Abstract
Various nonglycosylated analogs were designed in order to explore the role of glycosylation in formaecin I, an antibacterial glycopeptide of insect origin. The functional behavior of a designed nonglycosylated analog (P7,endo P8a,ΔT11)formaecin I was found to be similar to that of native glycosylated peptide. Both the peptides showed similar antibacterial activities against Escherichia coli and Salmonella strains. The designed nonglycosylated analog (P7,endo P8a,ΔT11)formaecin I has low binding affinity to LPS identical to that of native glycopeptide, formaecin I. Both the peptides have similar killing kinetics and are nontoxic to erythrocytes. Formaecin I and designed nonglycosylated (P7,endo P8a,ΔT11)formaecin I have no definite conformational features associated with them. The glycosylated residue of threonine in formaecin I and proline residues in designed peptide [(P7,endo P8a,ΔT11)formaecin I], possibly help in stabilizing the correct conformation that facilitates presentation of the peptide to its receptor. It is evident that a functionally equivalent nonglycosylated analog of native glycosylated antibacterial peptide can be designed by strategically modifying the sequence.
Keywords: glycopeptide antibiotic, rational design, antibacterial activity, insect immunity
The resistance of insects to bacterial infection is explained by their ability to synthesize a diverse range of antibacterial peptides (Hoffmann et al. 1993; Hultmark 1993; Cociancich et al. 1994). A remarkable feature of some proline-rich antibacterial peptides such as drosocin, pyrrhocoricin, formaecins, diptericin, and lebocins that have been isolated from insects, is the presence of a conserved glycosylated threonine residue (Hara and Yamakawa 1995; Otvos et al. 2000a). Formaecin I consists of 16 residues, nearly one-third of which are prolines. It is glycosylated on threonine 11 by a monosaccharide, 2-acetamido-2-deoxy-D-galactopyranosyl (GalNAc), and is active against Gram-negative bacteria at concentrations in the low micromolar range. Previous studies showed that the native glycosylated peptide, formaecin I, isolated from an ant, Myrmecia gulosa, had more activity than its synthetic deglycosylated form (Mackintosh et al. 1998). The integrity of the carbohydrate side chain is also necessary for maximum activity of other antimicrobial peptides such as drosocin, diptericin, and lebocins. The exact role of the sugar remains an enigma.
Formaecin I shows significant homology at its C-terminal end with the conserved residues of apidaecins (Casteels et al. 1994), a class of nonglycosylated peptides isolated from insects. Here we report designing of a nonglycosylated analog of formaecin I that shows the antibacterial and hemolytic activities and LPS-binding affinity similar to the glycopeptide formaecin I, indicating that carbohydrate function could be compensated by suitable modifications in the peptide sequence. The study provides insight into the possible role of glycosylation in the antibiotic activity of the glycopeptides of innate immune origin.
Results and Discussion
Design of an active nonglycosylated formaecin I analog
Formaecin I was earlier isolated from an ant, M. gulosa (Mackintosh et al. 1998), and belongs to the proline-rich class of antibacterial peptides. It has been shown that the glycopeptide formaecin I possesses higher antibacterial activity than its synthetically prepared deglycosylated form. We have chemically synthesized formaecin I and its nonglycosylated analog by using solid-phase chemistry. The mass spectrometry analysis gave molecular masses of 1997.43 Da and 1793.87 Da for formaecin I and its nonglycosylated analog, respectively, in full agreement with the theoretical values (1997.48 Da and 1794.28 Da). The antibacterial activity of formaecin I and its deglycosylated form against Escherichia coli BL21λD3 and Salmonella typhimurium were determined by radial diffusion assay (Fig. 1). The dose-dependent increase in the antibacterial activity was evident in both cases. The chemically synthesized glycopeptide formaecin I showed higher antibacterial activity as compared to that of the deglycosylated peptide against different strains of Gram-negative bacteria. This is consistent with the earlier observations (Mackintosh et al. 1998).
Figure 1.
Comparison of antibacterial activity of formaecin I with its deglycosylated form, (T11)formaecin I, against E. coli (A) and S. typhimurium (B) expressed in terms of inhibition zone area in the radial diffusion assay.
