Abstract
BTB-zinc finger (BTB-ZF) proteins are transcription regulators with roles in development, differentiation, and oncogenesis. In these proteins, the BTB domain (also known as the POZ domain) is a protein–protein interaction motif that contains a dimerization interface, a possible oligomerization surface, and surfaces for interactions with other factors, including nuclear co-repressors and histone deacetylases. The BTB-ZF protein LRF (also known as ZBTB7, FBI-1, OCZF, and Pokemon) is a master regulator of oncogenesis, and represses the transcription of a variety of important genes, including the ARF, c-fos, and c-myc oncogenes and extracellular matrix genes. We determined the crystal structure of the BTB domain from human LRF to 2.1 Å and observed the canonical BTB homodimer fold. However, novel features are apparent on the surface of the homodimer, including differences in the lateral groove and charged pocket regions. The residues that line the lateral groove have little similarity with the equivalent residues from the BCL6 BTB domain, and we show that the 17-residue BCL6 Binding Domain (BBD) from the SMRT co-repressor does not bind to the LRF BTB domain.
Keywords: BTB, POZ, LRF, ZBTB7, SMRT, transcription repression, X-ray crystallography
Proteins containing C2H2-type zinc finger (ZF) DNA-binding motifs constitute the largest family of sequence-specific transcription factors encoded in the human genome (for review, see Klug and Schwabe 1995). Many of these proteins can be further subdivided into groups defined by the presence of particular N-terminal domains, and the most commonly observed domains in ZF proteins are the BTB (bric-à-brac, tramtrack, broad-complex, also known as POZ), KRAB (Krüppel-associated box), and SCAN (also known as leucine-rich region, LeR) domains (Collins et al. 2001). The human genome encodes 44 BTB-ZF proteins, and many of these are regulators of key genes involved in cell proliferation and differentiation (Stogios et al. 2005). In typical BTB-ZF proteins, the BTB domain is found at the extreme N terminus, followed by a central section predicted to have a high degree of structural disorder, and is terminated by a C-terminal region containing multiple tandem ZF repeats. In these proteins, the BTB domains have two main functional roles: First, they drive the dimerization, and possibly oligomerization, of the protein (Ahmad et al. 1998, 2003; Li et al. 1999); second, they bind to other proteins, recruiting these to the regulatory sites of target genes. Consistent with their important role in transcription regulation, many BTB-ZF proteins, including BCL6, PLZF, Kaiso, HIC1, FAZF, and LRF/ZBTB7, are either known proto-oncogenes or have been strongly implicated in oncogenic processes (for review, see Kelly and Daniel 2006).
Many BTB-ZF proteins are transcription repressors, and this activity is usually due to the BTB-mediated recruitment of transcriptional co-repressors, such as N-CoR (nuclear co-repressor), SMRT (silencing mediator of retinoic acid and thyroid hormone receptor, also known as N-CoR2), mSin3A, or histone deacetylases to promotor regions (David et al. 1998; Melnick et al. 2002; Ahmad et al. 2003; Yoon et al. 2003). BTB domains have varying affinities for these interactions: The BCL6 domain strongly interacts with SMRT (Dhordain et al. 1998; Huynh and Bardwell 1998; Ahmad et al. 2003), Kaiso BTB selectively interacts with N-CoR but not SMRT (Yoon et al. 2003), while the HIC1 BTB domain does not interact with either (Deltour et al. 1999). Structural studies of BTB domains in complex with co-repressors have been important in the design of therapeutic interventions against the function of BTB-ZF proteins. For example, the 17-residue BCL6-binding domain (BBD) of SMRT binds to the lateral groove of the BCL6 BTB domain (Ahmad et al. 2003) and has been used to design a peptide inhibitor of BCL6 function (Polo et al. 2004). Further studies into the structure of the lateral groove and how it interacts with co-repressors and/or HDACs will provide valuable insights into designing similar targeted therapies against other BTB-ZF proteins.
