Abstract
The circadian input kinase (CikA) is a major element of the pathway that provides environmental information to the circadian clock of the cyanobacterium Synechococcus elongatus. CikA is a polypeptide of 754 residues and has three recognizable domains: GAF, histidine protein kinase, and receiver-like. This latter domain of CikA lacks the conserved phospho-accepting aspartyl residue of bona fide receiver domains and is thus a pseudo-receiver (PsR). Recently, it was shown that the PsR domain (1) attenuates the autokinase activity of CikA, (2) is necessary to localize CikA to the cell pole, and (3) is necessary for the destabilization of CikA in the presence of the quinone analog 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB). The solution structure of the PsR domain of CikA, CikAPsR, is presented here. A model of the interaction between the PsR domain and HPK portion of CikA provides a potential explanation for how the PsR domain attenuates the autokinase activity of CikA. Finally, a likely quinone-binding surface on CikAPsR is shown here.
Keywords: protein structure/folding, enzymes, heteronuclear NMR, circadian clock, cyanobacteria, histidine protein kinase, metabolism, photosynthesis, pseudo-receiver
Virtually all light-perceiving organisms display circadian (∼24-h) rhythms in their gene activity, metabolism, physiology, and behavior in anticipation of and in preparation for daily swings in sunlight and ambient temperature (Dunlap et al. 2004; Bell-Pedersen et al. 2005). These robust biological rhythms are the result of an endogenous biological circadian clock. Circadian clocks have three interdependent divisions: input pathways, output pathways, and central oscillators (Bell-Pedersen et al. 2005). The oscillator of a circadian clock can be reset by environmental cues, such as light and temperature, through clock input signal transduction pathways.
The simplest known circadian system is that of the cyanobacterium, Synechococcus elongatus (Ditty et al. 2003; Golden 2004). High-resolution structures of proteins of the central oscillator and output pathway components of S. elongatus and other cyanobacterial species have been determined by NMR and X-ray crystallography (Williams et al. 2002; Garces et al. 2004; Pattanayek et al. 2004; Uzumaki et al. 2004; Vakonakis and LiWang 2004; Vakonakis et al. 2004a,b; Xu et al. 2004; Ye et al. 2004; Hitomi et al. 2005; Iwase et al. 2005), but there are no solved structures for any member of the clock-input pathway. The circadian input kinase (CikA) protein of S. elongatus is a key part of the environmental input pathway for clock resetting (Schmitz et al. 2000). cikA-inactivated reporter strains fail to reset the phase of the clock in response to a 5-h dark pulse and display shortened period lengths. In both short- and long-period kai mutants, cikA inactivation changes the resulting period additively, which suggests that CikA and the Kai proteins have independent, nonoverlapping functions (Schmitz et al. 2000). Recently, it was shown that CikA forms a complex with the Kai proteins of the circadian oscillator and, during resetting of circadian phase by a dark pulse, affects the phosphorylation state of KaiC (Ivleva et al. 2006). CikA also copurifies with an iron-sulfur protein, LdpA, which is involved in light-dependent modulation of the circadian period (Katayama et al. 2003; Ivleva et al. 2005), which suggests that CikA and LdpA are part of the same input pathway to the clock. LdpA is redox-sensitive and proposed to confer sensitivity of the circadian period length to the metabolic state of the cell and thus implicates CikA as part of this redox-sensing mechanism as well.
CikA has limited similarity to phytochromes, which supports the notion of an evolutionary relationship between cyanobacterial and plant circadian systems (Schmitz et al. 2000; Vierstra and Davis 2000). The 754-residue CikA protein of S. elongatus has three distinct domains as predicted from bioinformatics: an N-terminal GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA)-like domain (residues 184–343), a central histidine-protein kinase (HPK) portion (residues 378–611), and a C-terminal pseudo-receiver (PsR) domain (residues 629–744). The central HPK portion is predicted to contain an N-terminal dimerization and histidine phosphotransfer (DHp) domain (residues 378–453) and a C-terminal catalytic and ATP-binding (CA) domain (residues 454–611); kinase activity and dimerization have been demonstrated experimentally (Mutsuda et al. 2003; Zhang et al. 2006).
GAF domains of phytochrome family members bind bilin chromophores by covalent linkage to a conserved cysteine residue (Lamparter 2004). However, the GAF domain of CikA lacks this conserved cysteine and has only a weak affinity for bilin in vitro (Mutsuda et al. 2003). Thus, the environmental signal and potential ligand to which the GAF domain of CikA is responsive are not known. Deletion of the GAF domain dramatically reduces the rate of autophosphorylation of CikA, which suggests a role for this domain in regulating HPK activity (Mutsuda et al. 2003).
The amino acid sequence of the C-terminal domain of CikA has limited sequence similarity to receiver domains of two-component response regulators but lacks the conserved phospho-accepting aspartyl residue of canonical receiver domains (Fig. 1). Assays for phosphorelay from the HPK portion of CikA to the C-terminal PsR domain were negative (Mutsuda et al. 2003), which supports the identification of the C terminus as a pseudo-receiver domain. Truncation of the PsR domain results in a 10-fold increase in autophosphorylation (Mutsuda et al. 2003), which indicates that the PsR domain attenuates the autokinase activity of CikA. A family of pseudo-response regulators (APRR) that lack the phospho-accepting aspartyl residue is also involved in circadian clock function in Arabidopsis thaliana (Nakamichi et al. 2005; Salome and McClung 2005).
Figure 1.
ClustalW (Thompson et al. 1994) sequence alignment of the PsR domain of CikA and receiver domains of PrrA, DrrD, FixJ, NtrC, Spo0F, PhoB, ArcA, and CheY. Dots, colons, and asterisks denote low and high sequence similarities and invariant residues, respectively. The phospho-accepting aspartyl residue of receiver domains is shown in boldface type. The integers along the top of the sequences correspond to the residue numbers of CikA.
PsR domains in some proteins regulate function through protein–protein interactions. For example, intermolecular contacts between the N-terminal PsR domain of AmiR and the protein AmiC in the bacterium Pseudomonas aeruginosa prevent the C-terminal domain of AmiR from binding nascent mRNA (O'Hara et al. 1999). The N-terminal PsR domain of the cyanobacterial clock oscillator protein KaiA is predicted to exert allosteric control over its C-terminal KaiC-binding domain, which is necessary and sufficient to increase the autokinase activity of KaiC (Williams et al. 2002); regulation of KaiC autophosphorylation should be important for resetting the circadian clock. These examples show that PsR domains, including that of CikA, regulate protein function through protein–protein interactions. The PsR domain is also necessary to localize CikA to the cell pole (Zhang et al. 2006). These observations are consistent with recent evidence linking CikA to cell division (Miyagishima et al. 2005). We previously proposed that a binding event between an unidentified cell-pole protein and the PsR domain of CikA affects the PsR-mediated inhibition of HPK activity and thereby helps determine when and where HPK activity is maximal. Here, we present the NMR structure of the PsR domain of CikA of S. elongatus and discuss structure–function relationships that are consistent with the model.
