Abstract
The pro-peptide of human nerve growth factor (NGF) functions as an intramolecular chaperone during oxidative renaturation of proNGF in vitro and interacts intramolecularly with the mature part of native proNGF. Here, we analyzed the structure formation and stability of the pro-peptide in the context of proNGF and its intramolecular interaction with the native mature part. Folding and unfolding of the NGF-coupled pro-peptide, as analyzed by fluorescence, were biphasic reactions with both phases depending on the interaction with the mature part. This interaction was characterized by an overall stability of ΔG = 20.9 kJ/mol that was subdivided into two reactions, native ↔ intermediate state (14.8 kJ/mol) and intermediate ↔ unfolded state (6.1 kJ/mol). An additional very fast unfolding reaction was observed using circular dichroism (CD), indicating the presence of at least two kinetically populated intermediates in the unfolding of proNGF. The part of the pro-peptide involved in the intramolecular association with mature NGF comprised the peptide Trp−83-Ala−63 as determined by H/D exchange experiments. Spectroscopic analyses revealed that on the NGF side, a surface area around Trp21 interacted with the pro-peptide. Trp21 also participates in binding to TrkA and p75 receptors. These overlapping binding sites of the pro-peptide and the NGF receptors might explain the previously observed lower affinity of proNGF to its receptors as compared to NGF.
Keywords: pro-peptide, proNGF, NGF, folding, receptor binding, TrkA, p75, Trp21
Nerve growth factor (NGF), a member of the neurotrophin family, as the neurotrophins 3–7 and brain-derived neurotrophic factor (BDNF) promotes growth, survival, and differentiation as well as apoptosis of neuronal cells (Barde 1990; Ernfors et al. 1990; Hohn et al. 1990; Bradshaw et al. 1993; Casaccia-Bonnefil et al. 1996; Frade et al. 1996). NGF contains a typical structure motif, the cystine knot: Two disulfide bridges form a loop of 14 amino acids, which is penetrated by a third disulfide bridge. In vivo, NGF is expressed as a pre-pro-protein. The N-terminal signal sequence mediates translocation into the endoplasmic reticulum. Secretion of the growth factor occurs in a cell-type-specific manner as a mixture of proNGF and mature NGF, in which the pro-peptide with 103 amino acids is removed by furin or related pro-protein convertases (Seidah et al. 1996; Hasan et al. 2003; Fahnestock et al. 2004). Both mature NGF and proNGF exist in their active forms as homodimers (Bothwell and Shooter 1977; Rattenholl et al. 2001a).
The pro-peptide acts in vitro as an intramolecular chaperone by facilitating and stimulating oxidative folding of the mature part (Rattenholl et al. 2001a,b). Folding-promoting functions of pro-peptides have been reported for several proteins, which are expressed as (pre-)pro-proteins (Shinde and Inouye 2000). Prominent examples are bovine pancreatic trypsin inhibitor (BPTI), guanylyl cyclase activating peptide (GCAP), or the macrophage inhibitory cytokine-1 (MIC-1), and proteases such as α-lytic protease, subtilisin, and carboxypeptidase Y (Ikemura et al. 1987; Winther and Sorensen 1991; Weissman and Kim 1992; Shinde et al. 1993; Hidaka et al. 1998; Fairlie et al. 2001). The pro-peptides of the α-conotoxins, toxins of marine snails, are not directly involved in the folding of the mature part, but can facilitate the structure formation by interacting with protein disulfide isomerase in the endoplasmic reticulum (Buczek et al. 2004). It has also been reported that pro-peptides guide the assembly of subunits into active protein complexes as in the case of the von Willebrand factor or caspase-3 (Wise et al. 1988; Feeney and Clark 2005). The pro-peptides of TGF-β and very likely also bone morphogenetic proteins retard the biological activities of the mature growth factors (Böttinger et al. 1996; Degnin et al. 2004; Hillger et al. 2005). While for some pro-peptides their biological activities are displayed even when they are not covalently linked to their mature parts (Gray and Mason 1990), in the case of NGF, the pro-peptide has to be covalently linked to the mature part to promote oxidative structure formation (Rattenholl et al. 2001a). Besides aiding in structure formation, the pro-peptide of proNGF confers a pro-apoptotic activity to the growth factor and thus responses that are completely opposite to the mature growth factor (Lee et al. 2001). Pro-apoptotic responses of proNGF are elicited by binding to a pro-form specific receptor, sortilin (Nykjaer et al. 2004).
Besides their biological relevance, little is known about the structures and stabilities of the various pro-peptides. In the case of the protease subtilisin, the pro-peptide loses the structure in the absence of the mature part (Ruvinov et al. 1997; Buevich et al. 2001). A similar observation was made for the pro-peptide of carboxypeptidase Y: Under conditions where the protein is functional, the pro-peptide is only partially folded. The pro-peptide contains secondary structural elements but a very low content of defined tertiary structure and exists in a molten globule state (Sorenson et al. 1993).
Recently, we demonstrated that the isolated pro-peptide of NGF is a monomeric protein with a distinct secondary structural content. In the isolated form the structure was significantly less stable than in its NGF-coupled form, suggesting an interaction of the pro-peptide and the mature part in native proNGF (Kliemannel et al. 2004). Here, we analyzed in detail the intramolecular interaction of the pro-peptide with the mature part of proNGF and the influence of the mature part on the structure formation process of the covalently linked pro-peptide.
Results
Thermodynamic and structural properties of the NGF-coupled pro-peptide
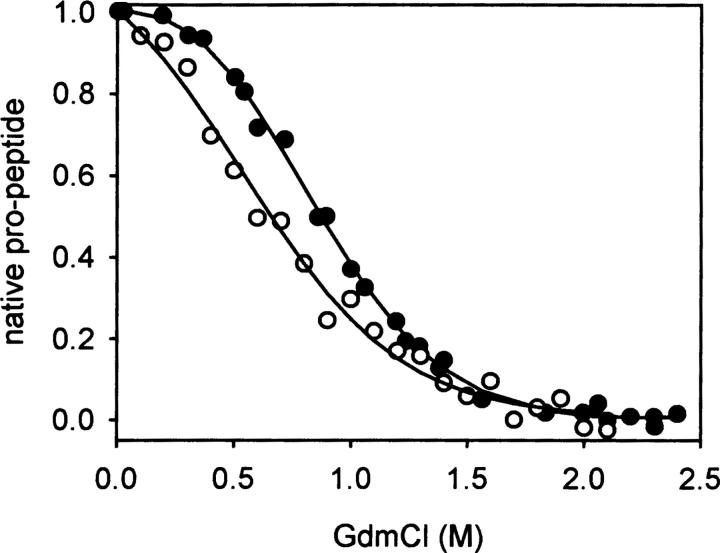
We have shown previously that the overall denaturation process of proNGF is biphasic with the first and second transition representing unfolding of the pro-peptide and NGF part, respectively (Kliemannel et al. 2004). Furthermore, we observed that the subtraction far-UV circular dichroism (CD) spectrum of proNGF and mature NGF was identical to that recorded for the isolated pro-peptide, indicating that the pro-peptide contains the same amount of secondary structure elements in the isolated form and in the NGF-coupled pro-peptide (“NGF-coupled” refers to the pro-peptide in the context of proNGF) (Rattenholl et al. 2001a; Kliemannel et al. 2004). This assumption could be confirmed by denaturation and renaturation transitions monitored by far-UV CD spectroscopy. It should be emphasized that the terms denaturation and renaturation as well as unfolding and folding refer in this work to the pro-peptide moiety, since at the denaturant concentrations used here the mature part retains its native structure (Kliemannel et al. 2004). The observed differences in ellipticities between native and unfolded pro-peptide were independent of whether the isolated pro-peptide or that coupled to the mature part was analyzed (Kliemannel et al. 2004). Equilibrium denaturation studies revealed a non-cooperative transition of the isolated pro-peptide, indicating that tertiary contacts are not involved in the stabilization of the pro-peptide. In contrast, the NGF-coupled pro-peptide showed a cooperative transition when unfolding was monitored by fluorescence measurements (Fig. 1).