The product of the Zbtb7 gene, which we will refer to as LRF, is known by several names and has been characterized in a variety of functional roles. The protein was first named FBI-1, or Factor Binding to IST-1, due to its ability to bind to sites on the HIV-1 genome known as the inducer of short transcripts (IST) (Pessler et al. 1997). FBI-1 was shown to interact with itself and the HIV-1 viral activator Tat (Morrison et al. 1999; Pendergrast et al. 2002). FBI-1 is also involved in the differentiation of preadipocyte cells (Laudes et al. 2004). The mouse homolog LRF, or Leukemia/Lymphoma Related Factor, was identified as a localization and heterodimerization partner of BCL6 (Davies et al. 1999). The rat homolog, OCZF or osteoclast-derived zinc finger, was shown to regulate differentiation of osteoclast cells and to be a transcription repressor that localizes to discrete nuclear foci (Kukita et al. 1999). LRF was shown to bind and regulate the expression of many genes containing a consensus binding sequence, which is often repeated in different orientations and spacing in many genes (Pessler and Hernandez 2003; Maeda et al. 2005a), including those encoding extracellular matrix collagen type I, II, IX, X, and XI, aggrecan, fibronectin, elastin, and human cartilage oligomeric matrix protein (COMP) (Widom et al. 2001; Liu et al. 2004), alcohol dehydrogenase ADH5/FDH (Lee et al. 2005), the ARF tumor suppressor (Maeda et al. 2005a), and the c-fos and c-myc oncoproteins (Pessler and Hernandez 2003). Furthermore, LRF interacts with other important transcription factors. LRF interferes with GC box recognition by SP-1, dependent on an interaction between LRF-BTB and the ZF region of SP-1 at the ADH5/FDH gene (Lee et al. 2002). LRF enhances transcription of NF-κB responsive genes by facilitating nuclear import, nuclear stabilization, and blocking nuclear export of this transcription factor (Lee et al. 2005). An interaction between LRF BTB domain and the Rel Homology Domain (RHD) of the p65 subunit of NF-κB or IκB was necessary for this activity.
LRF is also a repressor of the ARF tumor suppressor gene (p19Arf in the mouse, and p14ARF in humans), and is a central regulator in oncogenesis (Maeda et al. 2005a,b). LRF overexpression leads to reduced levels of ARF, resulting in the degradation of nuclear p53 and oncogenic transformation. Conversely, reduced levels of LRF results in senescence and apoptosis. Notably, LRF levels are often elevated in many human cancers, often in association with high levels of BCL6.
We report the crystal structure of the BTB domain from LRF. The structure shows a domain-swapped dimer that is similar to the previously determined structures of the PLZF and BCL6 BTB domains. However, distinct surface features in the LRF BTB domain suggest that LRF may have a different mechanism of co-repressor recruitment than BCL6, and may require the cooperation of other BTB-ZF proteins for its activities. Indeed, we show that unlike BCL6-BTB, LRF-BTB does not interact with the SMRT-BBD, and this is likely due to major sequence differences in the lateral groove.
Results and Discussion
General description of the LRF-BTB structure
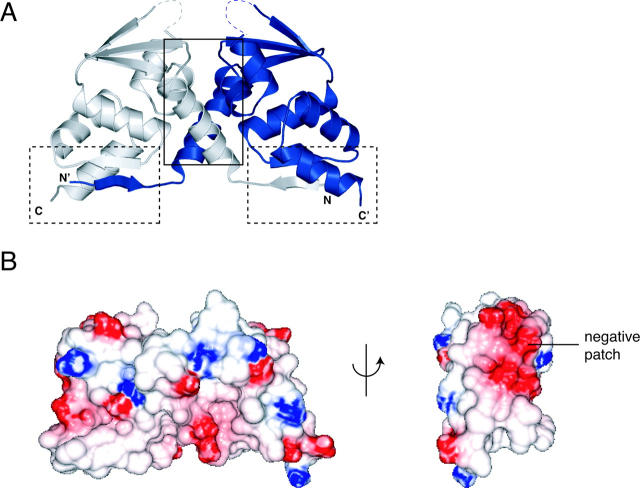
The BTB domain from LRF is a strand-exchanged homodimer (Fig. 1A; Table 1). The N-terminal β1 region of each monomer interacts exclusively with its partner chain, forming an interchain β1-β5 sheet. The second major contribution to the dimer interface is the tight packing of helices α1, α2, and α3 between the two subunits. Using the nomenclature of domain-swapped proteins (Liu and Eisenberg 2002), dimerization is mediated by an “open interface,” involving α1 from one chain plus α2 and α3 from the other, and a “closed interface,” comprised of β1 and α6 from one chain and β5 from the other. The extensive dimer interface involves 44 residues and 1647 Å2 in buried surface area. This is similar to what is observed in the other BTB domain structures. We have not observed the presence of monomers or any inter-subunit exchange between LRF-BTB dimers in solution, and as with PLZF and BCL6 (Li et al. 1997; Ahmad et al. 1998, 2003), and we describe the LRF BTB domain as an obligate homodimer. This is consistent with the result that LRF interacts with DNA as at least a dimeric molecule (Pessler and Hernandez 2003).