The attractive features of glycopeptide antibiotics compared to their nonglycosylated counterparts include increased solubility and oral availability (Poujade et al. 1983; Fisher et al. 1991), increased serum half-life (Powell et al. 1993), broader biological activity spectra (Biondi et al. 1993; Horvat et al. 1993), and stabilization of conformational variants (Laczko et al. 1992; Urge et al. 1992). The present study was aimed at exploring the functional role of glycosylation in formaecin I by designing various nonglycosylated analogs of the glycopeptide and screening for bioactivity. The apidaecin-type peptides (Casteels et al. 1989) are a group of nonglycosylated antibacterial peptides of the proline-rich class and consist of conserved regions of amino acids. The existence of conserved residues in apidaecin (HbIb) is represented as bold letters in Figure 2. Formaecin I can be best aligned with the apidaecin (HbIb) isoform (Casteels et al. 1994), which is an 18-residue-long peptide isolated from Apis mellifera (honeybee). We observed that the glycosylated peptide formaecin I shows a high degree of sequence homology at its C terminus to the conserved residues of apidaecin (HbIb) (Fig. 2). The removal of GalNAc from formaecin I resulted in (T11)formaecin I, which showed a significant decrease in its antibacterial activity. In order to compensate for the effect of sugar, the nonglycosylated analogs of formaecin I with minimal amino acid substitutions were designed on the basis of residue conservation with the apidaecin-type peptides. The glycosylated threonine was removed by comparison with apidaecin (HbIb), and it resulted in the nonglycosylated peptide (ΔT11)formaecin I. Apidaecin (HbIb) contains the conserved prolines at its ninth and eleventh positions that are not present in formaecin I. In further designing, these prolines were incorporated sequentially in the nonglycosylated analogs of formaecin I. In (endo P8a,ΔT11)formaecin I, one conserved proline residue was incorporated at the ninth position, whereas in (P7,endo P8a,ΔT11)formaecin I, the asparagine at the seventh position of nonglycosylated formaecin I was substituted with another conserved proline residue of apidaecin (HbIb), along with the above changes.
Figure 2.
Alignment of the sequences of designed analogs of formaecin I with reference to formaecin I and apidaecin.
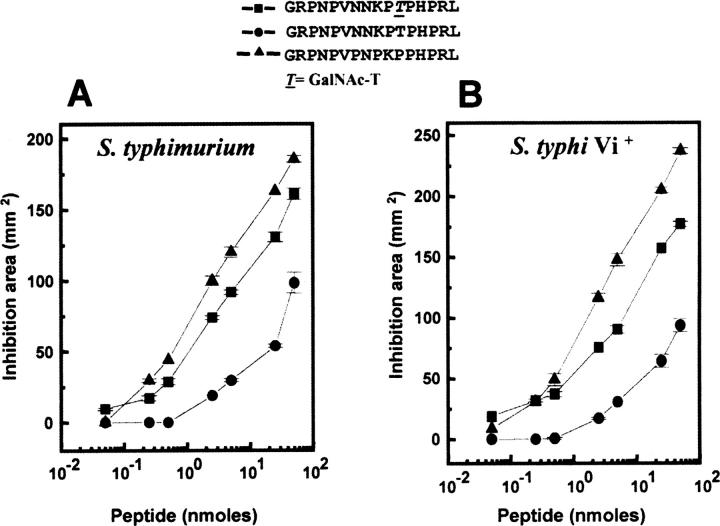
All these designed nonglycosylated analogs were synthesized and assayed for their comparative spectrum of antibacterial activities against E. coli BL21λD3 (Fig. 3). The (ΔT11)formaecin I peptide showed the lowest antibacterial activity, and insertion of one proline in (ΔT11)formaecin I resulted in the (endo P8a,ΔT11)formaecin I peptide, which improved its antibacterial activity to some extent. The (P7,endo P8a,ΔT11)formaecin I, containing all the conserved residues of apidaecin (HbIb), exhibited activity comparable to that of the native formaecin I (Fig. 3). The level of antibacterial activity was also found to be similar against the pathogenic strains of Salmonella, that is, S. typhimurium and S. typhi Vi+ (Fig. 4) for formaecin I and the designed nonglycosylated analog, (P7,endo P8a,ΔT11)formaecin I. Thus, the glycopeptide formaecin I and the nonglycosylated analog, (P7,endo P8a,ΔT11)formaecin I, showed comparable antibacterial activities against different Gram-negative bacterial strains. Therefore, comparison was also made in terms of other functional properties as well as structural features between the designed active nonglycosylated analog and native formaecin I.
Figure 3.

Dose-dependent activity curves of formaecin I and its designed nonglycosylated analogs showing antibacterial activity profiles against E. coli. The designed nonglycosylated analog, (P7,endo P8a,ΔT11)formaecin I exhibits activity comparable to that of the native formaecin I.
Figure 4.
Comparison of antibacterial activity of formaecin I with designed nonglycosylated analog, (P7,endo P8a,ΔT11)formaecin I against pathogenic strains of S. typhimurium (A) and S. typhi (Vi+) (B).
Conformational analyses
Peptides are believed to adopt specific structural features for effective binding to their putative targets. Accordingly, comparative structural analysis of modified analogs of formaecin I and glycopeptide formaecin I was carried out by circular dichroism studies.