Figure 1.
Equilibrium transitions of the NGF-coupled pro-peptide and the isolated pro-peptide. The proteins were incubated in 50 mM Na-phosphate, pH 7.0 at various GdmCl concentrations for 16 h at 20°C. Fluorescence signals of the NGF-coupled pro-peptide (•, 20 μg/mL, excitation: 280 nm, emission: 325 nm) and the isolated pro-peptide (○, 50 μg/mL, excitation: 250 nm, emission: 345 nm) were recorded.
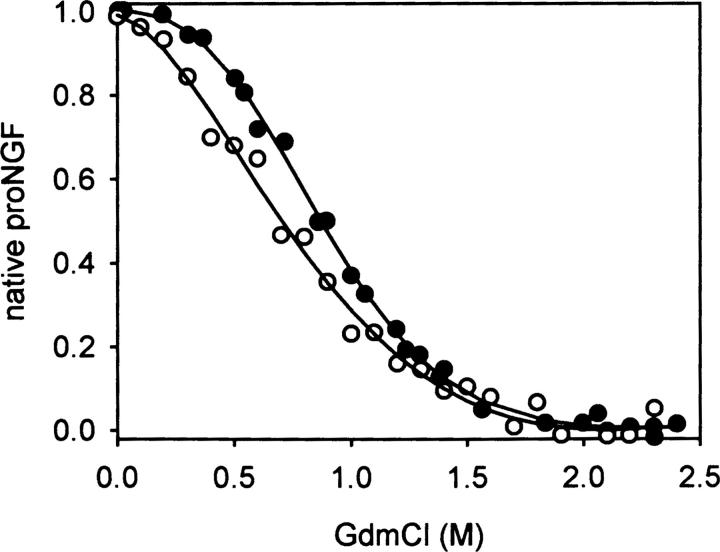
Denaturation and renaturation under equilibrium conditions of the NGF-coupled pro-peptide were reversible (Kliemannel et al. 2004). The transition midpoints occurred at a low guanidinium hydrochloride (GdmCl) concentration of 0.9 M, pointing to a low overall stabilization of the pro-peptide moiety. For a more comprehensive analysis of the unfolding reaction, a comparison of the transitions monitored by far-UV CD spectroscopy and fluorescence was carried out. Changes in ellipticities in the far-UV range reflect changes in secondary structure elements that are expected to occur during unfolding. Changes of the fluorescence represent alterations of solvent accessibilities of tryptophan residues and thus correspond to differences in tertiary contacts. Unexpectedly, the unfolding curves obtained by fluorescence and CD measurements did not coincide (Fig. 2). This result indicates that the loss of secondary structures and tertiary contacts are not coupled as would be expected. The transition midpoint measured by far-UV CD spectroscopy was at lower GdmCl concentrations than that measured by fluorescence. This unexpected fact predicts the existence of an intermediate where secondary structure is lost prior to the loss of specific tertiary contacts observed by tryptophan fluorescence.
Figure 2.
Comparison of the transitions of NGF-coupled pro-peptide measured by fluorescence (•) and far-UV CD spectroscopy (○). The fluorescence data were obtained as described in Figure 1. CD-signals were monitored at 220 nm.
On the basis of the obtained results, the intermediate could be characterized as a species with partly or fully unfolded secondary structure, though a tryptophan residue would still engage in tertiary interaction. In principle, the tryptophan residue(s) could either be localized in the mature part that contacts the pro-peptide or in the pro-peptide itself. Since a chimeric fusion protein consisting of NGF linked to the pro-peptide moiety of a related neurotrophin, NT-3, which lacks tryptophans, exhibited a change in tryptophan fluorescence upon unfolding, as well (A. Hauburger, M. Kliemannel, and E. Schwarz, in prep.), very likely part of the fluorescence signal is caused by a tryptophan residue in mature NGF.
Identification of pro-peptide regions involved in tertiary contacts
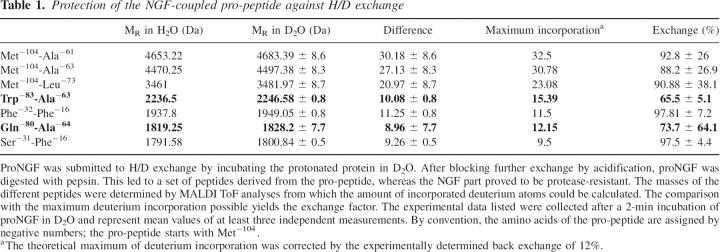
In order to identify those regions within the pro-peptide that could be involved in contacts with the mature part, mass spectrometry-coupled H/D exchange was employed. This method allows the identification of segments that are protected against H/D exchange due to their engagement in secondary and/or tertiary structure (Zhang and Smith 1993). For the analysis of the exchange of amide protons in the pro-peptide of proNGF, the protein was incubated in D2O. The exchange reaction was stopped by acidification to minimize a back exchange during the subsequent digestion with pepsin (Supplemental Fig. S1) and preparation for mass analysis (Bai et al. 1993). Analysis of the digestion products showed that the mature part of proNGF is completely intact under the applied experimental conditions. In contrast, defined proteolysis products were obtained from the pro-peptide moiety. The fragments were aligned with the program FindPept (http://www.expasy.ch/tools/findpept.html). The masses of the fragments were analyzed by MALDI-TOF, and exchange rates were corrected for the back exchange determined by control peptides (Table 1).
Table 1.
Protection of the NGF-coupled pro-peptide against H/D exchange
H/D exchange was impaired in peptides Trp−83-Ala−63 and Gln−80-Ala−64. To determine if the observed amide proton protection would result from hydrogen bonds present in the isolated pro-peptide as well, the same experiment was repeated with the isolated pro-peptide. Here, no defined fragments were obtained upon proteolysis. Thus, in contrast to its isolated form, the pro-peptide in proNGF is partially protease-protected when complexed with the mature part. Based on the H/D exchange data, it is very likely that a region of the pro-peptide containing the peptides Trp−83-Ala−63 and Gln−80-Ala−64 is involved in an interaction with the mature part itself or forms a structure that is stabilized by the interaction of the pro-peptide with the mature part.