We recently showed that CikA abundance varies inversely with light intensity (Ivleva et al. 2005) and that its stability decreases in the presence of the quinone analog DBMIB (2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone) (Ivleva et al. 2005, 2006). This finding suggests that CikA is part of an input pathway that entrains the circadian clock by synchronizing it with metabolism and particularly with photosynthesis. We further showed that CikA apparently binds DBMIB directly with its PsR domain (Ivleva et al. 2006). We show here evidence of the quinone binding surface of CikA.
Results and Discussion
The PsR domain of CikA is a monomer
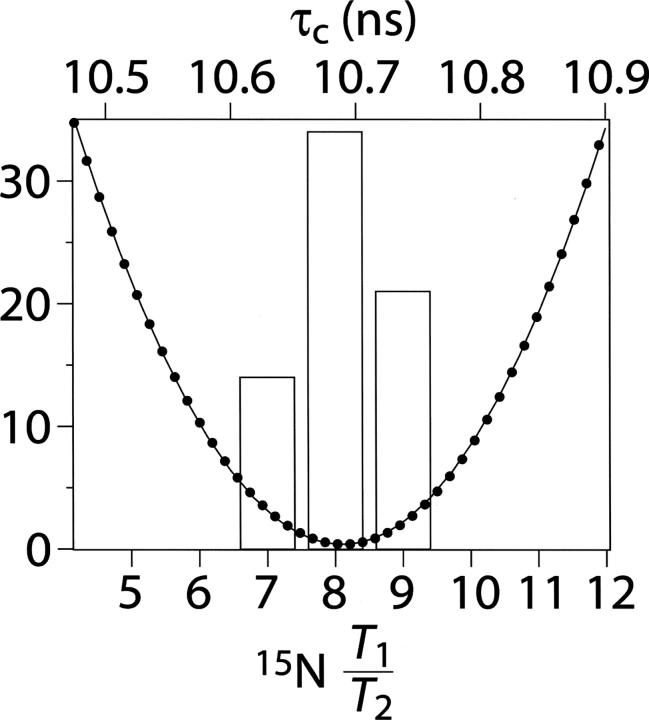
All data were collected on a 137-residue polypeptide containing the last 133 residues (622–754) of the CikA protein plus four residues introduced during cloning (Gao et al. 2005). Backbone amide 15N T 1, T 2, and 15N{-1H} NOE data were acquired to estimate the effective isotropic rotational correlation time, τ c, as described earlier (LiWang et al. 1999a). It has been shown previously that the 15N T 1/T 2 ratio can be used to estimate the overall rotational correlation time, τ c, of a protein provided that residues experiencing local motions that significantly affect either T 1 or T 2 are excluded (Farrow et al. 1994; Tjandra et al. 1996). As such, residues with 15N{-1H} NOE ≤ 0.60 were excluded from the calculation of τ c. Shown in Figure 2 are 15N T 1/T 2 ratios of nondegenerate residues with heteronuclear 15N{-1H} NOE values >0.60. The average T 1/T 2 ratio of these 78 residues is 8.6 ± 0.7. Of this set, residues experiencing conformational exchange on the microsecond to millisecond time scale have reduced T 2 but not T 1 values and also need to be removed from τ c analysis. They were identified using the following criterion (Tjandra et al. 1996): (<T 2> − T 2,i)/<T 2> − (<T 1> − T 1,i)/<T 1> > 1.5 SD, where <T 2> and <T 1> are the average 15N T 2 and T 1 values, respectively; T 2,i and T 1,i are the T 2 and T 1 values of residue i; and SD is the standard deviation of (<T 2> − T 2,i)/<T 2> − (<T 1> − T 1,i)/<T 1>, excluding residues with a 15N{-H} NOE ≤ 0.60. There were 69 residues remaining after applying this second criterion, which yielded an average T 1/T 2 ratio of 8.6 ± 0.6.
Figure 2.
Histogram of 15N T 1/T 2 ratios and estimation of τ c. Solid circles are E values (= Σi(T 1,i obs/T 2,i obs − T 1 calc /T 2 calc)2) plotted as a function of τ c. The values along the Y-axis correspond to the number of residues in the T 1/T 2 histogram.
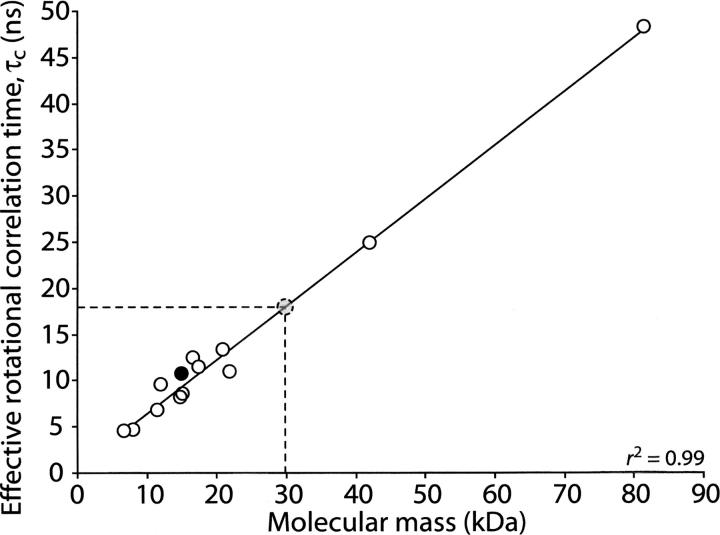
The T 1/T 2 ratios of the remaining residues were then used to estimate τ c by minimizing the following equation as a function of τ c: E = Σi(T 1,i obs/T 2,i obs − T 1 calc/T 2 calc)2. The model-free spectral density equations were used in the T 1 calc and T 2 calc equations with the order parameter, S 2, set at one. A one-dimensional grid search for the minimum in E as a function of τ c (Fig. 2) yielded a value of τ c = 10.7 ± 0.5 nsec. A comparison with the τ c values of several other proteins predicts that a dimeric 15N-enriched CikAPsR of 2 × 14.9 = 29.8 kDa would have τ c ∼ 17.9 nsec (Fig. 3; Table 1), which is inconsistent with our experimentally determined τ c. Instead, the 15N relaxation data are consistent with a monomeric CikAPsR even at the appreciable concentration of 1.3 mM. However, full-length CikA is a dimer (Zhang et al. 2006), which means that the dimerization interface lies within another domain. We think that CikA forms a homodimer through its DHp domain, as this is the case for other HPK proteins such as Spo0B, EnvZ, and TM0853 (Varughese et al. 1998; Tomomori et al. 1999; Marina et al. 2005).
Figure 3.
