Attachment of FmocProHyp-d-Ser(tBu)OH (7) to 2-chlorotrityl resin
Peptide 7 was loaded onto 2-chloritrityl resin in a fashion similar to that described for peptide 3. Loading was measured by UV spectroscopy using a reported protocol to be 0.44 mmol/g (Applied Biosystems Technical Note 123485, Rev 2).
Attachment of FmocProHypGlyOH to 2-chlorotrityl resin
FmocProHypGlyOH was loaded onto 2-chloritrityl resin in a fashion similar to that described for peptide 3. Loading was measured by UV spectroscopy using a reported protocol to be 0.66 mmol/g (Applied Biosystems Technical Note 123485, Rev 2).
Peptide synthesis and purification
All 21-mer peptides were synthesized by segment condensation of their corresponding Fmoc-tripeptides [FmocProHyp-d-AlaOH, FmocProHyp-d-Ser(tBu)OH, FmocProHypGlyOH] or Fmoc-protected amino acids on a 25-μmol scale by solid-phase methods. The synthesis was carried out by using HBTU-mediated coupling and standard reaction cycles on Applied Biosystems Model 432A automated peptide synthesizers in the University of Wisconsin–Madison Biotechnology Center. Use of a resin that was pre-loaded with trimer (as described above) generated a free C terminus following cleavage from the resin with trifluoroacetic acid (TFA)/triisopropylsilane/H2O (38:1:1). Each peptide has a free N terminus. Peptides were purified by reverse-phase HPLC with a Varian C18 semipreparative column. Water/acetonitrile gradients containing TFA [0.1% (v/v)] were used for the purification of POA, POdA, (POdA)7, POdS, (POdS)7, and (POG)7. All peptides were judged to be >90% pure by HPLC analysis. The identities of all peptides were confirmed by using matrix-assisted laser desorption ionization time of flight (MALDI–TOF) mass spectrometry. The calculated and observed molecular masses were: POA, calculated 1901.9, observed 1902.7; POdA, calculated 1901.9, observed 1902.4; (POdA)7, calculated 1986.0, observed 1986.5; POdS, calculated 1918.9, observed 1919.4; (POdS)7, calculated 2098.9, observed 2099.7; (POG)7, calculated 1887.9, observed 1888.5.
Circular dichroism spectroscopy
Far-UV CD spectra were obtained at 4°C in 50 mM acetic acid at pH 2.9 using a 1-mm pathlength quartz cuvette and a spectrometer bandwidth of 1 nm. Peptide concentrations (±20%) were determined by weighing peptides prior to their dissolution, and were 0.2 mg/mL (0.1 mM) for far-UV CD measurements and 0.4–0.8 mg/mL (0.2–0.4 mM) for thermal unfolding experiments. All samples were incubated at 4°C for ≥24 h before measurements. Thermal unfolding was monitored at 225 nm in a 1-mm pathlength quartz cuvette with a 5-min equilibration at each temperature. Values of T m in methanol were determined by fitting the molar ellipticity at 225 nm to a two-state model (Engel et al. 1977).
Acknowledgments
This work was supported by grant AR44276 (NIH). F.W.K. was supported by postdoctoral fellowship AR50881 (NIH). The University of Wisconsin—Madison Biophysics Instrumentation Facility was established with Grants BIR-9512577 (NSF) and RR13790 (NIH). NMRFAM was supported by Grant P41RR02301 (NIH).
Footnotes
Reprint requests to: Ronald T. Raines, Department of Biochemistry, University of Wisconsin–Madison, 433 Babcock Drive, Madison, WI 53706-1544, USA; e-mail: raines@biochem.wisc.edu; fax: (608) 262-3453.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062560107.
References
- Ananthanarayanan, V.S. and Cameron, T.S. 1988. Proline-containing β-turns. IV. Crystal and solution conformations of tert-butyloxycarbonyl-L-prolyl-D-alanine and tert-butyloxycarbonyl-L-prolyl-D-alanyl-L-alanine. Int. J. Pept. Protein Res. 31: 399–411. [PubMed] [Google Scholar]
- Anil, B., Song, B., Tang, Y., and Raleigh, D.P. 2004. Exploiting the right side of the Ramachandran plot: Substitution of glycines by D-alanine can significantly increase protein stability. J. Am. Chem. Soc. 126: 13194–13195. [DOI] [PubMed] [Google Scholar]
- Anil, B., Craig-Schapiro, R., and Raleigh, D.P. 2006. Design of a hyperstable protein by rational consideration of unfolded state interactions. J. Am. Chem. Soc. 128: 3144–3145. [DOI] [PubMed] [Google Scholar]
- Bang, D., Gribenko, A.V., Tereshko, V., Kossiakoff, A.A., Kent, S.B., and Makhatadze, G.I. 2006. Dissecting the energetics of protein α-helix C-cap termination through chemical protein synthesis. Nat. Chem. Biol. 2: 139–143. [DOI] [PubMed] [Google Scholar]
- Beck, K., Chan, V.C., Shenoy, N., Kirkpatrick, A., Ramshaw, J.A.M., and Brodsky, B. 2000. Destabilization of osteogenesis imperfecta collagen-like model peptides correlates with the identity of the residue replacing glycine. Proc. Natl. Acad. Sci. 97: 4273–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella, J., Eaton, M., Brodsky, B., and Berman, H.M. 1994. Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution. Science 266: 75–81. [DOI] [PubMed] [Google Scholar]