Kinetic analyses of the pro-peptide unfolding and refolding in proNGF
Since the isolated pro-peptide did not alter its tryptophan fluorescence upon denaturation, the change of tryptophan fluorescence of the NGF-coupled pro-peptide should reflect specific tertiary contacts between the pro-peptide and the mature part in proNGF. Folding and unfolding kinetics of the NGF-coupled pro-peptide were analyzed by fluorescence spectroscopy. Changes in spectroscopic signals obtained in a range from 0–2 M GdmCl originate solely from structural alterations of the pro-peptide, which has been shown previously to possess only a marginal stability (Fig. 1), while the mature part is stable up to 3 M GdmCl (Timm and Neet 1992; De Young et al. 1996; Kliemannel et al. 2004). Folding of the NGF-coupled pro-peptide exhibited a monophasic reaction (Fig. 3A). Since the pro-peptide contains nine prolyl residues, peptidyl prolyl cis–trans isomerization could be rate-limiting. However, the folding was neither accelerated by the addition of various peptidyl prolyl cis–trans isomerases nor in double-jump experiments (data not shown), indicating that peptidyl prolyl isomerization(s) may not to be a rate-limiting factor.
Figure 3.
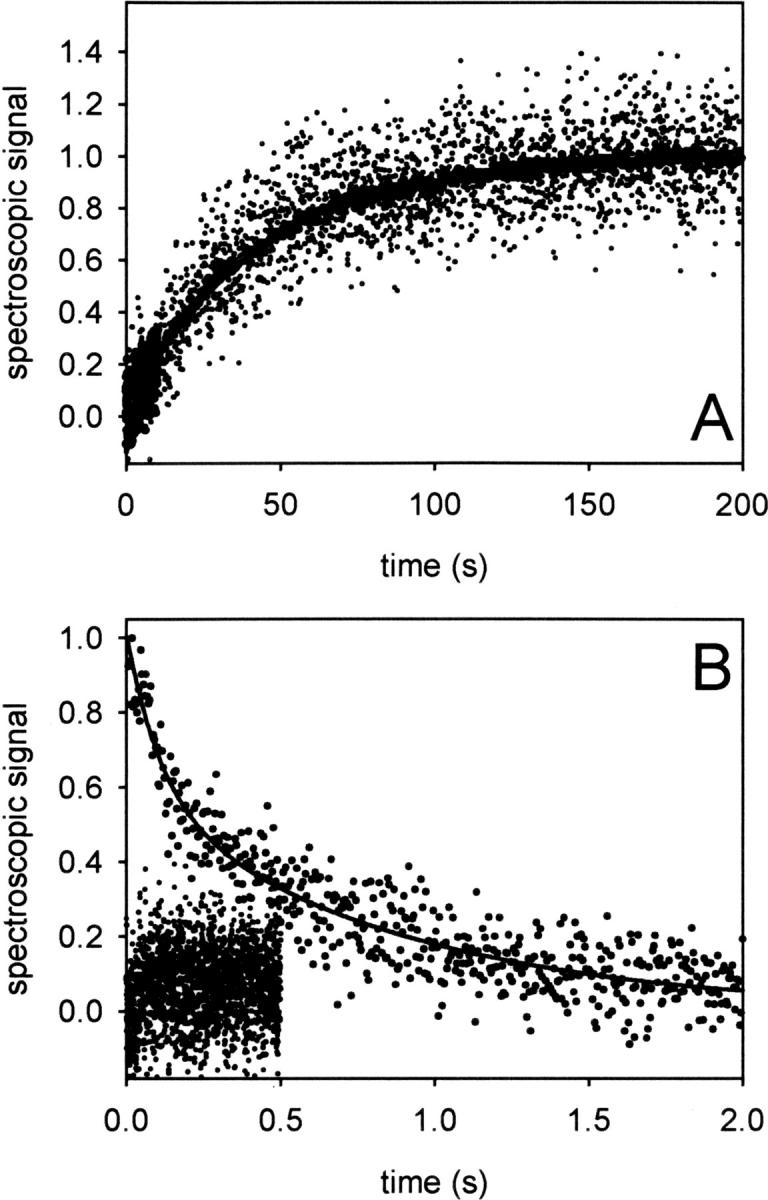
Kinetics of folding and unfolding of the NGF-coupled pro-peptide. (A) Folding of the NGF-coupled pro-peptide. ProNGF was denatured in 2 M GdmCl for at least 1 h at 20°C. Subsequently, refolding was initiated by a 1:11 dilution in 50 mM Na-phosphate, pH 7. Kinetics were monitored by stopped flow fluorescence (bold line) with an emission cut-off filter of 325 nm and stopped flow CD at 220 nm (dots). The data could be fitted to a single phase with k = 0.02 sec−1. (B) Unfolding of the NGF-coupled pro-peptide. Unfolding was started by a 1:11 dilution of proNGF in 50 mM Na-phosphate, pH 7, 2 M GdmCl at 20°C. Fluorescence and CD signals were measured as in A. The fluorescence data exhibited a biphasic reaction with k1 = 3.1 ± 0.4 sec−1 and k2 = 0.76 ± 0.08 sec−1 (n = 6) (solid line). The lower trace represents the final CD amplitude.
The same refolding kinetics as determined by fluorescence were observed by far-UV CD spectroscopy at 220 nm (Fig. 3A). The monophasic folding kinetics were surprising, because the equilibrium transitions clearly indicated the presence of an intermediate. Since the observed folding phase showed the full fluorescence amplitude, no fast reaction prior to the observed structure formation can be assumed; a fast phase following the observed folding reaction, however, would not be detectable. The GdmCl-dependent fluorescence amplitude of the folding phase fits well to the equilibrium transition measured by fluorescence (data not shown), indicating that the kinetic and equilibrium analyses describe the same process.
Denaturation kinetics of the NGF-coupled pro-peptide were more complex than refolding kinetics. When denaturation at 1.75 M GdmCl was monitored by fluorescence spectroscopy, a biphasic denaturation process was observed (Fig. 3B). Recording the unfolding by stopped flow CD measurements was not possible at this GdmCl concentration, since the process was too fast to be detectable (Fig. 3B). Thus, upon denaturation, secondary structure is lost first, followed by the disruption of tertiary contacts. This deduced order of reactions is unusual, but matches the data obtained from unfolding equilibria, which also pointed to a first loss of secondary structure(s) before disruption of tertiary contacts (Fig. 2).
In order to analyze whether the biphasic denaturation observed by fluorescence would reflect parallel or serial unfolding reactions with an intermediate accumulating, we performed double-jump experiments. In these experiments, the NGF-coupled pro-peptide was denatured for various times. Subsequently, the protein was transferred to native conditions, and refolding was monitored by fluorescence spectroscopy. When proNGF was denatured for 25 sec in 1.75 M GdmCl, monophasic refolding kinetics were observed. In contrast, upon shorter denaturation times, a biphasic refolding reaction was recorded, with the fast phase ∼20 times faster than the overall process (Fig. 4A). These data clearly indicate that an intermediate (I) is populated during denaturation. Together with the very fast CD kinetics, these results can be explained by a folding/unfolding scheme N ↔ I1 ↔ I2 ↔ U. In this scheme, the reaction N ↔ I1 is too fast to be quantified by stopped flow CD and is fluorescenctly silent, whereas the reactions I1 ↔ I2 and I2 ↔ U represent the fast and slow phases, respectively, observed by fluorescence.