The conformational properties of formaecin I, (T11)formaecin I, and (P7,endo P8a,ΔT11)formaecin I were analyzed by CD spectroscopy in water, 90% TFE, and 10 mM SDS (Fig. 5). In water, all three peptides showed the characteristics of an unordered structure (Fig. 5A). Formaecin I and (T11)formaecin I showed a negative band at 200 nm. In (P7,endo P8a,ΔT11)formaecin I, the negative broad band was somewhat red-shifted to 203 nm, probably because of the presence of seven prolines. In 90% aqueous trifluoroethanol, all the peptides exhibited CD spectra similar to the pattern that has been assigned to the presence of type I (III) β-turns or β-turn mixtures with significant type I character and earlier reported for drosocin and its deglycosylated form (Bulet et al. 1996). The structure-promoting effect of TFE was apparently less marked on formaecin I and its designed nonglycosylated analog (P7,endo P8a,ΔT11)formaecin I. Polyproline helices, however, exhibit a weak positive nπ* band above 220 nm, whereas formaecin I and (P7,endo P8a,ΔT11)formaecin I showed the broadening of the negative band. The broadening may indicate the presence of other types of turns, but only a low proportion of the peptide molecules acquired them (Fig. 5B). The micellar SDS has been used as a model of the negatively charged bacterial lipid membranes, with which cationic peptides first interact. In an SDS environment, all the three peptides exhibited very similar CD spectra that were characterized by an intense negative band at ~204 nm similar to that observed in TFE, suggesting mainly electrostatic peptide/micelles interaction without clear effects on the peptide conformation (Fig. 5C). The CD studies for formaecin I and its nonglycosylated analog (P7,endo P8a,ΔT11)formaecin I in water suggested that there is no significant difference between their secondary structures and the presence of unordered structure in these peptides. Indeed, the membrane-mimicking environments like TFE and SDS did not generate any appreciably ordered structure in both the peptides except the presence of β-turns as exhibited by their CD spectra. Thus, conformational studies by CD have shown that the glycopeptides formaecin I and (P7,endo P8a,ΔT11)formaecin I have similar secondary structures.
Figure 5.
Circular dichroism profiles of formaecin I, deglycosylated formaecin I [(T11)formaecin I] and designed nonglycosylated formaecin I [(P7,endo P8a,ΔT11)formaecin I] in water (A), in 90% TFE (B), and in 10 mM SDS (C).
Functional analyses
The designed nonglycosylated analog (P7,endo P8a,ΔT11)formaecin I showed similarity with the native glycopeptide formaecin I in terms of the antibacterial activity profiles against different bacterial strains as well as the structural features as observed by circular dichroism spectroscopy. In order to evaluate the equivalence of the glycopeptide and the nonglycoslylated analog, it was pertinent to address other functional attributes as well. Therefore, the kinetics of bacterial killing, hemolytic activity, and the ability to competitively displace Polymyxin B from endotoxin binding were compared for the two peptides.
The time-killing experiments were carried out to follow the kinetics of inactivation of E. coli BL21λD3 by formaecin I, (T11)formaecin I, and (P7,endo P8a,ΔT11)formaecin I and to establish whether these peptides are bacteriostatic or bactericidal. A time-course analysis of the inhibitory activity showed that it took an experimental period of ~2 h for glycopeptide and (P7,endo P8a,ΔT11)formaecin I at a minimum inhibitory concentration for either peptide to completely inhibit the subsequent growth of E. coli (Fig. 6A). After 80 min, the number of colony-forming units had dropped to ~0% and 5% for formaecin I and designed nonglycosylated analog (P7,endo P8a,ΔT11)formaecin I, respectively. The deglycosylated (T11)formaecin I acts more slowly. Similar results were found for deglycosylated drosocin (Cudic et al. 1999). The viable cell counts were 36% after 80 min, and all colony-forming potential disappeared only after 4 h in the case of deglycosylated formaecin I, (T11)formaecin I. The results show that all three peptides are bactericidal, and the kinetics of inactivation of E. coli is same for formaecin I and (P7,endo P8a,ΔT11)formaecin I. The similar killing kinetics could be ascribed to a similar mode of action of these peptides. It is also supported by the fact that both of these peptides are similar in structure. This could be further verified after the target receptors for these antimicrobial peptides are identified.
Figure 6.