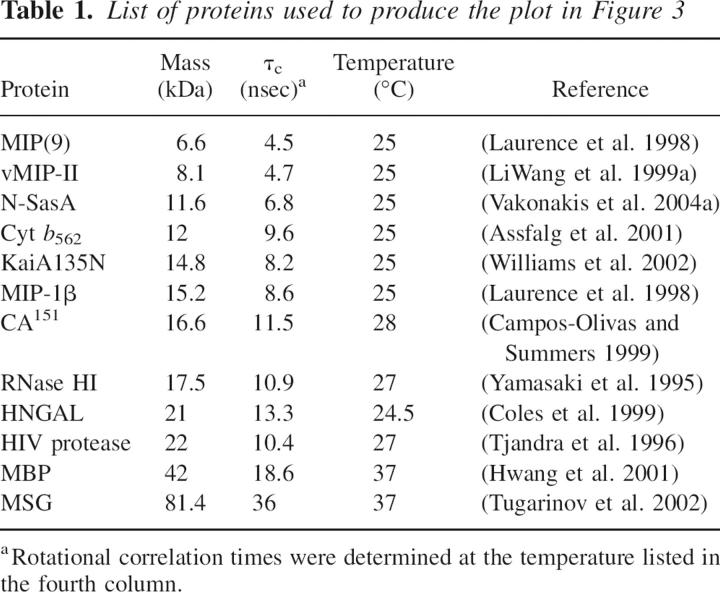
Plot of effective rotational correlation times of proteins as a function of molecular mass. The open circles are from the proteins listed in Table 2 with correlation times recalculated if necessary at 25°C using Stokes’ law: τ c = 4 π η r 3/(3 k B T). A linear regression fit to the open circles is shown by a solid line. The solid circle correlates the experimentally determined τ c of CikAPsR (10.7 nsec) to its calculated molecular mass (14.9 kDa for the 15N-enriched 137-residue polypeptide). The dashed lines and dashed circle indicate the predicted position of CikAPsR in this plot if it were a dimer with a molecular mass of 2 × 14.9 = 29.8 kDa. The correlation coefficient of the linear regression fit is given in the lower right corner.
Table 1.
List of proteins used to produce the plot in Figure 3
Overall structure of the PsR domain of CikA
Heteronuclear 15N{-1H} NOE values are ≤0.2 for the first eight and last 12 residues of our polypeptide construct, which indicates significant conformational dynamics at the termini. Subsequent analysis did not consider those residues, and hereafter, the PsR domain of CikA (CikAPsR) refers to residues 629–742.
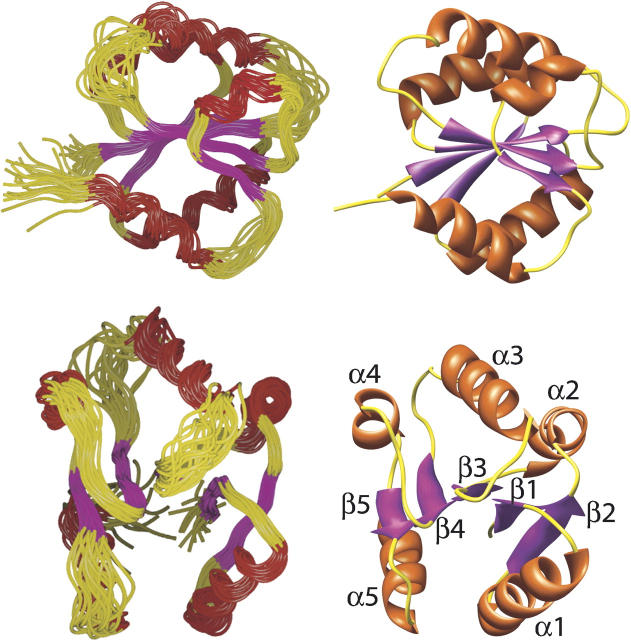
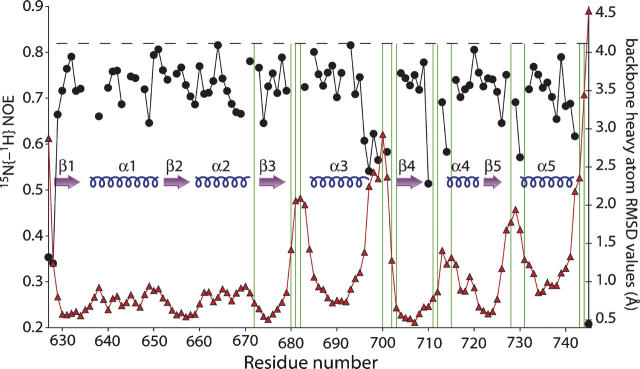
The structure of CikAPsR is shown in Figure 4, and the structural statistics are listed in Table 2. The fold of CikAPsR is a doubly wound, parallel, five-stranded β-sheet with a 2-1-3-4-5 topology. There are five α-helices, with α1 and α5 on one face of the β-sheet and α2, α3, and α4 on the other. The relative ratios of the principal components of the inertia tensor are 1.00:0.95:0.82, which predicts that rotational diffusion of CikAPsR is close to isotropic. The average 15N{-1H} NOE of the secondary structure elements is 0.74 (±0.4), which is 10% lower than the theoretical maximum of 0.82 for τ c = 10.7 nsec at 50.5 MHz 15N resonance frequency (assuming 15N (σ || − σ ⊥) = –170 ppm and r NH = 1.04 Å). Figure 5 shows that the loops connecting secondary elements α3-β4, β4-α4, and β5-α5 experience significant dynamics as indicted by the lower 15N{-1H} values. Consistent with these localized motions are the high backbone RMSDs of these loops (Fig. 5). The β3-α3 loop, which also has high backbone RMSDs due to the scarcity of long-range 1H-1H NOEs, contains three consecutive proline residues (P680–P682).
Figure 4.
NMR structure of CikAPsR (PDB no. 2j48). On the left are superpositions of the 20 lowest-energy structures (out of 100 total) in two orthogonal views. The restrained regularized mean structure is shown on the right in the same orthogonal orientations. The stretches of secondary structure elements are β1 (629–633), α1 (636–650), β2 (653–657), α2 (659–671), β3 (674–678), α3 (685–696), β4 (705–708), α4 (714–721), β5 (723–724), and α5 (731–740).
Table 2.