Figure 4.
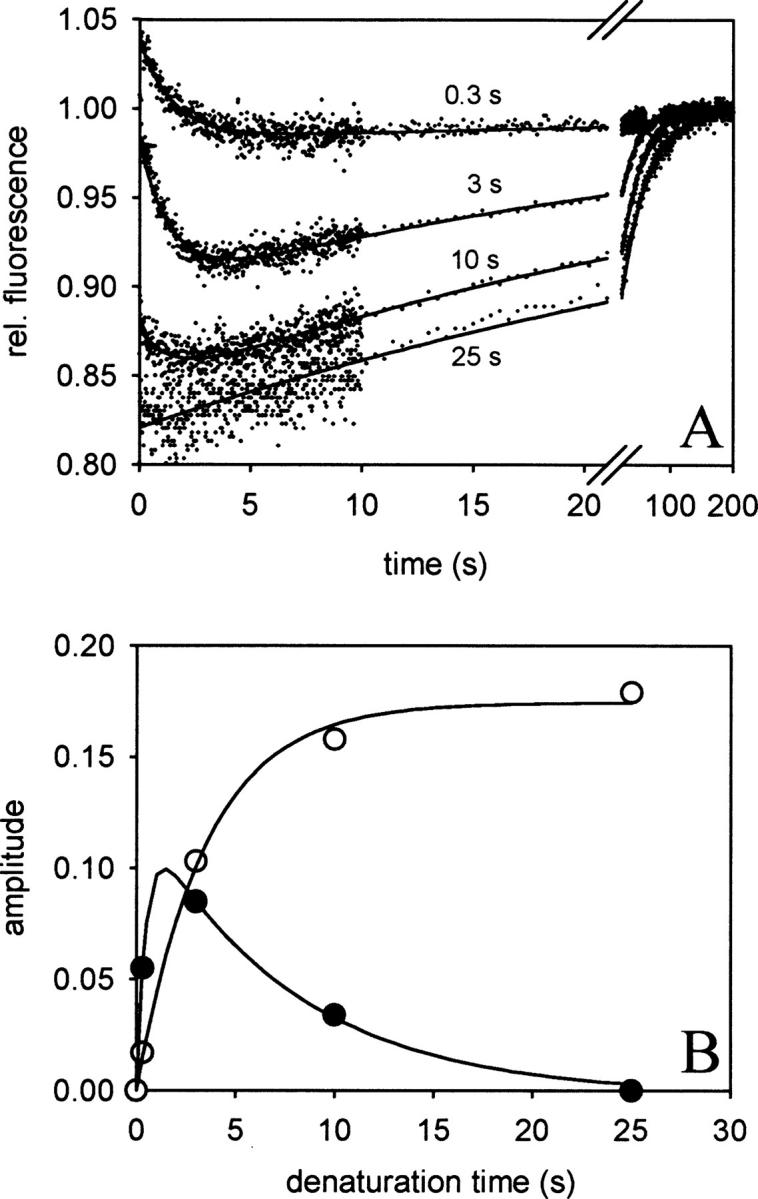
Time-dependent accumulation of a folding intermediate upon unfolding of the NGF-coupled pro-peptide. ProNGF was denatured in 1.7 M GdmCl for 0.3, 3, 10, and 25 sec. Subsequently, refolding was initiated by a 1:11 dilution in 50 mM Na-phosphate, pH 7, and the change in fluorescence was analyzed (280 nm excitation, 325 nm cut off for emission). (A) Refolding kinetics after different times of denaturation. The traces could be described by a biphasic reaction with rate constants of k1 = 0.71 sec−1 and k2 = 0.03 sec−1. (B) Analyses of the amplitudes of the folding reactions. The amplitudes of the fast (•) and slow (○) phase correspond to the population of a folding intermediate and the denatured state, respectively. The time-dependent distribution of both species could be fitted by a serial first order reaction with k1 = 2.2 sec−1 and k2 = 0.3 sec−1, values similar to those of the corresponding unfolding kinetic (see Fig. 3B).
Upon denaturation of the native protein at 1.75 M GdmCl, I2 is accumulating during the first 1 sec of denaturation. Subsequently, I2 unfolds to the fully denatured state. The time-dependent population of I2 can be visualized directly by varying the time of denaturation in double-jump experiments (Fig. 4A). In the subsequent refolding reactions, the amplitudes of the fast and slow kinetics correspond to the accumulation of I2 and U, respectively. Taking these amplitudes, the kinetics of accumulation and disappearance of I2 and the accumulation of U could be analyzed (Fig. 4B). The resulting rate constants of the two processes fit well to the constants obtained from the biphasic denaturation kinetics measured by stopped flow fluorescence (Fig. 3B).
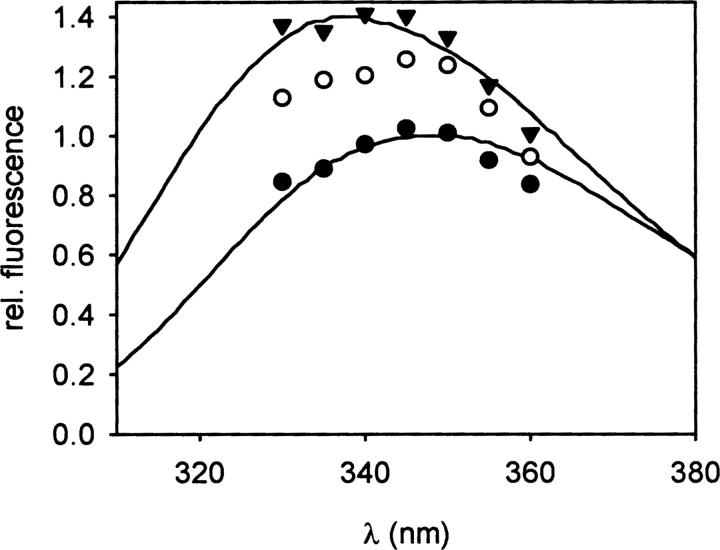
A fluorescence spectrum of the intermediate I2 could not be recorded during the unfolding kinetics because the reaction I2 → U was too fast. Therefore, denaturation kinetics at 1.8 M GdmCl were measured at different emission wavelengths between 330 and 360 nm. From these kinetic data, the specific fluorescence of the native, intermediate, and denatured state could be calculated and the spectra reconstituted (Fig. 5). The data revealed a wavelength shift of the maximum emission from 340 nm (native state) to 345–350 nm (denatured state) already in the intermediate I2. Thus, the solvent accessibility of the tryptophan that is responsible for the change of the fluorescence signal must be similar in I2 and U. It should be noted that the fluorescence intensities of the observed spectra are valid only under the given conditions (1.8 M GdmCl). Since the fluorescence intensity of the spectrum of the intermediate I2 decreases with increasing GdmCl concentrations while that of the native state does not (data not shown), the intermediate possesses a lower fluorescence than the native state at denaturing conditions but a slightly higher fluorescence in the absence of GdmCl. The fluorescence of isolated tryptophan molecules decreases with increasing GdmCl concentrations in a range of 0–3 M (Schmid 1997). Thus, the decrease in intrinsic fluorescence of the intermediate I2 with increasing GdmCl concentrations indicates the solvent exposure of a tryptophan residue in the intermediate structure. This result is confirmed by the red-shift of the fluorescence maximum of the corresponding spectrum (Fig. 5). Similar fluorescence characteristics of folding intermediates have been observed for other proteins such as antibody fragments (Lilie et al. 1995).