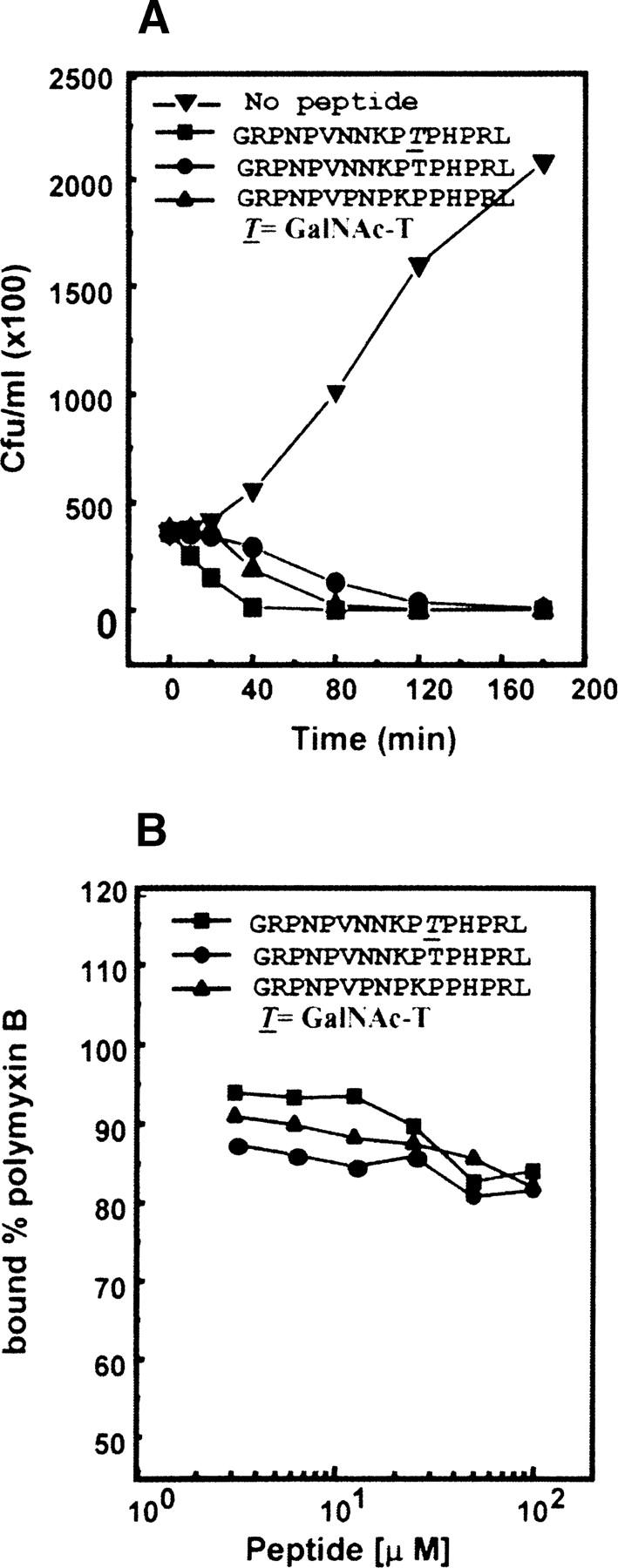
Comparison of functional properties of formaecin I and designed nonglycosylated active analog. (A) Kinetics of bactericidal activity of formaecin I (50 μM), (T11) formaecin I (500 μM), and (P7,endo P8a,ΔT11)formaecin I (50 μM), on mid-log-phase cultures of E. coli BL21λD3 at minimum inhibitory concentration of each peptide. (B) Competitive displacement of Dansyl Polymyxin B in a dose-dependent manner by different peptides for endotoxin binding.
The effect of formaecin I and its designed analogs was also tested on 1% (v/v) suspensions of rat erythrocytes. When tested in a conventional hemolytic test, formaecin I and designed nonglycosylated peptide, (P7,endo P8a,ΔT11)formaecin I, and (T11)formaecin I had no hemolytic activity on rat erythrocytes at a concentration of up to 100 μM (data not shown). All these peptides show nontoxicity to rat red blood cells. Thus, hemolytic activity was absent in formaecin I as well as in (P7,endo P8a,ΔT11)formaecin I.
It has been suggested that in Gram-negative bacteria, the initial interaction of these peptides with LPS occurs through cation-binding sites on the LPS surface. The fluorescent probe Dansyl Polymyxin B can be used to assess the relative binding affinities of various compounds to LPS. The competitive displacement of Polymyxin B from endotoxin by formaecin I was analyzed and compared with the nonglycosylated analogs. It was observed that the behavior of displacement of Polymyxin B by formaecin I, its deglycosylated form (T11)formaecin I, and designed nonglycosylated analog (P7,endo P8a,ΔT11)formaecin I was almost similar. All these peptides could displace only ~10% of the bound Polymyxin B even at the maximum concentration of 100 μM used (Fig. 6B). Formaecin I and its designed nonglycosylated analog exhibited low binding affinity to LPS unlike apidaecin, drosocin, and pyrrhocoricin. The other than LPS molecules may be involved to facilitate the uptake of formaecin I peptides into the cell, followed by its binding to target molecules.