NMR structure statistics and restraints for CikAPsR
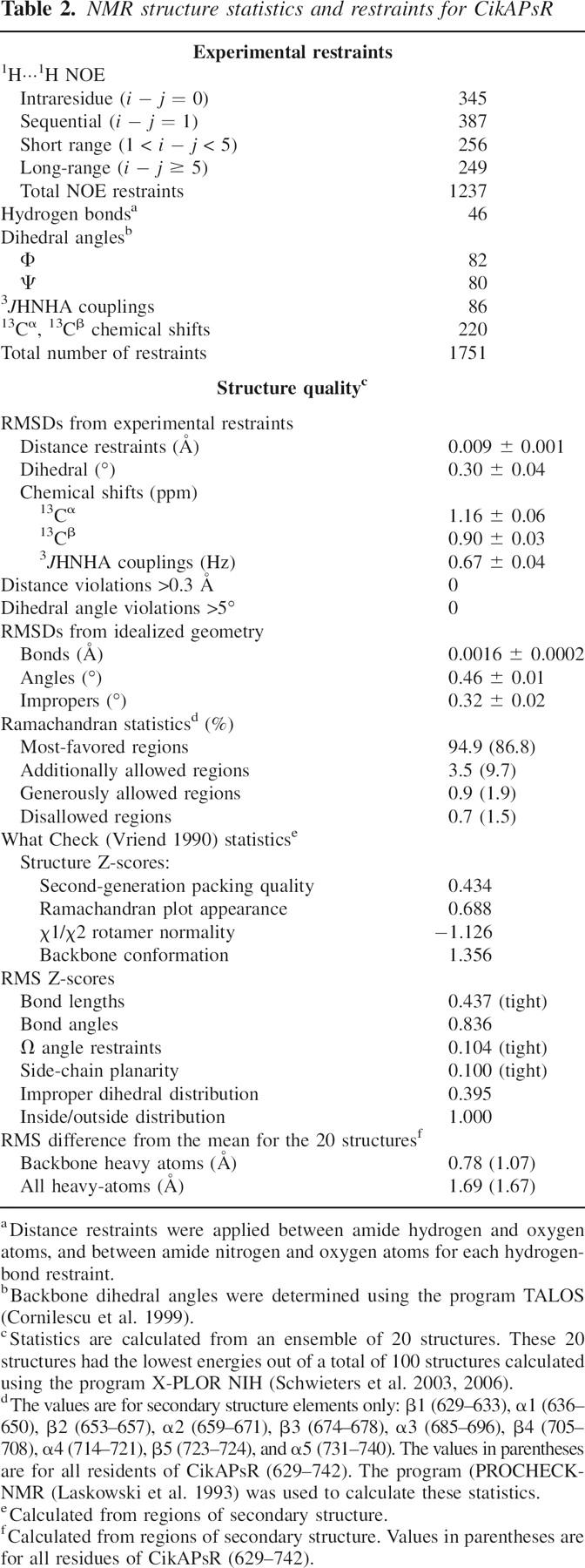
Figure 5.
15N{-1H} NOE and backbone heavy atom RMSD values for residues 627–745 of CikA. The RMSD values are shown as red triangles, and the NOE values are represented by solid circles. Values of adjacent residues are connected by red or black lines. The dashed horizontal line at 0.82 indicates the theoretical maximum of 15N{-1H} NOE for τ c = 10.7 nsec at 50.5-MHz 15N resonance frequency (assuming 15N (σ ‖ – σ ⊥) = –170 ppm and r NH = 1.04 Å). The green vertical lines indicate the positions of the 14 proline residues in this stretch of CikA: P672, P680, P681, P682, P700, P702, P703, P711, P712, P715, P728, P731, P743, and P744.
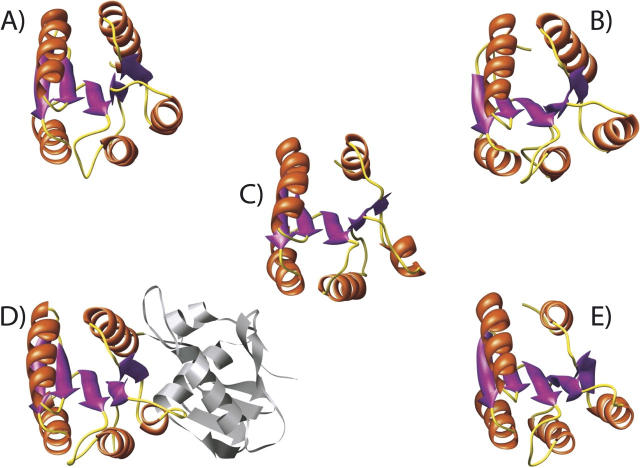
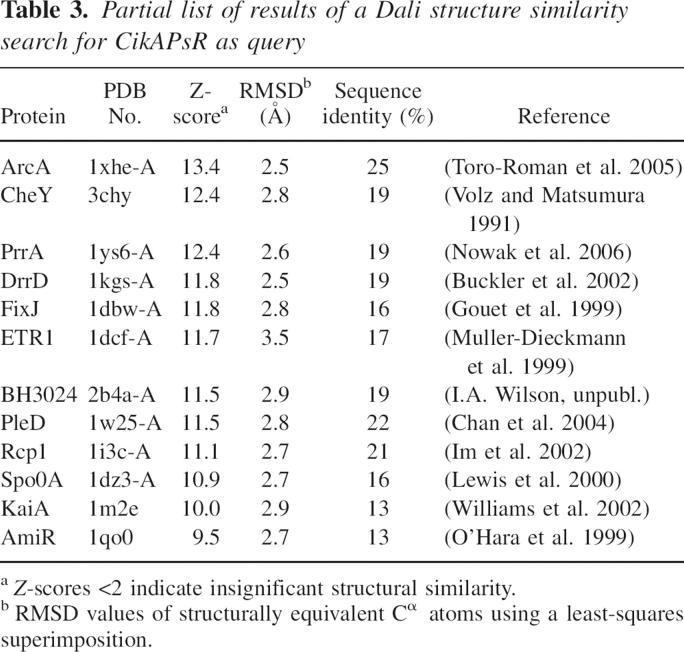
A search of similar protein structures using the Dali search engine (Holm and Sander 1998) yielded predominantly receiver domains of response regulators of bacterial two-component signaling systems (Fig. 6; Table 3; Stock et al. 2000). The largest difference between CikAPsR and these receiver domains is with the orientation of the fourth α helix, α4, which is most similar in angle to that of the phosphorylated form of the receiver domain of NtrC (Kern et al. 1999).
Figure 6.
Structures of (A) ArcA (PDB no. 1xhe-A), (B) CheY (3chy), (C) CikAPsR, (D) PrrA (1ys6-A), and (E) Spo0F (1nat). These structures are displayed in the same orientation. The N-terminal receiver and C-terminal effector domains of PrrA are colored and gray, respectively.
Table 3.
Partial list of results of a Dali structure similarity search for CikAPsR as query
Model of an interaction between the PsR domain and HPK portion of CikA
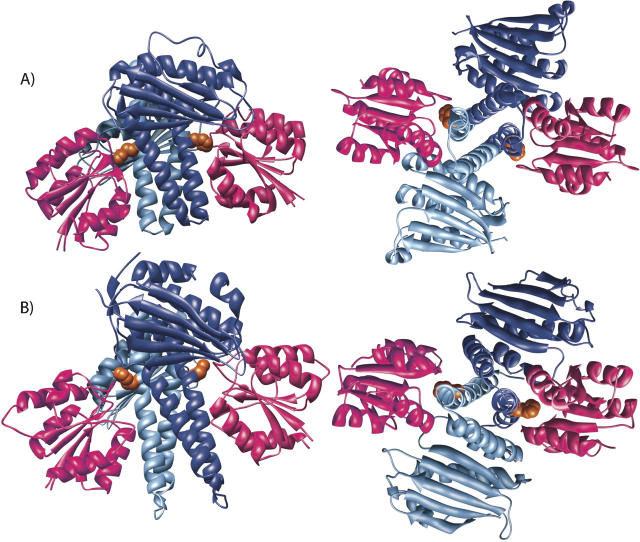
A truncation mutant of CikA lacking the PsR domain has 10-fold greater autokinase activity than wild type (Mutsuda et al. 2003). Thus, the PsR domain attenuates autophosphorylation of the HPK portion. In order to gain insight into how this attenuation may be achieved, we turn to the X-ray crystal structure of the complex between the Spo0F response regulator and the histidine protein kinase Spo0B (Fig. 7A; Zapf et al. 2000; Varughese et al. 2006).