Figure 1.
Structure of the LRF BTB domain. (A) Ribbon representation. (Dashed lines) Regions of the turn between α3 and β4 with missing electron density; (solid box) open dimerization interface; (dashed boxes) closed dimerization interfaces. (B) Electrostatic surface representation of the LRF-BTB homodimer, shown in two views. A large negatively charged patch is labeled.
Table 1.
Data collection and refinement statistics
There is a negatively charged patch on each side of the homodimer, in a region that is far from the dimerization interface (Fig. 1B). This region is close to but separate from the predicted surface by which some BTB domains may interact with Cul3 (Stogios et al. 2005). The role, if any, of this patch is unknown, but it could represent a binding surface for proteins such as the RHD domain or the ZFs of SP-1 (Lee et al. 2002, 2005).
No electron density was observed in either of the two chains in the region connecting α3 and β4 (Fig. 1). The loop has the sequence GAVVDQQ, and forms the lining of the charged pocket region of the domain. The lack of density in these residues indicates that this region is likely to be flexible and unstructured in solution, but it may adopt a fixed conformation if they are involved in a protein-binding interaction.
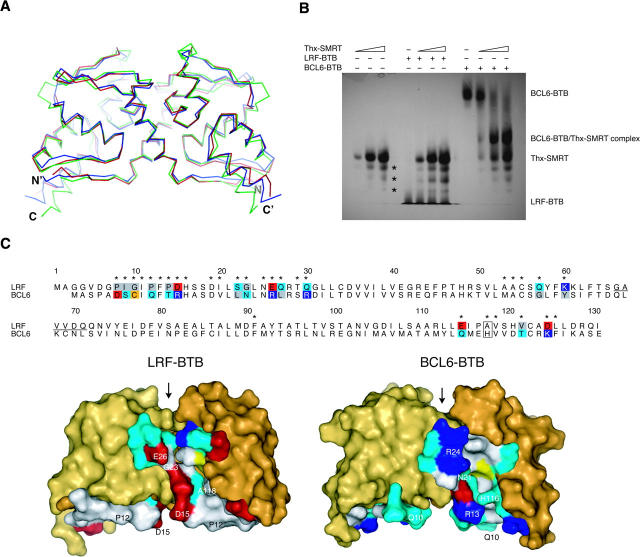
The LRF-BTB structure closely resembles the other known BTB domains, and the RMSD between equivalent Cα atoms is 1.5 Å between LRF and PLZF (37% identical at sequence level), and 2.2 Å between LRF and BCL6 (32% identical) (Fig. 2). There are minor differences in the structures, including the missing loop in LRF-BTB, the length of the α3-β4 loop, the β4-α4 turn, and a slight rotation of α6 at the C terminus of the domain.
Figure 2.