Figure 5.
Fluorescence spectra of proNGF. The specific fluorescence of the native (▾), intermediate (○), and denatured state (•) of proNGF was calculated from biphasic denaturation kinetics, measured at different wavelengths. For comparison, spectra of native (upper spectrum) and denatured (2 M GdmCl, lower spectrum) proNGF are shown (Kliemannel et al. 2004).
Thermodynamics of pro-peptide folding
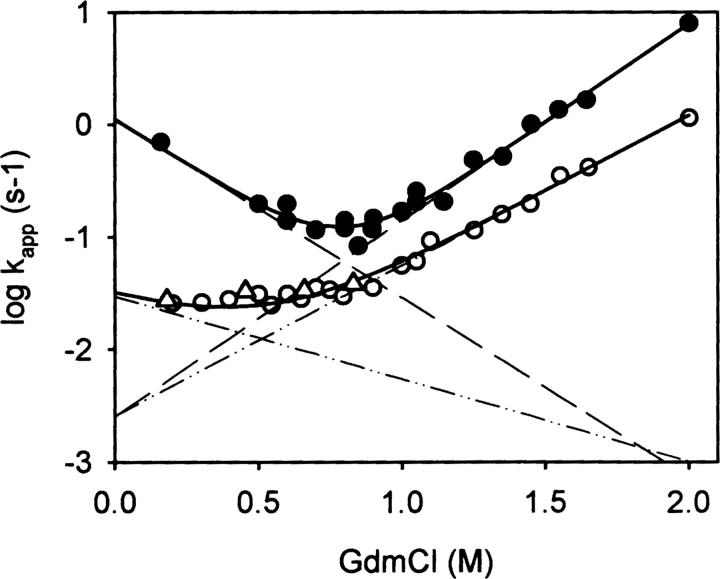
In order to understand the kinetic and energetic relationship of the different states of the pro-peptide moiety folding, the microscopic rate constants of folding and unfolding were determined. To this end, the GdmCl-dependent folding and unfolding kinetics were analyzed by fluorescence spectroscopy. Whereas the change in tryptophan fluorescence upon denaturation permitted the observation of both the transient accumulation of the intermediate and the formation of the denatured state, the same method revealed just the reaction U → I upon refolding. Folding of the intermediate to the native state was measured separately by double-jump experiments, in which native proNGF was submitted to 1.8 M GdmCl for only 1 sec (or 1.2 M GdmCl for 4 sec) to populate the intermediate I2. In the following refolding reaction, the reaction I2 → I1/N could be quantified. As mentioned before, the states I1 and N could not be discerned by fluorescence. Therefore, in the following section, the species I1 and N are not distinguished. The results of folding/unfolding are depicted in Figure 6. The global fit of the data yielded microscopic rates of folding and unfolding of N/I1, I2, and U under native conditions as well as the thermodynamic stability of the native and intermediate state I2 (Fig. 6). The overall stability of N/I1 ↔ U of the NGF-coupled pro-peptide is 20.9 kJ/mol, which is subdivided in the two transitions N/I1 ↔ I2 (14.8 kJ/mol) and I2 ↔ U (6.1 kJ/mol). The microscopic unfolding rate of both N/I1 and I2 was identical (ku = 0.0025 sec−1); the activation energy was 86.3 kJ/mol. In contrast, the activation energy of folding showed a difference of 8.3 kJ/mol for folding of U and I2, respectively (Supplemental Fig. S2).
Figure 6.
Unfolding/refolding kinetics of the NGF-coupled pro-peptide. Unfolding and refolding at different GdmCl concentrations of the NGF-coupled pro-peptide were measured as described in Figure 3. Unfolding and refolding were monitored by fluorescence. The transitions of N/I1 ↔ I2 (•) and I2 ↔ U (○) are depicted. CD measurements were used to observe refolding (△). (Solid lines) Fit of the complete data set, (dashed lines) GdmCl dependence of the microscopic rate constants of folding and unfolding.
It should be stressed that the Chevron plot does not describe the complete folding/unfolding process of the pro-peptide of NGF, since unfolding measured by far-UV CD spectroscopy was too fast to be quantified. Thus, only changes in the environment of the tryptophans could be monitored; changes on the secondary structural level are not considered in the interpretation of the Chevron plot.
Discussion
Despite the increasing knowledge about the biological function(s) of pro-peptides, little information is available about the structural properties of pro-peptides and how they exert their roles on a structural basis. Only for some serine and cysteine proteases has the X-ray structure of the pro-form been solved (Gallagher et al. 1995; Podobnik et al. 1997; Jain et al. 1998; LaLonde et al. 1999) and a molecular model predicted how the pro-peptide serves as an intramolecular chaperone (Gallagher et al. 1995). Here, we analyzed structure formation, stability, and intramolecular interaction of the NGF pro-peptide in the context of the mature part. Unexpectedly, kinetic and thermodynamic analyses revealed a loss of at least a major part of the secondary structures before disruption of tertiary contacts. These tertiary contacts reflect an intramolecular interaction of the pro-peptide with the mature part.
It has been shown previously that the pro-peptide acts as an intramolecular chaperone during oxidative folding of proNGF (Rattenholl et al. 2001a,b). This function suggests an interaction of the pro-peptide with folding intermediates of the NGF part based primarily on the properties of the solvent-exposed surface of these intermediates, such as hydrophobicity, rather than well defined structural features. The pro-peptide needs to be covalently attached to NGF in order to exhibit this folding-enhancing function, as the addition of the isolated pro-peptide to folding NGF molecules in trans does not promote folding. This finding indicates a very weak interaction of the pro-peptide with folding intermediates of the mature part (Rattenholl et al. 2001a). In contrast, the intramolecular interaction of the pro-peptide with native NGF is surprisingly stable: It is characterized by an overall energy of ΔG = 20.9 kJ/mol. Furthermore, one of the transitions reflecting this interaction is highly cooperative (cf. fast phases in Fig. 6), indicating a well defined structural interface between the pro-peptide and NGF. Thus, the intramolecular interaction of the pro-peptide and NGF seems to be quite different in the native state and during folding regarding both structural and thermodynamic parameters.