Figure 7.
A model of the interaction between the PsR domain and HPK portion of CikA. (A) The X-ray crystal structure of the Spo0F–Spo0B complex in two orthogonal orientations. (B) A model of the interaction between the PsR domain and HPK portion of CikA in the same two orthogonal orientations. The generation of the model is described in the main text. The HPK protomers are shown in dark and light blue. Spo0F and CikAPsR are shown in pink. Side-chains of H30 of Spo0B and H393 of CikA are shown in orange.
Sporulation in Bacillus subtilis is mediated by a phosphorelay signal transduction pathway, of which Spo0B and Spo0F are key components (Tzeng et al. 1998). Spo0F is a 124-residue response regulator that adopts the fold typical of receiver domains. The sensor histidine kinase KinA donates a phosphoryl group to D54 of Spo0F, which then transfers the phosphoryl group to H30 of Spo0B. Finally, Spo0B transfers the group to the transcription factor of sporulation genes, Spo0A. Recent X-ray crystal structures of the Spo0F–Spo0B complex show that the surface of Spo0F that interacts with the HPK portion of Spo0B is largely composed of the five loops around the active site. In this complex, D54 of Spo0F and H30 of Spo0B are brought into close proximity for phosphoryl transfer (Zapf et al. 2000; Varughese et al. 2006). Perhaps the PsR domain and HPK portion of CikA physically interact similarly. Such an interaction would physically block H393 from phosphoryl transfer reactions and could explain how the PsR domain attenuates autophosphorylation of CikA.
In order to model possible interactions between the PsR domain and the HPK portion of CikA, we first needed a model of the structure of the HPK portion. We used the structure prediction programs LOOPP (Learning, Observing, and Outputting Protein Patterns) (Meller and Elber 2001; Teodorescu et al. 2004) and, as an independent check, SAM-T02 (Sequence Alignment and Modeling Software System) (Karchin et al. 2003; Karplus et al. 2003), with the CikA HPK portion (residues 378–611) as query. LOOPP is a fold-recognition program that uses threading to generate atomic coordinates. SAM T02 uses a linear hidden Markov model approach to protein modeling. For both programs, the HPK domain of CikA was predicted to be most similar to the X-ray crystal structure of the entire cytoplasmic portion of a sensor histidine-kinase protein from the thermophilic bacterium Thermotoga maritima (Marina et al. 2005) (Protein Data Bank [PDB] no. 2c2a). The LOOPP program gave a Z-score of 9.7 for the prediction, which means that the energy of alignment of the sequence of the HPK portion of CikA onto the 2c2a structure is 9.7 standard deviations lower than that of a random alignment. The SAM-T02 prediction yielded an E-value of 9 × 10−11; an E-value <10−5 means that the query sequence is very likely to have the same fold as the target. Thus, these two different programs both predicted with high confidence that the structure of the HPK portion of CikA is most similar to that of 2c2a.
The atomic coordinates of the HPK portion of CikA generated by LOOPP were then superimposed on Spo0B in the structure of the Spo0F–Spo0B complex (PDB no. 1f51), and the NMR structure of CikAPsR was superimposed on Spo0F. The resulting qualitative model of the physical interaction between the PsR and HPK of CikA is shown in Figure 7B. In this model the PsR domain would physically block H393 of the HPK from participating in phosphoryl transfer, which provides a possible explanation for the 10-fold increase in autophosphorylation of CikA upon deletion of PsR (Mutsuda et al. 2003). Receiver domains can also play inhibitory roles, as is the case for NarL (Eldridge et al. 2002; Zhang et al. 2003), CheB (Djordjevic et al. 1998), and Spo0A (Grimsley et al. 1994). Phosphorylation of these receiver domains causes a conformational change that lifts the inhibition of their effector domains; perhaps a similar conformational change is effected in CikAPsR through protein–protein interactions (with a membrane-bound partner protein instead of phosphorylation [Zhang et al. 2006]).
Pseudo-receiver domains also use the α4-β5-α5 surface for protein–protein interactions. In the homodimeric circadian clock protein KaiA of S. elongatus, the α4-β5-α5 surface of the N-terminal pseudo-receiver domain forms intermolecular contacts with the domain-swapped C-terminal domain of the other protomer (Ye et al. 2004). The homodimeric protein AmiR from Pseudomonas aeruginosa forms intermolecular contacts across the dimer interface using the α4-β5-α5 surface of its pseudo-receiver domain (O'Hara et al. 1999). In a similar fashion, we anticipate that CikAPsR uses its α4-β5-α5 surface for intermolecular protein–protein interactions. As was shown earlier, the PsR domain of CikA is required for the localization of CikA to the cell pole; it may be the α4-β5-α5 surface of CikAPsR that regulates the cellular localization (Zhang et al. 2006). Whether this notion is correct or not can be tested through in vivo experiments using CikA variants with mutations along the α4-β5-α5 surface. This surface and that which interacts with the HPK portion of CikA may be coupled such that autokinase activity depends on cellular localization.
CikA abundance and the light-dependent redox state of the quinone pool are correlated, which suggests that CikA is part of a clock input pathway that entrains the circadian clock by synchronizing it with photosynthetic activity (Ivleva et al. 2005). Furthermore, the stabilities of full-length CikA and the isolated PsR domain are decreased in the presence of a plastoquinone analog DBMIB, but that of the variant lacking the PsR domain is not. Additional experiments showed that DBMIB directly binds to the PsR domain: At an equimolar mixture of DBMIB and CikAPsR, although several CikAPsR NMR signals are perturbed, a majority of peaks appear relatively unaffected (Ivleva et al. 2006). This observation is consistent with an interaction between DBMIB and a specific site on CikAPsR.
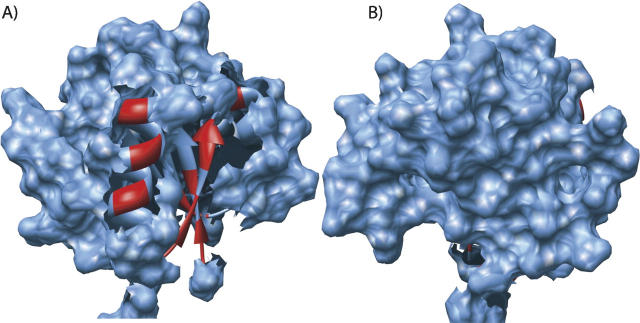
Now we can map the DBMIB-induced spectral perturbations of CikAPsR observed earlier (Ivleva et al. 2006) onto the three-dimensional structure. As seen in Figure 8A, it appears as though DBMIB interacts with a surface formed by α1 and β2. In contrast, this qualitative analysis suggests that the opposite side of CikAPsR is not affected by the presence of 0.2 mM DBMIB (Fig. 8B). The fact that CikAPsR physically interacts with DBMIB is consistent with the notion that CikA is an essential component of the input pathway that synchronizes the circadian clock with photosynthesis by directly sensing the redox state of the plastoquinone pool. At this point, it is unknown how quinone binding affects the interaction between the PsR and HPK portion of CikA. It is also unknown how quinone binding affects the PsR-mediated cellular localization of CikA.