Comparison of the LRF and BCL6 BTB domains. (A) Superposition of Cα atoms of the LRF, PLZF, and BCL6 BTB homodimers (PDB ID codes 2NN2, 1BUO, 1R29). (Red) LRF, (blue) PLZF, (green) BCL6. (B) Binding of SMRT-BBD to BTB domains. LRF and BCL6 BTB domain were titrated with increasing amounts of Thioredoxin-SMRT (Thx-SMRT) fusion protein and separated by native gel electrophoresis. Lanes 1–3 contained Thx-SMRT alone (2.5, 8, and 13 μg), lanes 4–7 and 8–11 contained 10 μg LRF-BTB or BCL6-BTB, respectively, and 0, 2.5, 8, or 13 μg of Thx-SMRT. (Asterisks) Three minor impurities in the Thx-SMRT sample. (C) Comparison of the lateral grooves of the LRF and BCL6 BTB domains. A sequence alignment of LRF and BCL6-BTB is shown. (Asterisks) Residues located in the lateral groove region. Lateral groove residues that show non-conservative substitutions between the two BTB domains are shaded by residue type: (blue) basic, (red) acidic, (gray) hydrophobic, (cyan) polar, and (yellow) sulfur-containing. Conservative substitutions are not shaded, except for His 116/Ala 118 (boxed). Residues are numbered according to the LRF sequence. The region of the disordered loop in the LRF structure is underlined. The structures of the LRF and BCL6 BTB domains are shown in surface representation below the sequence alignment, with the lateral groove residues colored as in the alignment. Residues that have the largest buried surface areas in the BCL6-SMRT complex, and their equivalents in LRF, are labeled. Residues not involved in the lateral groove are colored shades of yellow. (Arrow) The charged pocket region of the BTB domains.
The crystal packing of the two structures of PLZF-BTB showed a possible mechanism of dimer oligomerization involving interactions between β1 of different homodimers (Ahmad et al. 1998; Li et al. 1999); however, this feature is not present in these LRF BTB domain crystals.
The lateral grooves of LRF and BCL6 show many residue differences
Two BBD peptides of the SMRT co-repressor were shown to interact with the lateral grooves of the BCL6-BTB homodimer (Ahmad et al. 2003). Each of the two grooves is at the dimer interface and involves residues from both chains. We compared the residues of the lateral grooves of the BCL6 and LRF BTB domains to gain insights into the possibility of BBD binding to LRF. Interestingly, of the 30 BCL6 residues involved in the BCL6/BBD interface, only nine residues are identical in LRF, and 17 positions show non-conservative substitutions, including many charge-reversal changes (Fig. 2C). This results in a very different residue composition and charge distribution in the LRF lateral groove. Notably, the most important BTB residues in the BCL6/SMRT interface are residues Gln 10, Arg 13, Arg 24, and His 116, and the equivalent residues in LRF are Pro 12, Asp 15, Glu 26, and Ala 118. The absence of the His 116 side chain is notable as this residue forms a clasp and makes many interactions with the SMRT peptide (Ahmad et al. 2003). As well, β1 is primarily hydrophobic in LRF-BTB, but polar in BCL6-BTB. The many differences suggest that the SMRT-BBD does not bind to the LRF BTB domain, at least in the same mode as seen in the BCL6-BTB/SMRT-BBD complex.
LRF does not interact with the SMRT-BBD
To investigate the effect of the extensive sequence differences in the lateral groove of the LRF BTB domain, we tested for binding to the SMRT-BBD (Fig. 2B) via native PAGE. As predicted, the SMRT-BBD fusion protein (Thx-SMRT) did not form a complex with the LRF BTB domain. The positive control of the BCL6 BTB domain did form a complex, as evidenced by the loss of the BCL6-BTB band and the appearance of a new band due to the complex. Therefore, if the LRF BTB domain is to recruit the SMRT co-repressor for transcription repression, it does so at a region distinct from the BBD. This observation suggests that the BBD peptide is specific for the BCL6 BTB domain (Polo et al. 2004) and does not directly interfere with the functions of the LRF BTB domain. This would be presumably true as well in any putative BCL6-BTB/LRF-BTB heterodimer, since the two lateral grooves are located at the dimer interface, each with contributions from both BTB chains.