The experimental data suggest that that the interaction between NGF and its pro-peptide is mediated by a surface area of the mature NGF comprising a tryptophan residue and the peptide Trp−83-Ala−63 of the pro-peptide. Unfortunately, currently no detailed structural data for the pro-peptide are available. Even the CD spectrum of the NGF-coupled pro-peptide hardly allows a prediction of the overall secondary structural content (Kliemannel et al. 2004). A sequence analysis of the pro-peptide according to the hydrophobic moment method of Eisenberg (Eisenberg et al. 1984) shows that the peptide Trp−83-Ala−63 possesses the highest hydrophobic moment of the whole pro-peptide. In a helical wheel representation, this peptide is highly amphiphilic with a perfect segregation of hydrophobic and polar/charged amino acids (not shown). However, without structural data of the pro-peptide at the atomic level, a structural interpretation of the pro-peptide region involved in the intramolecular interaction with NGF remains speculative.
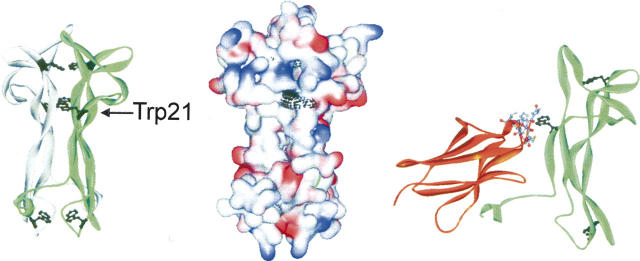
As mentioned above, the intramolecular association of NGF with its pro-peptide is mainly of a hydrophobic nature and involves tryptophan residues. The fluorescence change obtained upon unfolding of the NGF-coupled pro-peptide is related to the environmental change of one or more tryptophans. Since a chimeric construct of NGF and the related heterologous pro-peptide of NT-3, which lacks tryptophans, also showed a change in tryptophan fluorescence upon unfolding (A. Hauburger, M. Kliemannel, and E. Schwarz, in prep.), at least part of the fluorescence signal must originate from an altered solvent accessibility of tryptophans of the NGF part. The NGF monomer contains three tryptophans, two of which are located in the dimer interface and thus are buried in the hydrophobic core. These tryptophan residues would presumably not change their spectroscopic properties upon unfolding of the pro-peptide. The third tryptophan of NGF, Trp21, has been identified as the most solvent-exposed and reactive tryptophan residue that is readily modified by N-bromosuccinimide (Cohen et al. 1980). Solvent exposure of this residue has been confirmed by X-ray structure analysis (McDonald et al. 1991; Fig. 7). Our fluorescence data suggest that the pro-peptide shields Trp21 of NGF from solvent since unfolding of the NGF-coupled pro-peptide alters the solvent accessibility of Trp21 of mature NGF. Assuming that the presence of the pro-peptide does not affect the structure of the mature part (especially the dimer interface where the other tryptophans are located), the spectroscopic properties of proNGF would indicate a direct interaction of the pro-peptide with Trp21 of mature NGF.
Figure 7.
Structure of NGF and its complex with the extracellular domain 5 of the TrkA receptor (PDB file 1www.pdb [Wiesmann et al. 1999]). The panels show a scheme of dimeric NGF with the Trp residues marked (black, left), the surface of dimeric NGF with the surface of Trp residues marked (black, middle), and the complex of a monomeric subunit of NGF with TrkA (right), where Trp21 of NGF and the surrounding residues of domain 5 of TrkA (8 Å distance) are shown in detail.
Trp21 of NGF is also part of the interface of the NGF-receptor complex in both the TrkA and p75 receptors (Cohen et al. 1980; Drinkwater et al. 1991; Wiesmann et al. 1999; He and Garcia 2004). In the case of TrkA, for example, seven amino acids of the receptor are <8 Å apart from Trp21 of NGF (Fig. 7). Thus, from the structural point of view it seems that the pro-peptide competes for the binding site of the mature part to both the TrkA and p75 receptors. This should result in a substantially decreased affinity of proNGF to its receptors compared with mature NGF, which indeed has been observed experimentally (Nykjaer et al. 2004). The spectroscopic and thermodynamic analyses of the NGF-coupled pro-peptide showed that Trp21 of NGF, which is buried in native proNGF, becomes solvent-accessible in the reaction N/I1 → I2 as demonstrated by the fluorescence spectrum of the intermediate I2 (Fig. 5). This reaction and thus the loss of interaction of the pro-peptide with NGF via a region involving Trp21 require an energy input of 14.8 kJ/mol under the given buffer conditions. Assuming a simple model of competition, this energy must be provided by the binding of the receptors to proNGF in order to form a proNGF–receptor complex. Another possibility would be a partial overlap of the binding sites for the pro-peptide and the receptors. In this case it would be conceivable that receptor binding may occur without displacing the pro-peptide. In this scheme the receptor affinity to proNGF would be decreased as well compared with mature NGF. However, depending on the size and the overlapping part of the contact sites for receptor and pro-peptide, the loss of energy in receptor binding due to the presence of the pro-peptide may be smaller than in the displacing model. Although a competition of the pro-peptide with NGF for binding to the receptors TrkA or p75 appears plausible, the precise mode of proNGF receptor interaction remains to be deduced by structural analysis at atomic resolution of proNGF and its receptor complex.
Materials and methods
Preparation of recombinant human proNGF and the isolated pro-peptide
All recombinant constructs were cloned in the vector pET11a. Escherichia coli BL21(DE3) was employed as a bacterial host. Recombinant production of proNGF resulted in formation of inclusion bodies. Processing of inclusion bodies of proNGF and protein renaturation was performed as described previously (Rattenholl et al. 2001a). The isolated pro-peptide of NGF was obtained in soluble form. The fully folded protein was obtained as published previously (Kliemannel et al. 2004).
Fluorescence spectroscopy
Fluorescence measurements were carried out at a FluoroMax-2 (Jobin-Yvon). Slit widths for both excitation and emission wavelengths were 5 nm. Experiments were performed in 50 mM Na-phosphate, pH 7.0, and 1 mM EDTA at 20°C. Equilibrium transitions of the NGF-coupled pro-peptide (10 μg/mL) were monitored at an excitation wavelength of 280 nm and 325 nm emission. Fluorescence of the isolated pro-peptide (50 μg/mL) was analyzed using an excitation of phenylalanine at 250 nm and after an energy transfer to tryptophan an emission at 345 nm (Kliemannel et al. 2004).
Fluorescence kinetics were carried out either on a FluoroMax 2 using conditions mentioned above or by stopped flow experiments on a SX.18MV spectrometer (Applied Photophysics). In the latter case, fluorescence was detected using a cut-off filter of 320 nm (cp = 0.4 mg/mL) or by single wavelength detection (cp = 1.5 mg/mL). Fluorescence spectra from denaturation kinetics at different emission wavelengths were reconstituted by calculating the specific fluorescence of the native, intermediate, and denatured state using an equation for a serial first-order reaction.