Figure 8.
Mapping of DBMIB-induced spectral perturbations onto the structure of CikAPsR. The surface of CikAPsR is shown in blue, and residues spectrally perturbed by DBMIB are highlighted in red on the ribbon diagram. The α1-β2-α2 and α4-β5-α5 sides of CikAPsR are shown in A and B, respectively.
The function of receiver domains is regulated by phosphorylation. However, the function and regulation of pseudo-receiver domains, which are present in many bacterial and plant proteins, is much less clear. Quinone binding by CikAPsR shows that pseudo-receiver domains can function as sensors of small molecules.
Materials and methods
The last 133 residues (622–754) of CikA plus four residues introduced during cloning were overexpressed and isotopically enriched as described earlier (Gao et al. 2005). The calculated molecular mass for this polypeptide uniformly enriched with 15N is 14.9 kDa.
NMR spectroscopy
All experiments were performed at 25 °C using Bruker 800-MHz and Varian Inova 600-MHz and 500-MHz spectrometers equipped with triple-axis gradient probes. Sequential 13C, 15N, and 1H, backbone and side-chain assignments were described earlier (Gao et al. 2005). NOE interproton distance restraints were derived from 15N, 13C-edited, and 13C, 13C-edited four-dimensional NOESY spectra. Hydrogen-bond restraints were applied based on hydrogen-exchange protection data collected by NMR, as well as the existence of expected regular secondary structure 1H···1H NOEs as described previously (LiWang et al. 1999b). The isomerization states of 12 out of 14 proline residues were experimentally determined as trans by the observation of NOESY cross-peaks between the δ protons of the proline side-chain and the α proton of the preceding residue (Wüthrich 1986); P703 and P728 are also trans in the calculated structure but without experimental restraints. Backbone dynamics measurements were collected on and analyzed from a single set of 15N T 1, 15N T 2, and 15N{-1H} NOE relaxation data collected at 11.7 T as described previously (LiWang et al. 1999a).
Structure calculations
The ϕ and ψ dihedral angle values were derived from the program TALOS (Cornilescu et al. 1999). 1H···1H distance restraints were grouped into four ranges depending on the NOESY cross-peak intensity: ranges 1.8–2.9 Å, 1.8–3.5 Å, 1.8–5.0 Å, and 1.8–6.0 Å correspond to strong, medium, weak, and very weak NOESY cross-peaks. The upper distance bound was increased by 0.5 Å for NOESY cross-peaks involving methyl groups. The XPLOR-NIH software package (Schwieters et al. 2003, 2006) was used for all stages of NMR structure calculations. Only NOE, dihedral angle, hydrogen bond, and 3 J HNHA potential energy terms were used as restraints during simulated annealing. Additional potential energy terms were applied during the structure refinement process: radius of gyration (Kuszewski et al. 1999), conformational database (Kuszewski et al. 1996), and 13Cα and 13Cβ chemical shifts potentials (Kuszewski et al. 1995). The final ensemble consists of the 20 lowest-energy structures out of 100 total (PDB ID: 2j48).
We used the software program QUEEN to assess our NOE distance restraints during the structure calculation process (Nabuurs et al. 2003, 2006). QUEEN allowed us to sort the restraints by unique information content for systematic evaluation.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM064576 to A.L.; GM62419 and NS39546 to S.S.G.). We are grateful for NMR support from Drs. Xiangming Kong and Youlin Xia. We also thank Elihu Ihms for assistance on molecular modeling. The NMR instrumentation in the Biomolecular NMR Laboratory at Texas A&M University was supported by the National Science Foundation grant DBI-9970232.
Footnotes
Reprint requests to: Andy LiWang, Department of Biochemistry & Biophysics, Texas A&M University College Station, TX 77843, USA; e-mail: andy-liwang@tamu.edu; fax: (979) 845-9274.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062532007.
References
- Assfalg M., Banci, L., Bertini, I., Ciofi-Baffoni, S., and Barker, P.D. 2001. 15N backbone dynamics of ferricytochrome b 562: Comparison with the reduced protein and the R98C variant. Biochemistry 40: 12761–12771. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone, V.M., Earnest, D.J., Golden, S.S., Hardin, P.E., Thomas, T.L., and Zoran, M.J. 2005. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 6: 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler D.R., Zhou, Y., and Stock, A.M. 2002. Evidence of intradomain and interdomain flexibility in an OmpR/PhoB homolog from Thermotoga maritima . Structure 10: 153–164. [DOI] [PubMed] [Google Scholar]
- Campos-Olivas R. and Summers, M.F. 1999. Backbone dynamics of the N-terminal domain of the HIV-1 capsid protein and comparison with the G94D mutant conferring cyclosporin resistance/dependence. Biochemistry 38: 10262–10271. [DOI] [PubMed] [Google Scholar]
- Chan C., Paul, R., Samoray, D., Amiot, N.C., Giese, B., Jenal, U., and Schirmer, T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. 101: 17084–17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles M., Diercks, T., Muehlenweg, B., Bartsch, S., Zölzer, V., Tschesche, H., and Kessler, H. 1999. The solution structure and dynamics of human neutrophil gelatinase-associated lipocalin. J. Mol. Biol. 289: 139–157. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13: 289–302. [DOI] [PubMed] [Google Scholar]
- Ditty J.L., Williams, S.B., and Golden, S.S. 2003. A cyanobacterial circadian timing mechanism. Annu. Rev. Genet. 37: 513–543. [DOI] [PubMed] [Google Scholar]
- Djordjevic S., Goudreau, P.N., Xu, Q., Stock, A.M., and West, A.H. 1998. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl. Acad. Sci. 95: 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J.C., Loros, J.J., and DeCoursey, P.J. 2004. Chronobiology biological timekeeping. Sinauer Associates, Inc, Sunderland, MA.