Comparison of the charged pocket of BTB domains
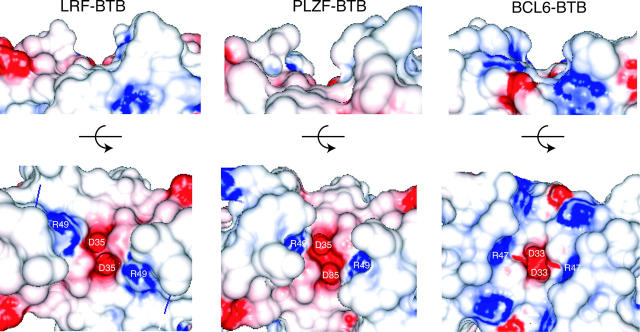
The loop between α3 and β4 of the BTB domains forms the edge of a charged pocket, containing two Asp and two Arg residues, that is thought to be functionally important for transcription repression even though it does not contact the SMRT-BBD (Melnick et al. 2000, 2002; Ahmad et al. 2003; Puccetti et al. 2005). The charged pocket is different in depth and contains different residues in each BTB domain structure (Fig. 3). In all three structures, the bottom of the charged pocket is made up of Asp 33/35 and Arg 47/49 in nearly identical positions. The LRF and PLZF charged pockets most closely resemble each other, as Asp 35 and Arg 49 are the only charged features and there are neutral features that extend away from the pocket. The BCL6 charged pocket shows additional basic features, and the width of the pocket is larger than the other BTB domains. Notably, the PLZF BTB domain interacts only weakly with SMRT via regions distinct from the SMRT-BBD. Therefore, the residue composition in the charged pockets of PLZF and LRF is consistent with weak or no SMRT interactions.
Figure 3.
Comparison of the charged pocket of the LRF, PLZF, and BCL6 BTB homodimers (PDB ID codes 2NN2, 1BUO, 1R29), shown as electrostatic surface representations. Two views are shown, the first in the same perspective as Fig. 2, the second rotated 90° toward the reader. Residues Asp 33/35 and Arg 47/49 are labeled.
The lack of electron density for the α3-β4 loop is not unusual given that other BTB structures have unresolved loops. The recently solved structure of the BTB domain from Bach1, a BTB-leucine zipper transcription factor, does not show electron density for this same loop at the charged pocket (PDB code 2ihc). Furthermore, the BTB-ZF protein Hic-1, whose structure has not been solved, contains a 13-residue alanine-rich insertion in another region of the BTB domain that is predicted to be disordered (Deltour et al. 1999). The significance of flexibility of the α3-β4 loop, the variability in the depth and residue composition of the pocket, and the Hic-1 insertion have not been experimentally clarified, but could be important selective determinants for binding of corepressors or HDACs to BTB domains. Given that the LRF BTB domain does not interact with the SMRT-BBD, other regions of this BTB domain, such as the charged pocket, could be important for co-repressor interactions.
While this manuscript was in preparation, a structure of another LRF BTB domain has been published (Schubot et al. 2006). The features we observe are consistent with this structure, including missing density in the α3-β4 loop and residue changes in the lateral groove.
Summary
The crystal structure of the BTB domain from LRF closely resembles the previously determined structures of domain-swapped BTB homodimers. The extensive dimerization interface is primarily hydrophobic. However, the surface-exposed residues of the LRF BTB homodimer differ from the other BTB structures, as expected from our earlier analysis of the sequences of BTB domains (Stogios et al. 2005). There is little sequence conservation in the lateral groove, a region experimentally shown to interact with co-repressor proteins in BCL6, and, as a consequence, the LRF BTB domain does not interact with the SMRT-BBD. As well, there are some changes in the vicinity of the charged pocket of LRF-BTB, a region thought to be important for transcription repression function of other BTB domains. In light of recent research findings showing the significant and central role of LRF in oncogenesis, the structure of the LRF BTB domain will be important for understanding the molecular basis of transcription repression and for the rational design of therapies.