A diverse array of mechanisms by which proline-rich antimicrobial peptides act on the bacterial cell have been proposed. The mammalian proline-rich peptide PR-39 kills bacteria by interrupting both DNA and protein synthesis (Skerlavaj et al. 1990), and the bactenecins are suggested to cause loss of macromolecular synthesis ability (Boman et al. 1993). Unlike the α-helical and most of the cysteine-stabilized antimicrobial peptides that kill bacteria through a nonstereospecific fashion (Bessalle et al. 1990; Wade et al. 1990), the all-D-enantiomer of (P7,endo P8a,ΔT11)formaecin I is totally ineffective against E. coli and Salmonella strains (data not shown) similar to apidaecin, drosocin, and pyrrhocoricin, suggesting that these peptides are bactericidal through a mechanism that involves a stereoselective recognition of a chiral cellular target, most likely a protein (Casteels and Tempst 1994; Bulet et al. 1996; Otvos et al. 2000b). Castle et al. (1999) proposed a model of a permease/transporter-mediated mechanism for the mode of action of apidaecin on the Gram-negative bacterium E. coli. In an attempt to identify the processes and molecules that might be targeted by apidaecin after entering E. coli cells, it was shown that the peptide rapidly blocked the protein synthesis machinery. Otvos et al. (2000a) observed that some of the proline-rich peptides bind to the bacterial heat-shock protein DnaK and chaperonin GroEL. The antibacterial peptides formaecin I, drosocin, apidaecins, and pyrrhocoricin, which exhibit sequence similarities, are likely to have similar modes of action.
Thus, the killing mechanism of bacteria involving interventions at the level other than membrane, which may include protein/nucleic acid synthesis, is evident. Also, certain proteins/nucleic acid targets to which proline-rich glycopeptide antibiotics bind have been identified. However, precise description of the glycopeptide receptor binding site is yet to be elucidated.
Conclusion
Many peptide antibiotics kill bacteria by membrane pore formation. A characteristic property of membrane pores involving peptide antibiotics has been suggested to be that it is stereo nonspecific as it involves aggregation of amphipathic α-helices (Bessalle et al. 1990; Wade et al. 1990). However, certain antibiotic peptides such as apidaecin, drosocin, and pyrrhocoricin have been shown to kill bacteria in a stereospecific manner, suggesting the possibility of alternate modes of action such as the presence of an intracellular target biopolymer with chiral properties (Casteels and Tempst 1994; Bulet et al. 1996; Otvos et al. 2000b). We have found that the formaecin I analog (P7,endo P8a,ΔT11)formaecin I was inactive when made of D-amino acid residues, implying the likely mode of action to be similar to that of apidaecin, drosocin, and pyrrhocoricin.
If the formaecin I functions via a mechanism that requires stereospecific recognition of a target molecule, it must adopt a certain functional conformation, and considering that glycan moiety was shown to affect activity, it may influence the functionally relevant conformation of the glycopeptide antibiotic. Indeed, other biochemical evidence exists indicating that glycosylation influences peptide and protein structure (Rose et al. 1984; Shogren et al. 1989; Chapman et al. 1996). It has been suggested that peptide glycosylation influences conformation by simply reducing the available conformational repertoire of the otherwise flexible peptide (Andreotti and Kahne 1993; Rickert and Imperiali 1995; O'Conner and Imperiali 1998).
We have shown that the designed nonglycosylated analog (P7,endo P8a,ΔT11)formaecin I could mimic the effect of O-glycosylation in formaecin I. The (P7,endo P8a,ΔT11)formaecin I, which is a nonglycoslyated analog, shows antibacterial activity comparable to that of the native glycopeptide. Both of these peptides have similar functional properties such as killing kinetics and binding affinity to LPS; the nonglycosylated (P7,endo P8a,ΔT11)formaecin I must adopt conformational properties that are similar to those of formaecin I. In other words, the sequence modifications in this peptide may bring about similar constraints as those by the glycan moiety. Probably, O-linked glycosylation of threonine stabilized the bioactive conformation of formaecin I, and the replacement of the glycosylated-amino acid and incorporation of highly turn-forming proline residues in (P7,endo P8a,ΔT11)formaecin I resulted in an unaltered conformational variant. The glycosylated threonine in formaecin I and proline residues in designed peptide [P7,endo P8a,ΔT11)formaecin I], possibly, help in stabilizing the correct conformation that facilitates presentation of the peptide to its stereospecific receptor. Therefore, the designed nonglycosylated peptide could mimic the functional behavior of glycosylated peptide. The high-resolution conformational analysis of these peptides is needed to specifically address the precise effect of O-linked sugar and insertions of proline residues in the peptide.
Thus, the effect of sugar can be mimicked by making strategically designed amino acid substitutions in the nonglycosylated analog of glycosylated peptide. Indeed, physiologically relevant sugar–peptide mimicry between these two topologically similar but chemically independent molecules has been demonstrated (Jain et al. 2000). The reverse of this approach would also be interesting since glycosylated peptides are much better candidates for therapy. Indeed, such an attempt has been made by Gobbo et al. (2002), where an extra glycosylated threonine residue was inserted in the native apidaecin. Although the resulting analog in this case exhibited lower antibacterial activity, improved design strategies in a variety of such systems may enable the design of effective glycopeptide analogs of a peptide antibiotic.