- Eldridge A.M., Kang, H.-S., Johnson, E., Gunsalus, R., and Dahlquist, F.W. 2002. Effect of phosphorylation on the interdomain interaction of the response regulator, NarL. Biochemistry 41: 15173–15180. [DOI] [PubMed] [Google Scholar]
- Farrow N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Forman-Kay, J.D., and Kay, L.E. 1994. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain by 15N NMR relaxation. Biochemistry 33: 5984–6003. [DOI] [PubMed] [Google Scholar]
- Gao T., Zhang, X., Xia, Y., Cho, Y., Sacchettini, J.C., Golden, S.S., and Li Wang, A.C. 2005. 1H, 13C and 15N chemical shift assignments of the C-terminal, 133-residue pseudo-receiver domain of circadian input kinase (CikA) in Synechococcus elongatus . J. Biomol. NMR 32: 259. [DOI] [PubMed] [Google Scholar]
- Garces R.G., Wu, N., Gillon, W., and Pai, E.F. 2004. Anabaena circadian clock proteins KaiA and KaiB reveal a potential common binding site to their partner KaiC. EMBO J. 23: 1688–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden S.S.. 2004. Meshing the gears of the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. 101: 13697–13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P., Fabry, B., Guillet, V., Birck, C., Mourey, L., Kahn, D., and Samama, J.-P. 1999. Structural transitions in the FixJ receiver domain. Structure 7: 1517–1526. [DOI] [PubMed] [Google Scholar]
- Grimsley J., Tjalkens, R., Strauch, M., Bird, T., Spiegelman, G., Hostomsky, Z., Whiteley, J., and Hoch, J. 1994. Subunit composition and domain structure of the Spo0A sporulation transcription factor of Bacillus subtilis . J. Biol. Chem. 269: 16977–16982. [PubMed] [Google Scholar]
- Hitomi K., Oyama, T., Han, S., Arvai, A.S., and Getzoff, E.D. 2005. Tetrameric architecture of the circadian clock protein KaiB: A novel interface for intermolecular interactions and its impact on the circadian rhythm. J. Biol. Chem. 280: 19127–19135. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander, C. 1998. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26: 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang P.M., Skrynnikov, N.R., and Kay, L.E. 2001. Domain orientation in β-cyclodextrin-loaded maltose binding protein: Diffusion anisotropy measurements confirm the results of a dipolar coupling study. J. Biomol. NMR 20: 83–88. [DOI] [PubMed] [Google Scholar]
- Im Y.J., Rho, S.-H., Park, C.-M., Yang, S.-S., Kang, J.-G., Lee, J.Y., Song, P.-S., and Eom, S.H. 2002. Crystal structure of a cyanobacterial phytochrome response regulator. Protein Sci. 11: 614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva N.B., Bramlett, M.R., Lindahl, P.A., and Golden, S.S. 2005. LdpA: A component of the circadian clock senses redox state of the cell. EMBO J. 24: 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva N.B., Gao, T., LiWang, A., and Golden, S.S. 2006. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. 103: 17468–17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase R., Imada, K., Hayashi, F., Uzumaki, T., Morishita, M., Onai, K., Furukawa, Y., Namba, K., and Ishiura, M. 2005. Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex. J. Biol. Chem. 280: 43141–43149. [DOI] [PubMed] [Google Scholar]
- Karchin R., Cline, M., Mandel-Gutfreund, Y., and Karplus, K. 2003. Hidden Markov models that use predicted local structure for fold recognition: Alphabets of backbone geometry. Proteins 51: 504–514. [DOI] [PubMed] [Google Scholar]
- Karplus K., Karchin, R., Draper, J., Casper, J., Mandel-Gutfreund, Y., Diekhans, M., and Hughey, R. 2003. Combining local-structure, fold-recognition, and new-fold methods for protein structure prediction. Proteins 53: 491–496. [DOI] [PubMed] [Google Scholar]
- Katayama M., Kondo, T., Xiong, J., and Golden, S.S. 2003. ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. J. Bacteriol. 185: 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D., Volkman, B.F., Luginbuhl, P., Nohaile, M.J., Kustu, S., and Wemmer, D.E. 1999. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature 402: 894–898. [DOI] [PubMed] [Google Scholar]
- Kuszewski J., Qin, J., Gronenborn, A.M., and Clore, G.M. 1995. The impact of direct refinement against 13Cα and 13Cβ chemical shifts on protein structure determination by NMR. J. Magn. Reson. B. 106: 92–96. [DOI] [PubMed] [Google Scholar]
- Kuszewski J., Gronenborn, A.M., and Clore, G.M. 1996. Improving the quality of NMR and crystallographic protein structures by means of a conformational database potential derived from structure databases. Protein Sci. 5: 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuszewski J., Gronenborn, A.M., and Clore, G.M. 1999. Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J. Am. Chem. Soc. 121: 2337–2338. [Google Scholar]
- Lamparter T.. 2004. Evolution of cyanobacterial and plant phytochromes. FEBS Lett. 573: 1–5. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.W. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Laurence J.S., LiWang, A.C., and LiWang, P.J. 1998. Effect of N-terminal truncation and solution conditions on chemokine dimer stability: Nuclear magnetic resonance structural analysis of macrophage inflammatory protein 1-βmutants. Biochemistry 37: 9346–9354. [DOI] [PubMed] [Google Scholar]
- Lewis R.J., Muchová, K., Brannigan, J.A., Barák, I., Leonard, G., and Wilkinson, A.J. 2000. Domain swapping in the sporulation response regulator Spo0A. J. Mol. Biol. 297: 757–770. [DOI] [PubMed] [Google Scholar]
- LiWang A.C., Cao, J.J., Zheng, H., Lu, Z., Peiper, S.C., and LiWang, P.J. 1999a. Dynamics study on the anti-human immunodeficiency virus chemokine viral macrophage-inflammatory protein-II (vmip-II) reveals a fully monomeric protein. Biochemistry 38: 442–453. [DOI] [PubMed] [Google Scholar]
- LiWang A.C., Wang, Z.W., Peiper, S.C., and Liwang, P.J. 1999b. Solution structure of the anti-HIV chemokine vMIP-II. Protein Sci. 8: 2270–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marina A., Waldburger, C.D., and Hendrickson, W.A. 2005. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 24: 4247–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller J. and Elber, R. 2001. Linear optimization and a double statistical filter for protein threading protocols. Proteins 45: 241–261. [DOI] [PubMed] [Google Scholar]
- Miyagishima S.-y., Wolk, C.P., and Osteryoung, K.W. 2005. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 56: 126–143. [DOI] [PubMed] [Google Scholar]
- Muller-Dieckmann H.-J., Grantz, A.A., and Kim, S.-H. 1999. The structure of the signal receiver domain of the Arabidopsis thaliana ethylene receptor ETR1. Structure 7: 1547–1556. [DOI] [PubMed] [Google Scholar]
- Mutsuda M., Michel, K.-P., Zhang, X., Montgomery, B.L., and Golden, S.S. 2003. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J. Biol. Chem. 278: 19102–19110. [DOI] [PubMed] [Google Scholar]