Materials and methods
Protein expression and purification
A cDNA encoding full-length LRF was obtained as a Mammalian Gene Collection (MGC)-compliant clone (MGC ID 99631) from American Type Culture Collection. A segment corresponding to residues 1–131 was amplified by PCR and subcloned into a modified pET-32(a) (Novagen) vector. The expression vector was transformed into Escherichia coli BL21(DE3) codon plus cells (Stratagene) that were then grown at 37°C in LB medium in the presence of 100 mg/L ampicillin to an A600 of 0.5. Protein expression was induced with 100 μM IPTG for 3.5 h at 23°C. Cells were harvested and homogenized in 500 mM NaCl, 20 mM Tris (pH 8.5), 10 mM imidazole, and 5 mM β-mercaptoethanol in an Emulsiflex (Avestin). The 6×His fusion protein was bound to a NiNTA column (Qiagen), washed with lysis buffer, and eluted with lysis buffer containing 100 mM imidazole. The fusion protein was further purified by size exclusion chromatography on a Superdex 200 column (Amersham Pharmacia Biotech) equilibrated with thrombin cleavage buffer (150 mM NaCl, 20 mM Tris at pH 8, 10 mM β-mercaptoethanol). The fusion protein was digested with four units of thrombin (human plasma thrombin, high activity; Calbiochem) per milligram of protein for 24–48 h at 4°C. The reaction was stopped by the addition of 50 μL of benzamidine-sepharose beads (GE Healthcare). Free LRF BTB domain was further purified by size exclusion chromatography on a Superdex 200 column into the final crystallization buffer (300 mL NaCl, 20 mM Tris at pH 8.0, 1 mM TCEP). The protein concentration of the LRF BTB domain was determined by UV spectrophotometry using an extinction coefficient of 4470 M−1 cm−1. The LRF BTB domain was stored at 4°C until use, or flash frozen into liquid nitrogen and stored at −70°C in buffer containing 20% glycerol. A SMRT-BBD (residues 1414–1430) fusion protein with thioredoxin (Thx-SMRT) was expressed and purified as described previously (Ahmad et al. 2003).
Crystallization and X-ray data collection
Crystals were grown at 23°C using the hanging drop method, by mixing 1 μL of protein at 6.5 mg/mL with 1 μL of reservoir solution containing 0.7 M sodium citrate (pH 5.4). Crystals were cryoprotected with 20% ethylene glycol prior to flash freezing. Native data were collected at 100 K at the Cornell High Energy Synchrotron Source (CHESS), beamline A1. Diffraction data were reduced with the HKL package, SCALEPACK, and DENZO (Otwinowski and Minor 1997).
Structure determination and refinement
The structure was solved by molecular replacement using Phaser (McCoy et al. 2005), and the search model was the PLZF BTB dimer (PDB ID 1BUO) (Ahmad et al. 1998). The LRF model was rebuilt using ARP/wARP (Perrakis et al. 2001), Refmac5 (Murshudov et al. 1997), and O (Jones et al. 1991). The final atomic model includes residues 7–129 of LRF, except residues 66–71. Side chains for residues Arg 129 of chain A and Gln 72 of chain B have missing electron density.
Structural analysis
Structure superpositions were calculated with the SuperPose server (Maiti et al. 2004). Buried surface area was calculated using the program NACCESS using the default probe size (http://wolf.bms.emist.ac.uk/naccess). Electrostatic potential surfaces were calculated using GRASP2 (Petrey and Honig 2003). Red is negative, white is neutral, blue is positively charged, and surfaces were contoured between −10 and +10 k B T/e and −10 k B T/e, where k B is the Boltzmann constant, T is temperature, and e is the electronic charge. Protein structure images were produced with PyMOL (DeLano 2002).
Native PAGE
LRF and BCL6 BTB domains were mixed with Thx-SMRT and run at 4°C on a 5% native PAGE gel in Tris-glycine buffer (pH 8.8). Equal amounts (10 μg) of LRF and BCL6 BTB domains were loaded in all lanes, while the amount of added Thx-SMRT ranged from 2.5 to 13 μg. The calculated isoelectric points are 4.4 and 6.3 for LRF-BTB and BCL6-BTB, and 5.4 for Thx-SMRT.
Data deposition
Atomic coordinates and diffraction data have been deposited with the PDB, with access code 2NN2.
Acknowledgments
We thank Doug Kuntz for data collection and the staff at the A1 beamline at CHESS. This research is supported by the Canadian Cancer Society by an NCIC grant to G.G.P. P.J.S. is a research student of The Terry Fox Foundation through an award from the National Cancer Institute of Canada.
Footnotes
Reprint requests to: Gilbert G. Privé, Division of Cancer Genomics and Proteomics, Ontario Cancer Institute, Departments of Medical Biophysics and Biochemistry, University of Toronto, 101 College Street, Toronto, Ontario, M5G 1L7, Canada; e-mail: prive@uhnres.utoronto.ca; fax: (416) 581-7562.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062660907.