Materials and methods
HMP (4-hydroxymethyl phenoxymethyl polystyrene) resin, solvents, and reagents used for synthesis were supplied by Applied Biosystems Inc. Fmoc amino acid derivatives were procured from Bachem Feinchemikalein AG. Other chemicals were purchased from Sigma Chemical Co. The Gram-negative bacterial strains S. typhimurium, S. typhi Vi+ (clinical isolates), and E. coli BL21(λD3) were used for the radial diffusion assay. Agarose I (Biotechnology grade) was obtained from Amresco, and tryptic soy broth (TSB) was from Himedia Laboratories Pvt. Ltd.
Peptide synthesis, purification, and characterization
The Fmoc-Thr(GalNAc)-OH was synthesized by following earlier reported protocols (Kuduk et al. 1998; Winans et al. 1999). Peptides were synthesized by the solid-phase method using an automated peptide synthesizer Model 431A (Applied Biosystems Inc.), employing standard Fmoc-methodology. The peptides were cleaved from the resin by treatment with TFA/thioanisole/phenol/water/EDT in ratio as recommended by Applied Biosystems Inc. The crude peptides were purified using a C-18 column (Deltapak-100 Å, 15 μ, spherical, 19 × 300 mm, Waters), and peptide purity was verified using a C-18 analytical column (Deltapak-300 Å, 15 μ, spherical, 7.8 × 300 mm, Waters). The sugar residue of the glycosylated peptide formaecin I was deacetylated by its treatment with 5% hydrazine-hydrate and finally again purified by reverse phase column chromatography using a C-18 column. Characterization was performed by molecular mass determination using a single Quadruple Mass Analyser (Fisons Instruments).
Antibacterial assays
The radial diffusion assays were performed using double-layered agarose as described previously (Nagpal et al. 1999).
The kinetics of inactivation of the E. coli by peptides was performed in 10% LB. Mid-log-phase bacteria (~5 × 104 colony forming units/mL) were incubated at 37°C for up to 300 min in the absence (control) or the presence of the peptides at a concentration equal to their MIC values. Aliquots of the suspension were withdrawn at the selected time intervals and plated. The number of colony-forming units was counted after 24 h of incubation at 37°C.
Hemolytic assay
Hemolytic activity of the peptides was determined on freshly isolated rat erythrocytes. Heparinized rat blood was initially centrifuged at 3000 rpm. The erythrocyte suspension was further washed thrice with phosphate-saline buffer (10 mM PB at pH 7.4, 150 mM NaCl). Finally, the erythrocytes were suspended in phosphate-buffered saline. The different concentrations of peptide solutions dissolved in 250 μL of PBS were added to 250 μL of 2% erythrocyte solution. The resulting cell suspensions in Eppendorf tubes were incubated with the peptide in duplicate for 30 min at 37°C with gentle mixing. The tubes were then centrifuged, and the absorbance of the supernatants was measured at 540 nm. The hemolysis effected by only PBS and 1% Triton X-100 was considered as zero and 100% hemolysis, respectively.
Dansyl Polymyxin B displacement assay
Dansyl Polymyxin B, a fluorescent derivative of Polymyxin B, was prepared by condensing Polymyxin B sulfate with dansyl chloride as described by Schindler and Teuber (1975). The comparative binding affinity of various antibiotic peptides for endotoxin was investigated by a Dansyl Polymyxin B displacement assay (Moore et al. 1986). The data were expressed as the % Dansyl Polymyxin B bound to the endotoxin as a function of peptide concentration.
Circular dichroism
The circular dichroism (CD) experiments were carried out on a JASCO 710 spectropolarimeter with a 1.0-nm bandwidth at 0.1 nm of resolution, and 1 sec of response time using a 10-mm pathlength cell at 25°C. Twenty to 25 scans with a speed of 200 nm/min in the range of 250–190 nm were accumulated and averaged. The spectra were recorded at the peptide concentration of 10 μM in water, 10 mM phosphate buffer (pH 7), 90% trifluoroethanol, and micelles of 10 mM sodium dodecyl sulfate in 10 mM PB (pH 7). Results were expressed as mean residue molar ellipticity in deg cm/dmol.
Acknowledgments
We thank Ayub Qadri for providing clinical isolates of S. typhimurium and S. typhi Vi+. This work was supported by a Centre of Excellence grant from the Department of Biotechnology and an extramural grant from the Department of Science and Technology, Government of India.
Footnotes
Reprint requests to: Kanwal J. Kaur, Structural Biology Unit, National Institute of Immunology, Aruna Asaf Ali Marg, New Delhi 110 067, India; e-mail: kanwal@nii.res.in; fax: 00-91-11-26717113.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062581707.