- Nabuurs S.B., Spronk, C.A.E.M., Krieger, E., Maassen, H., Vriend, G., and Vuister, G.W. 2003. Quantitative evaluation of experimental NMR restraints. J. Am. Chem. Soc. 125: 12026–12034. [DOI] [PubMed] [Google Scholar]
- Nabuurs S.B., Spronk, C.A.E.M., Vuister, G.W., and Vriend, G. 2006. Traditional biomolecular structure determination by NMR spectroscopy allows for major errors. PLoS Comput. Biol. 2: 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kita, M., Ito, S., Yamashino, T., and Mizuno, T. 2005. Pseudo-response regulators, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana . Plant Cell Physiol. 46: 686–698. [DOI] [PubMed] [Google Scholar]
- Nowak E., Panjikar, S., Konarev, P., Svergun, D.I., and Tucker, P.A. 2006. The structural basis of signal transduction for the response regulator PrrA from Mycobacterium tuberculosis. J. Biol. Chem. 281: 9659–9666. [DOI] [PubMed] [Google Scholar]
- O'Hara B.P., Norman, R.A., Wan, P.T.C., Roe, S.M., Barrett, T.E., Drew, R.E., and Pearl, L.H. 1999. Crystal structure and induction mechanism of AmiC-AmiR: A ligand-regulated transcription antitermination complex. EMBO J. 18: 5175–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R., Wang, J., Mori, T., Xu, Y., Johnson, C.H., and Egli, M. 2004. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol. Cell 15: 375–388. [DOI] [PubMed] [Google Scholar]
- Salome P.A. and McClung, C.R. 2005. Pseudo-response regulator 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz O., Katayama, M., Williams, S.B., Kondo, T., and Golden, S.S. 2000. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289: 765–768. [DOI] [PubMed] [Google Scholar]
- Schwieters C.D., Kuszewski, J.J., Tjandra, N., and Clore, G.M. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160: 66–74. [DOI] [PubMed] [Google Scholar]
- Schwieters C.D., Kuszewski, J.J., and Marius Clore, G. 2006. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 48: 47–62. [Google Scholar]
- Stock A.M., Robinson, V.L., and Goudreau, P.N. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69: 183–215. [DOI] [PubMed] [Google Scholar]
- Teodorescu O., Galor, T., Pillardy, J., and Elber, R. 2004. Enriching the sequence substitution matrix by structural information. Proteins 54: 41–48. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra N., Wingfield, P., Stahl, S., and Bax, A. 1996. Anisotropic rotational diffusion of perdeuterated HIV protease from 15N NMR relaxation measurements at two magnetic fields. J. Biomol. NMR 8: 273–284. [DOI] [PubMed] [Google Scholar]
- Tomomori C., Tanaka, T., Dutta, R., Park, H., Saha, S.K., Zhu, Y., Ishima, R., Liu, D., Tong, K.I., and Kurokawa, H., et al. 1999. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nature 6: 729–734. [DOI] [PubMed] [Google Scholar]
- Toro-Roman A., Mack, T.R., and Stock, A.M. 2005. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: A symmetric dimer mediated by the α4-β5-α5 face. J. Mol. Biol. 349: 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugarinov V., Muhandiram, R., Ayed, A., and Kay, L.E. 2002. Four-dimensional NMR spectroscopy of a 723-residue protein: Chemical shift assignments and secondary structure of Malate Synthase G. J. Am. Chem. Soc. 124: 10025–10035. [DOI] [PubMed] [Google Scholar]
- Tzeng Y.-L., Zhou, X.Z., and Hoch, J.A. 1998. Phosphorylation of the Spo0B response regulator phosphotransferase of the phosphorelay initiating development in Bacillus subtilis . J. Biol. Chem. 273: 23849–23855. [DOI] [PubMed] [Google Scholar]
- Uzumaki T., Fujita, M., Nakatsu, T., Hayashi, F., Shibata, H., Itoh, N., Kato, H., and Ishiura, M. 2004. Crystal structure of the C-terminal clock-oscillator domain of the cyanobacterial KaiA protein. Nat. Struct. Mol. Biol. 11: 623–631. [DOI] [PubMed] [Google Scholar]
- Vakonakis I. and LiWang, A.C. 2004. Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: Implications for KaiC regulation. Proc. Natl. Acad. Sci. 101: 10925–10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakonakis I., Klewer, D.A., Williams, S.B., Golden, S.S., and LiWang, A.C. 2004a. Structure of the N-terminal domain of the circadian clock-associated histidine kinase SasA. J. Mol. Biol. 342: 9–17. [DOI] [PubMed] [Google Scholar]
- Vakonakis I., Sun, J., Wu, T., Holzenburg, A., Golden, S.S., and LiWang, A.C. 2004b. NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: Implications for the KaiA-KaiC interaction. Proc. Natl. Acad. Sci. 101: 1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varughese K.I., Madhusudan, Zhou, X.Z., Whiteley, J.M., and Hoch, J.A. 1998. Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol. Cell 2: 485–493. [DOI] [PubMed] [Google Scholar]
- Varughese K.I., Tsigelny, I., and Zhao, H. 2006. The crystal structure of beryllofluoride Spo0F in complex with the phosphotransferase Spo0B represents a phosphotransfer pretransition state. J. Bacteriol. 188: 4970–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. and Davis, S.J. 2000. Bacteriophytochromes: New tools for understanding phytochrome signal transduction. Semin. Cell Dev. Biol. 11: 511–521. [DOI] [PubMed] [Google Scholar]
- Volz K. and Matsumura, P. 1991. Crystal structure of Escherichia coli CheY refined at 1.7-Å resolution. J. Biol. Chem. 266: 15511–15519. [DOI] [PubMed] [Google Scholar]
- Vriend G.. 1990. WHAT IF: A molecular modelling and drug design program. J. Mol. Graph. 8: 52–56. [DOI] [PubMed] [Google Scholar]
- Williams S.B., Vakonakis, I., Golden, S.S., and LiWang, A.C. 2002. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: A potential clock input mechanism. Proc. Natl. Acad. Sci. 99: 15357–15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K.. 1986. NMR of proteins and nucleic acids. Wiley, New York.
- Xu Y., Mori, T., Pattanayek, R., Pattanayek, S., Egli, M., and Johnson, C.H. 2004. Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc. Natl. Acad. Sci. 101: 13933–13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K., Saito, M., Oobatake, M., and Kanaya, S. 1995. Characterization of the internal motions of Escherichia coli ribonuclease HI by a combination of 15N-NMR relaxation analysis and molecular dynamics simulation: Examination of dynamic models. Biochemistry 34: 6587–6601. [DOI] [PubMed] [Google Scholar]
- Ye S., Vakonakis, I., Ioerger, T.R., LiWang, A.C., and Sacchettini, J.C. 2004. Crystal structure of circadian clock protein KaiA from Synechococcus elongatus . J. Biol. Chem. 279: 20511–20518. [DOI] [PubMed] [Google Scholar]
- Zapf J., Sen, U., Madhusudan, Hoch, J.A., and Varughese, K.I. 2000. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure 8: 851–862. [DOI] [PubMed] [Google Scholar]
- Zhang J.H., Xiao, G., Gunsalus, R.P., and Hubbell, W.L. 2003. Phosphorylation triggers domain separation in the DNA binding response regulator NarL. Biochemistry 42: 2552–2559. [DOI] [PubMed] [Google Scholar]
- Zhang X., Dong, G., and Golden, S.S. 2006. The pseudo-receiver domain of CikA regulates the cyanobacterial circadian input pathway. Mol. Microbiol. 60: 658–668. [DOI] [PubMed] [Google Scholar]