References
- Ahmad, K.F., Engel, C.K., and Privé, G.G. 1998. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. 95: 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, K.F., Melnick, A., Lax, S., Bouchard, D., Liu, J., Kiang, C.L., Mayer, S., Takahashi, S., Licht, J.D., and Privé, G.G. 2003. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell 12: 1551–1564. [DOI] [PubMed] [Google Scholar]
- Collins, T., Stone, J.R., and Williams, A.J. 2001. All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 21: 3609–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, G., Alland, L., Hong, S.H., Wong, C.W., DePinho, R.A., and Dejean, A. 1998. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene 16: 2549–2556. [DOI] [PubMed] [Google Scholar]
- Davies, J.M., Hawe, N., Kabarowski, J., Huang, Q.H., Zhu, J., Brand, N.J., Leprince, D., Dhordain, P., Cook, M., and Morriss-Kay, G., et al. 1999. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene 18: 365–375. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. 2002. In The PyMOL molecular graphics system. Delano Scientific, San Carlos, CA.
- Deltour, S., Guerardel, C., and Leprince, D. 1999. Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: The case of HIC-1 and γFBP-B. Proc. Natl. Acad. Sci. 96: 14831–14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhordain, P., Lin, R.J., Quief, S., Lantoine, D., Kerckaert, J.P., Evans, R.M., and Albagli, O. 1998. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 26: 4645–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, K.D. and Bardwell, V.J. 1998. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene 17: 2473–2484. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kelly, K.F. and Daniel, J.M. 2006. POZ for effect—POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 16: 578–587. [DOI] [PubMed] [Google Scholar]
- Klug, A. and Schwabe, J.W. 1995. Protein motifs 5. Zinc fingers. FASEB J. 9: 597–604. [PubMed] [Google Scholar]
- Kukita, A., Kukita, T., Ouchida, M., Maeda, H., Yatsuki, H., and Kohashi, O. 1999. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood 94: 1987–1997. [PubMed] [Google Scholar]
- Laudes, M., Christodoulides, C., Sewter, C., Rochford, J.J., Considine, R.V., Sethi, J.K., Vidal-Puig, A., and O'Rahilly, S. 2004. Role of the POZ zinc finger transcription factor FBI-1 in human and murine adipogenesis. J. Biol. Chem. 279: 11711–11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.K., Suh, D., Edenberg, H.J., and Hur, M.W. 2002. POZ domain transcription factor, FBI-1, represses transcription of ADH5/FDH by interacting with the zinc finger and interfering with DNA binding activity of Sp1. J. Biol. Chem. 277: 26761–26768. [DOI] [PubMed] [Google Scholar]
- Lee, D.K., Kang, J.E., Park, H.J., Kim, M.H., Yim, T.H., Kim, J.M., Heo, M.K., Kim, K.Y., Kwon, H.J., and Hur, M.W. 2005. FBI-1 enhances transcription of the nuclear factor-κB (NF-κB)-responsive E-selectin gene by nuclear localization of the p65 subunit of NF-κB. J. Biol. Chem. 280: 27783–27791. [DOI] [PubMed] [Google Scholar]
- Li, X., Lopez-Guisa, J.M., Ninan, N., Weiner, E.J., Rauscher III, F.J., and Marmorstein, R. 1997. Overexpression, purification, characterization, and crystallization of the BTB/POZ domain from the PLZF oncoprotein. J. Biol. Chem. 272: 27324–27329. [DOI] [PubMed] [Google Scholar]
- Li, X., Peng, H., Schultz, D.C., Lopez-Guisa, J.M., Rauscher III, F.J., and Marmorstein, R. 1999. Structure–function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 59: 5275–5282. [PubMed] [Google Scholar]
- Liu, Y. and Eisenberg, D. 2002. 3D domain swapping: As domains continue to swap. Protein Sci. 11: 1285–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.J., Prazak, L., Fajardo, M., Yu, S., Tyagi, N., and Di Cesare, P.E. 2004. Leukemia/lymphoma-related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. J. Biol. Chem. 279: 47081–47091. [DOI] [PubMed] [Google Scholar]