References
- Andreotti, A.H. and Kahne, D. 1993. Effect of glycosylation on peptide backbone conformation. J. Am. Chem. Soc. 115: 3352–3353. [Google Scholar]
- Bessalle, R., Kapitkovsky, A., Gorea, A., Shalit, I., and Fridkin, M. 1990. All-D-magainin: Chirality, antimicrobial activity and proteolytic resistance. FEBS Lett. 274: 151–155. [DOI] [PubMed] [Google Scholar]
- Biondi, L., Filira, F., Rocchi, R., Tzehoval, E., and Fridkin, M. 1993. Synthesis and biological activity of [L-hydroxyproline]3-tuftsin analogue and its α- or β-O-D-glucosylated derivatives. Int. J. Pept. Protein Res. 41: 43–51. [DOI] [PubMed] [Google Scholar]
- Boman, H.G., Agerberth, B., and Boman, A. 1993. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect. Immun. 61: 2978–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulet, P., Urge, L., Ohresser, S., Hetru, C., and Otvos Jr., L. 1996. Enlarged scale synthesis and range of activity of drosocin, an O-glycosylated antibacterial peptide of Drosophila. Eur. J. Biochem. 238: 64–69. [DOI] [PubMed] [Google Scholar]
- Casteels, P. and Tempst, P. 1994. Apidaecin-type peptide antibiotics function through a non-poreforming mechanism involving stereospecificity. Biochem. Biophys. Res. Commun. 199: 339–345. [DOI] [PubMed] [Google Scholar]
- Casteels, P., Ampe, C., Jacobs, F., Vaeck, M., and Tempst, P. 1989. Apidaecins: Antibacterial peptides from honeybees. EMBO J. 8: 2387–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteels, P., Romagnolo, J., Castle, M., Casteel-Josson, K., Erdjument-Bromage, H., and Tempst, P. 1994. Biodiversity of apidaecin-type peptide antibiotics. J. Biol. Chem. 269: 26107–26115. [PubMed] [Google Scholar]
- Castle, M., Nazarian, A., Yi, S.S., and Tempst, P. 1999. Lethal effects of apidaecin on Escherichia coli involve sequential molecular interactions with diverse targets. J. Biol. Chem. 274: 32555–32564. [DOI] [PubMed] [Google Scholar]
- Chapman, B.S., Eckart, M.R., Kaufman, S.E., and Lapointe, G.R. 1996. O-linked oligosaccharide on the 75-kDa neurotrophin receptor. J. Neurochem. 66: 1707–1716. [DOI] [PubMed] [Google Scholar]
- Cociancich, S., Bulet, P., Hetru, C., and Hoffmann, J.A. 1994. The inducible antibacterial peptides of insects. Parasitol. Today 10: 132–139. [DOI] [PubMed] [Google Scholar]
- Cudic, M., Bulet, P., Hoffmamm, R., Craik, D.J., and Otvos Jr., L. 1999. Chemical synthesis, antibacterial activity and conformation of diptericin, an 82-mer peptide originally isolated from insects. Eur. J. Biochem. 266: 549–558. [DOI] [PubMed] [Google Scholar]
- Fisher, J.F., Harrison, A.W., Bundy, G.L., Wilkinson, K.F., Rush, B.D., and Ruwart, M.J. 1991. Peptide to glycopeptide: glycosylated oligopeptide renin inhibitors with attenuated in vivo clearance properties. J. Med. Chem. 34: 3140–3143. [DOI] [PubMed] [Google Scholar]
- Gobbo, M., Biondi, L., Filira, F., Gennaro, R., Benincasa, M., Scolaro, B., and Rocchi, R. 2002. Antimicrobial peptides: Synthesis and antibacterial activity of linear and cyclic drosocin and apidaecin 1B analogues. J. Med. Chem. 45: 4494–4504. [DOI] [PubMed] [Google Scholar]
- Hara, S. and Yamakawa, M. 1995. A novel antibacterial peptide family isolated from the silkworm, Bombyx mori . Biochem. J. 310: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, J.A., Hetru, C., and Reichhart, J.-M. 1993. The humoral antibacterial response of Drosophila . FEBS Lett. 325: 63–66. [DOI] [PubMed] [Google Scholar]
- Horvat, S., Horvat, J., Varga-Defterdarovic, L., Pavelic, K., Chung, N.N., and Schiller, P.W. 1993. Methionine-enkephalin related glycoconjugates. Synthesis and biological activity. Int. J. Pept. Protein Res. 41: 399–404. [PubMed] [Google Scholar]
- Hultmark, D. 1993. Immune reactions in Drosophila and other insects: A model for innate immunity. Trends Genet. 9: 178–183. [DOI] [PubMed] [Google Scholar]
- Jain, D., Kaur, K.J., Goel, M., and Salunke, D.M. 2000. Structural basis of functional mimicry between carbohydrate and peptide ligands of Con A. Biochem. Biophys. Res. Commun. 272: 843–849. [DOI] [PubMed] [Google Scholar]
- Kuduk, S.D., Schwarz, J.B., Chen, X.T., Glunz, P.W., Sames, D., Ragupathi, G., Livingston, P.O., and Danishefsky, S.J. 1998. Synthetic and immunological studies on clustered modes of mucin-related Tn and TF O-linked antigens: The preparation of a glycopeptide-based vaccine for clinical trials against prostrate cancer. J. Am. Chem. Soc. 120: 12474–12485. [Google Scholar]