Circular dichroism (CD) spectroscopy
Far UV-CD spectra were recorded on a Jasco J710 spectropolarimeter. Equilibrium transitions of the NGF-coupled pro-peptide (100 μg/mL) were recorded at 220 nm in 50 mM Na-phosphate, pH 7.0 at 20°C in a 1-cm cuvette. Rapid structural changes were followed at 220 nm by stopped flow experiments on a π-star spectropolarimeter (Applied Photophysics). The final protein concentrations of both folding and unfolding reactions were 1 mg/mL.
H/D exchange-coupled MALDI-TOF mass spectrometry
Determination of the deuterium incorporation by exchange of the amide protons of peptide bonds was carried out by MALDI-TOF mass spectrometry using a BRUKER Reflex II mass spectrometer (BRUKER Daltonik GmbH). Experimental details are described in the Supplemental Material. In short, proNGF was incubated in D2O for 2 min. Subsequently, it was digested with pepsin, and the resulting fragments were analyzed by MALDI-ToF. For the alignments of the obtained proteolytic fragments, the program FindPept (http://www.expasy.ch/tools/findpept.html) was used.
Acknowledgments
We thank Renate Nitsch for technical assistance and Angelika Schierhorn for help in mass spectroscopy. This work was supported by the Deutsche Forschungsgemeinschaft (grant SCHW 375/4 to E.S. and R.R.).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Elisabeth Schwarz, Martin-Luther-Universität Halle-Wittenberg, Institut für Biotechnologie, Kurt-Mothes-Str. 3, D-06120 Halle, Germany; e-mail: Elisabeth.Schwarz@biochemtech.uni-halle.de; fax: + 49 345 55 27 013.
Abbreviations: CD, circular dichroism; GdmCl, guanidinium hydrochloride; NGF, human nerve growth factor; proNGF, pro-form of NGF containing the 102-amino-acid-comprising pro-peptide and a start methionine at the N terminus.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062376207.
References
- Bai Y., Milne, J.S., Mayne, L., and Englander, S.W. 1993. Primary structure effects on peptide group hydrogen exchange. Proteins 17: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barde Y.A.. 1990. The nerve growth factor family. Prog. Growth Factor Res. 2: 237–248. [DOI] [PubMed] [Google Scholar]
- Bothwell M.A. and Shooter, E.M. 1977. Dissociation equilibrium constant of β nerve growth factor. J. Biol. Chem. 252: 8532–8536. [PubMed] [Google Scholar]
- Böttinger E.P., Factor, V.M., Tsang, M.L., Weatherbee, J.A., Kopp, J.B., Qian, S.W., Wakefield, L.M., Roberts, A.B., Thorgeirsson, S.S., and Sporn, M.B. 1996. The recombinant proregion of transforming growth factor β1 (latency-associated peptide) inhibits active transforming growth factor β1 in transgenic mice. Proc. Natl. Acad. Sci. 93: 5877–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw R.A., Blundell, T.L., Lapatto, R., McDonald, N.Q., and Murray-Rust, J. 1993. Nerve growth factor revisited. Trends Biochem. Sci. 18: 48–52. [DOI] [PubMed] [Google Scholar]
- Buczek O., Olivera, B.M., and Bulaj, G. 2004. Propeptide does not act as an intramolecular chaperone but facilitates protein disulfide isomerase-assisted folding of a conotoxin precursor. Biochemistry 43: 1093–1101. [DOI] [PubMed] [Google Scholar]
- Buevich A.V., Shinde, U.P., Inouye, M., and Baum, J. 2001. Backbone dynamics of the natively unfolded pro-peptide of subtilisin by heteronuclear NMR relaxation studies. J. Biomol. NMR 20: 233–249. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P., Carter, B.D., Dobrowsky, R.T., and Chao, M.V. 1996. Death of oligodendrocytes mediated by the interaction of nerve growth factor with its receptor p75. Nature 383: 716–719. [DOI] [PubMed] [Google Scholar]
- Cohen P., Sutter, A., Landreth, G., Zimmermann, A., and Shooter, E.M. 1980. Oxidation of tryptophan-21 alters the biological activity and receptor binding characteristics of mouse nerve growth factor. J. Biol. Chem. 255: 2949–2954. [PubMed] [Google Scholar]
- De Young L.R., Burton, L.E., Liu, J., Powell, M.F., Schmelzer, C.H., and Skelton, N.J. 1996. RhNGF slow unfolding is not due to proline isomerization: Possibility of a cystine knot loop-threading mechanism. Protein Sci. 5: 1554–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnin C., Jean, F., Thomas, G., and Christian, J.L. 2004. Cleavages within the prodomain direct intracellular trafficking and degradation of mature bone morphogenetic protein-4. Mol. Biol. Cell 15: 5012–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinkwater C.C., Suter, U., Angst, C., and Shooter, E.M. 1991. Mutation of tryptophan-21 in mouse nerve growth factor (NGF) affects binding to the fast NGF receptor but not induction of neurites on PC12 cells. Proc. Biol. Sci. 246: 307–313. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss, R.M., and Terwilliger, T.C. 1984. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. 81: 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernfors P., Ibanez, C.F., Ebendal, T., Olson, L., and Persson, H. 1990. Molecular cloning and neurotrophic activities of a protein with structural similarities to nerve growth factor: Developmental and topographical expression in the brain. Proc. Natl. Acad. Sci. 87: 5454–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahnestock M., Yu, G., and Coughlin, M.D. 2004. ProNGF: A neurotrophic or an apoptotic molecule? Prog. Brain Res. 146: 107–110. [DOI] [PubMed] [Google Scholar]
- Fairlie W.D., Zhang, H.P., Wu, W.M., Pankhurst, S.L., Bauskin, A.R., Russell, P.K., Brown, P.K., and Breit, S.N. 2001. The propeptide of the transforming growth factor-β superfamily member, macrophage inhibitory cytokine-1 (MIC-1), is a multifunctional domain that can facilitate protein folding and secretion. J. Biol. Chem. 276: 16911–16918. [DOI] [PubMed] [Google Scholar]
- Feeney B. and Clark, A.C. 2005. Reassembly of active caspase-3 is facilitated by the propeptide. J. Biol. Chem. 280: 39772–39785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade J.M., Rodriguez-Tebar, A., and Barde, Y.A. 1996. Induction of cell death by endogenous nerve growth factor through its p75 receptor. Nature 383: 166–168. [DOI] [PubMed] [Google Scholar]
- Gallagher T., Gilliland, G., Wang, L., and Bryan, P. 1995. The prosegment-subtilisin BPN′ complex: Crystal structure of a specific ‘foldase.’. Structure 3: 907–914. [DOI] [PubMed] [Google Scholar]