- Bretscher, L.E., Jenkins, C.L., Taylor, K.M., DeRider, M.L., and Raines, R.T. 2001. Conformational stability of collagen relies on a stereoelectronic effect. J. Am. Chem. Soc. 123: 777–778. [DOI] [PubMed] [Google Scholar]
- Engel, J., Chen, H.-T., Prockop, D.J., and Klump, H. 1977. The triple helix–coil conversion of collagen-like polytripeptides in aqueous and nonaqueous solvents. Comparison of the thermodynamic parameters and the binding of water to (L-Pro–L-Pro–Gly)n and (L-Pro–L-Hyp–Gly)n . Biopolymers 16: 601–622. [DOI] [PubMed] [Google Scholar]
- Fairman, R., Anthony-Cahill, S.J., and DeGrado, W.F. 1992. The helix-forming propensity of D-alanine in a right-handed α-helix. J. Am. Chem. Soc. 114: 5458–5459. [Google Scholar]
- Feng, Y., Melacini, G., Taulane, J.P., and Goodman, M. 1996. Acetyl-terminated and template-assembled collagen-based polypeptides composed of Gly-Pro-Hyp sequences. 2. Synthesis and conformational analysis by circular dichroism, ultraviolet absorbance, and optical rotation. J. Am. Chem. Soc. 118: 10351–10358. [Google Scholar]
- Fischer, P.M. 2003. The design, synthesis and application of stereochemical and directional peptide isomers: A critical review. Curr. Protein Pept. Sci. 4: 339–356. [DOI] [PubMed] [Google Scholar]
- Frau, J. and Price, S.L. 1996. On the electrostatic and steric similarity of lactam compounds and the natural substrate for bacterial cell-wall biosynthesis. J. Comput. Aided Mol. Des. 10: 107–122. [DOI] [PubMed] [Google Scholar]
- Ganter, C. and Plückthun, A. 1990. Glycine to alanine substitutions in helices of glyceraldehyde-3-phosphate dehydrogenase: Effects on stability. Biochemistry 29: 9395–9402. [DOI] [PubMed] [Google Scholar]
- Haque, T.S. and Gellman, S.H. 1997. Insights on β-hairpin stability in aqueous solution from peptides with enforced type I′ and type II′ β-turns. J. Am. Chem. Soc. 119: 2303–2304. [Google Scholar]
- Imperiali, B., Fisher, S.L., Moats, R.A., and Prins, T.J. 1992. A conformational study of peptides with the general structure Ac-L-Xaa-Pro-D-Xaa-L-Xaa-NH2: Spectroscopic evidence for a peptide with significant β-turn character in water and in dimethyl sulfoxide. J. Am. Chem. Soc. 114: 3182–3188. [Google Scholar]
- Krause, E., Bienert, M., Schmieder, P., and Wenschuh, H. 2000. The helix-destabilizing propensity scale of D-amino acids: The influence of side chain steric effects. J. Am. Chem. Soc. 122: 4865–4870. [Google Scholar]
- Matthews, B.W., Nicholson, H., and Becktel, W.J. 1987. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc. Natl. Acad. Sci. 84: 6663–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.B.O. and Smith, J. 2003. D-Amino acid residues in peptides and proteins. Proteins 50: 563–571. [DOI] [PubMed] [Google Scholar]
- Pohl, E., Sheldrick, G.M., Fischer, S., and Lackner, H. 1994. Structure of cyclo-[L-threonyl-D-valyl-L-prolyl-sarcosyl-N-methyl-L-valyl-OThr] at 153 K. Acta Crystallogr. C 50: 100–103. [Google Scholar]
- Ramshaw, J.A.M., Shah, N.K., and Brodsky, B. 1998. Gly–X–Y tripeptide frequencies in collagen: A context for host–guest triple-helical peptides. J. Struct. Biol. 122: 86–91. [DOI] [PubMed] [Google Scholar]
- Schumacher, M., Mizuno, K., and Bächinger, H.P. 2005. The crystal structure of the collagen-like polypeptide (glycyl-4(R)-hydroxyprolyl-4(R)-hydroxyprolyl)9 at 1.55 Å resolution shows up-puckering of the proline ring in the Xaa position. J. Biol. Chem. 280: 20397–20403. [DOI] [PubMed] [Google Scholar]
- Stites, W.E. and Pranata, J. 1995. Empirical evaluation of the influence of side chains on the conformational entropy of the polypeptide backbone. Proteins 22: 132–140. [DOI] [PubMed] [Google Scholar]
- Struthers, M.D., Cheng, R.P., and Imperiali, B. 1996. Economy in protein design: Evolution of a metal-independent ββα motif based on the zinc finger domains. J. Am. Chem. Soc. 118: 3073–3081. [Google Scholar]
- Tsai, M., Xu, Y., and Dannenberg, J.J. 2005. Completely geometrically optimized DFT/ONIOM triple-helical collagen-like structures containing the ProProGly, ProProAla, ProProDAla, and ProProDSer triads. J. Am. Chem. Soc. 127: 14130–14131. [DOI] [PubMed] [Google Scholar]
- Valiyaveetil, F.I., Sekedat, M., MacKinnon, R., and Muir, T.W. 2004. Glycine as a D-amino acid surrogate in the K+-selectivity filter. Proc. Natl. Acad. Sci. 101: 17045–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, C., Prahl, A., Ericken, B., Wu, Z., Zeng, P., Li, X., Lu, W.-Y., Lubkowski, J., and Lu, W. 2005. Reconstruction of the conserved β-bulge in mammalian defensins using D-amino acids. J. Biol. Chem. 280: 32921–32929. [DOI] [PubMed] [Google Scholar]