- Laczko, I., Hollosi, M., Urge, L., Ugen, K.E., Weiner, D.B., Mantsch, H.H., Thurin, J., and Otvos Jr., L. 1992. Synthesis and conformational studies of N-glycosylated analogues of the HIV-1 principal neutralizing determinant. Biochemistry 31: 4282–4288. [DOI] [PubMed] [Google Scholar]
- Mackintosh, J.A., Veal, D.A., Beattie, A.J., and Gooley, A.A. 1998. Isolation from an ant Myrmecia gulosa of two inducible O-glycosylated proline-rich antibacterial peptides. J. Biol. Chem. 273: 6139–6143. [DOI] [PubMed] [Google Scholar]
- Moore, R.A., Bates, N.C., and Hancock, R.E.W. 1986. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob. Agents Chemother. 29: 496–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, S., Gupta, V., Kaur, K.J., and Salunke, D.M. 1999. Structure–function analysis of tritrypticin, an antibacterial peptide of innate immune origin. J. Biol. Chem. 274: 23296–23304. [DOI] [PubMed] [Google Scholar]
- O'Conner, S.E. and Imperiali, B. 1998. A molecular basis for glycosylation-induced conformational switching. Chem. Biol. 5: 427–437. [DOI] [PubMed] [Google Scholar]
- Otvos Jr., L., Rogers, M.E., Consolvo, P.J., Condie, B.A., Lovas, S., Bulet, P., and Blaszczyk-Thurin, M. 2000a. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 39: 14150–14159. [DOI] [PubMed] [Google Scholar]
- Otvos Jr., L., Bokonyi, K., Varga, I., Otvos, B.I., Hoffmann, R., Ertl, H.C.J., Wade, J.D., McManus, A.M., Craik, D.J., and Bulet, P. 2000b. Insect peptides with improved protease-resistance protect mice against bacterial infection. Protein Sci. 9: 742–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poujade, C., Lavielle, S., Torrens, Y., and Marquet, A. 1983. Synthesis and biological activity of glycosylated analogs of the C-terminal hexapeptide and heptapeptide of substance P. Int. J. Pept. Protein Res. 21: 254–257. [DOI] [PubMed] [Google Scholar]
- Powell, M.F., Stewart, T., Otvos Jr., L., Urge, L., Gaeta, F.C., Sette, A., Arrhenius, T., Thomson, D., Soda, K., and Colon, S.M. 1993. Peptide stability in drug development. II. Effect of single amino acid substitution and glycosylation on peptide reactivity in human serum. Pharm. Res. 10: 1268–1273. [DOI] [PubMed] [Google Scholar]
- Rickert, K.W. and Imperiali, B. 1995. Analysis of the conserved glycosylation site in the nicotinic acetylcholine receptor: Potential roles in complex assembly. Chem. Biol. 2: 751–759. [DOI] [PubMed] [Google Scholar]
- Rose, M.C., Voter, W.A., Sage, H., Brown, C.F., and Kaufman, B. 1984. Effects of deglycosylation on the architecture of ovine submaxillary mucin glycoprotein. J. Biol. Chem. 261: 14307–14312. [PubMed] [Google Scholar]
- Schindler, P.R.G. and Teuber, M. 1975. Action of polymyxin B on bacterial membranes: Morphological changes in the cytoplasm and in the outer membrane of Salmonella typhimurium and Escherichia coli B. Antimicrob. Agents Chemother. 8: 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren, R., Gerken, T.A., and Jentoft, N. 1989. Role of glycosylation on the conformation and chain dimensions of O-linked glycoproteins: Light-scattering studies of ovine submaxillary mucin. Biochemistry 28: 5525–5536. [DOI] [PubMed] [Google Scholar]
- Skerlavaj, B., Romeo, D., and Gennaro, R. 1990. Rapid membrane permeabilization and inhibition of vital functions of Gram-negative bacteria by bactenecins. Infect. Immun. 58: 3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urge, L., Gorbics, L., and Otvos Jr., L. 1992. Chemical glycosylation of peptide T at natural and artificial glycosylation sites stabilizes or rearranges the dominant reverse turn structure. Biochem. Biophys. Res. Commun. 184: 1125–1132. [DOI] [PubMed] [Google Scholar]
- Wade, D., Boman, A., Wahlin, B., Drain, C.M., Andreu, D., Boman, H.G., and Merrifield, R.B. 1990. All-D amino acid-containing channel-forming antibiotic peptides. Proc. Natl. Acad. Sci. 87: 4761–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans, K.A., King, D.S., Rao, V.R., and Bertozzi, C.R. 1999. A chemically synthesized version of the insect antibacterial glycopeptide, diptericin, distrupts bacterial membrane integrity. Biochemistry 38: 11700–11710. [DOI] [PubMed] [Google Scholar]