- Maeda, T., Hobbs, R.M., Merghoub, T., Guernah, I., Zelent, A., Cordon-Cardo, C., Teruya-Feldstein, J., and Pandolfi, P.P. 2005a. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature 433: 278–285. [DOI] [PubMed] [Google Scholar]
- Maeda, T., Hobbs, R.M., and Pandolfi, P.P. 2005b. The transcription factor Pokemon: A new key player in cancer pathogenesis. Cancer Res. 65: 8575–8578. [DOI] [PubMed] [Google Scholar]
- Maiti, R., Van Domselaar, G.H., Zhang, H., and Wishart, D.S. 2004. SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Res. 32: W590–W594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C., and Read, R.J. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61: 458–464. [DOI] [PubMed] [Google Scholar]
- Melnick, A., Ahmad, K.F., Arai, S., Polinger, A., Ball, H., Borden, K.L., Carlile, G.W., Privé, G.G., and Licht, J.D. 2000. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 20: 6550–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick, A., Carlile, G., Ahmad, K.F., Kiang, C.L., Corcoran, C., Bardwell, V., Privé, G.G., and Licht, J.D. 2002. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol. Cell. Biol. 22: 1804–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, D.J., Pendergrast, P.S., Stavropoulos, P., Colmenares, S.U., Kobayashi, R., and Hernandez, N. 1999. FBI-1, a factor that binds to the HIV-1 inducer of short transcripts (IST), is a POZ domain protein. Nucleic Acids Res. 27: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pendergrast, P.S., Wang, C., Hernandez, N., and Huang, S. 2002. FBI-1 can stimulate HIV-1 Tat activity and is targeted to a novel subnuclear domain that includes the Tat-P-TEFb-containing nuclear speckles. Mol. Biol. Cell 13: 915–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis, A., Harkiolaki, M., Wilson, K.S., and Lamzin, V.S. 2001. ARP/wARP and molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 57: 1445–1450. [DOI] [PubMed] [Google Scholar]
- Pessler, F. and Hernandez, N. 2003. Flexible DNA binding of the BTB/POZ-domain protein FBI-1. J. Biol. Chem. 278: 29327–29335. [DOI] [PubMed] [Google Scholar]
- Pessler, F., Pendergrast, P.S., and Hernandez, N. 1997. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol. Cell. Biol. 17: 3786–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrey, D. and Honig, B. 2003. GRASP2: Visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 374: 492–509. [DOI] [PubMed] [Google Scholar]
- Polo, J.M., Dell'Oso, T., Ranuncolo, S.M., Cerchietti, L., Beck, D., Da Silva, G.F., Privé, G.G., Licht, J.D., and Melnick, A. 2004. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat. Med. 10: 1329–1335. [DOI] [PubMed] [Google Scholar]
- Puccetti, E., Zheng, X., Brambilla, D., Seshire, A., Beissert, T., Boehrer, S., Nurnberger, H., Hoelzer, D., Ottmann, O.G., and Nervi, C., et al. 2005. The integrity of the charged pocket in the BTB/POZ domain is essential for the phenotype induced by the leukemia-associated t(11;17) fusion protein PLZF/RARα. Cancer Res. 65: 6080–6088. [DOI] [PubMed] [Google Scholar]
- Schubot, F.D., Tropea, J.E., and Waugh, D.S. 2006. Structure of the POZ domain of human LRF, a master regulator of oncogenesis. Biochem. Biophys. Res. Commun. 351: 1–6. [DOI] [PubMed] [Google Scholar]
- Stogios, P.J., Downs, G.S., Jauhal, J.J., Nandra, S.K., and Privé, G.G. 2005. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom, R.L., Lee, J.Y., Joseph, C., Gordon-Froome, I., and Korn, J.H. 2001. The hcKrox gene family regulates multiple extracellular matrix genes. Matrix Biol. 20: 451–462. [DOI] [PubMed] [Google Scholar]
- Yoon, H.G., Chan, D.W., Reynolds, A.B., Qin, J., and Wong, J. 2003. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 12: 723–734. [DOI] [PubMed] [Google Scholar]