- Gray A.M. and Mason, A.J. 1990. Requirement for activin A and transforming growth factor–β 1 pro-regions in homodimer assembly. Science 247: 1328–1330. [DOI] [PubMed] [Google Scholar]
- Hasan W., Pedchenko, T., Krizsan-Agbas, D., Baum, L., and Smith, P.G. 2003. Sympathetic neurons synthesize and secrete pro-nerve growth factor protein. J. Neurobiol. 57: 38–53. [DOI] [PubMed] [Google Scholar]
- He X.L. and Garcia, K.C. 2004. Structure of nerve growth factor complexed with the shared neurotrophin receptor p75. Science 304: 870–875. [DOI] [PubMed] [Google Scholar]
- Hidaka Y., Ohno, M., Hemmasi, B., Hill, O., Forssmann, W.G., and Shimonishi, Y. 1998. In vitro disulfide-coupled folding of guanylyl cyclase-activating peptide and its precursor protein. Biochemistry 37: 8498–8507. [DOI] [PubMed] [Google Scholar]
- Hillger F., Herr, G., Rudolph, R., and Schwarz, E. 2005. Biophysical comparison of BMP-2, ProBMP-2, and the free pro-peptide reveals stabilization of the pro-peptide by the mature growth factor. J. Biol. Chem. 280: 14974–14980. [DOI] [PubMed] [Google Scholar]
- Hohn A., Leibrock, J., Bailey, K., and Barde, Y.A. 1990. Identification and characterization of a novel member of the nerve growth factor/brain-derived neurotrophic factor family. Nature 344: 339–341. [DOI] [PubMed] [Google Scholar]
- Ikemura H., Takagi, H., and Inouye, M. 1987. Requirement of pro-sequence for the production of active subtilisin E in Escherichia coli . J. Biol. Chem. 262: 7859–7864. [PubMed] [Google Scholar]
- Jain S.C., Shinde, U., Li, Y., Inouye, M., and Berman, H.M. 1998. The crystal structure of an autoprocessed Ser221Cys-subtilisin E-propeptide complex at 2.0 Å resolution. J. Mol. Biol. 284: 137–144. [DOI] [PubMed] [Google Scholar]
- Kliemannel M., Rattenholl, A., Golbik, R., Balbach, J., Lilie, H., Rudolph, R., and Schwarz, E. 2004. The mature part of proNGF induces the structure of its pro-peptide. FEBS Lett. 566: 207–212. [DOI] [PubMed] [Google Scholar]
- LaLonde J.M., Zhao, B., Janson, C.A., D'Alessio, K.J., McQueney, M.S., Orsini, M.J., Debouck, C.M., and Smith, W.W. 1999. The crystal structure of human procathepsin K. Biochemistry 38: 862–869. [DOI] [PubMed] [Google Scholar]
- Lee R., Kermani, P., Teng, K.K., and Hempstead, B.L. 2001. Regulation of cell survival by secreted proneurotrophins. Science 294: 1945–1948. [DOI] [PubMed] [Google Scholar]
- Lilie H., Jaenicke, R., and Buchner, J. 1995. Characterization of a quaternary-structured folding intermediate of an antibody Fab-fragment. Protein Sci. 4: 917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald N.Q., Lapatto, R., Murray-Rust, J., Gunning, J., Wlodawer, A., and Blundell, T.L. 1991. New protein fold revealed by a 2.3-Å resolution crystal structure of nerve growth factor. Nature 354: 411–414. [DOI] [PubMed] [Google Scholar]
- Nykjaer A., Lee, R., Teng, K.K., Jansen, P., Madsen, P., Nielsen, M.S., Jacobsen, C., Kliemannel, M., Schwarz, E., and Willnow, T.E., et al. 2004. Sortilin is essential for proNGF-induced neuronal cell death. Nature 427: 843–848. [DOI] [PubMed] [Google Scholar]
- Podobnik M., Kuhelj, R., Turk, V., and Turk, D. 1997. Crystal structure of the wild-type human procathepsin B at 2.5 Å resolution reveals the native active site of a papain-like cysteine protease zymogen. J. Mol. Biol. 271: 774–788. [DOI] [PubMed] [Google Scholar]
- Rattenholl A., Lilie, H., Grossmann, A., Stern, A., Schwarz, E., and Rudolph, R. 2001a. The pro-sequence facilitates folding of human nerve growth factor from Escherichia coli inclusion bodies. Eur. J. Biochem. 268: 3296–3303. [DOI] [PubMed] [Google Scholar]
- Rattenholl A., Ruoppolo, M., Flagiello, A., Monti, M., Vinci, F., Marino, G., Lilie, H., Schwarz, E., and Rudolph, R. 2001b. Pro-sequence assisted folding and disulfide bond formation of human nerve growth factor. J. Mol. Biol. 305: 523–533. [DOI] [PubMed] [Google Scholar]
- Ruvinov S., Wang, L., Ruan, B., Almog, O., Gilliland, G.L., Eisenstein, E., and Bryan, P.N. 1997. Engineering the independent folding of the subtilisin BPN′ prodomain: Analysis of two-state folding versus protein stability. Biochemistry 36: 10414–10421. [DOI] [PubMed] [Google Scholar]
- Schmid F.X.. 1997. Spectral methods of characterizing protein conformation and conformational changes. In Protein structure: A practical approach (ed. T.E. Creighton), pp. 261–297. IRL-Press, Oxford, United Kingdom.
- Seidah N.G., Benjannet, S., Pareek, S., Savaria, D., Hamelin, J., Goulet, B., Laliberte, J., Lazure, C., Chretien, M., and Murphy, R.A. 1996. Cellular processing of the nerve growth factor precursor by the mammalian pro-protein convertases. Biochem. J. 314: 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde U. and Inouye, M. 2000. Intramolecular chaperones: Polypeptide extensions that modulate protein folding. Semin. Cell Dev. Biol. 11: 35–44. [DOI] [PubMed] [Google Scholar]
- Shinde U., Li, Y., Chatterjee, S., and Inouye, M. 1993. Folding pathway mediated by an intramolecular chaperone. Proc. Natl. Acad. Sci. 90: 6924–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson P., Winther, J.R., Kaarsholm, N.C., and Poulsen, F.M. 1993. The pro region required for folding of carboxypeptidase Y is a partially folded domain with little regular structural core. Biochemistry 32: 12160–12166. [DOI] [PubMed] [Google Scholar]
- Timm D.E. and Neet, K.E. 1992. Equilibrium denaturation studies of mouse β-nerve growth factor. Protein Sci. 1: 236–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman J.S. and Kim, P.S. 1992. The pro region of BPTI facilitates folding. Cell 71: 841–851. [DOI] [PubMed] [Google Scholar]
- Wiesmann C., Ultsch, M.H., Bass, S.H., and de Vos, A.M. 1999. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature 401: 184–188. [DOI] [PubMed] [Google Scholar]
- Winther J.R. and Sorensen, P. 1991. Propeptide of carboxypeptidase Y provides a chaperone-like function as well as inhibition of the enzymatic activity. Proc. Natl. Acad. Sci. 88: 9330–9334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise R.J., Pittman, D.D., Handin, R.I., Kaufman, R.J., and Orkin, S.H. 1988. The propeptide of von Willebrand factor independently mediates the assembly of von Willebrand multimers. Cell 52: 229–236. [DOI] [PubMed] [Google Scholar]
- Zhang Z. and Smith, D.L. 1993. Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 2: 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]