Abstract
The coxsackievirus and adenovirus receptor (CAR) mediates entry of coxsackievirus and adenovirus. CAR possesses an extracellular region that is comprised of 2 immunoglobulin domains termed CAR–D1 and CAR–D2. In the present work, the solution structure of CAR–D2, consisting of residues 142–235 of human CAR, has been determined by NMR spectroscopy. CAR–D2 is shown to be a β-sandwich motif comprised of two β-sheets, which are stabilized by two disulfide bonds. The first β-sheet is comprised of β-strands A, B, and E, and the second β-sheet is comprised of β-strands C, F, and G. A relatively hydrophobic helix is found between β-strands C and E, which replaces β-strand D of the classical c-type immunoglobulin fold.
Keywords: adenovirus, CAR, cell adhesion, coxsackievirus, NMR
The coxsackievirus and adenovirus receptor (CAR) mediates entry of coxsackievirus (CVB), a cause of viral cardiac infections and viral meningitis (Abelmann 1973), and adenovirus (Ad), a cause of respiratory, gastroenteric, and ocular infections, as well as a popular vector for gene therapy (Lukashok and Horwitz 1998). The CAR receptor is expressed in a wide variety of tissue types and thought to mediate cell adhesion and signal transduction (Bergelson et al. 1997; Tomko et al. 1997; Honda and Kuwano 2000; Philipson and Pettersson 2004; Hauwel et al. 2005). Interestingly, CAR exhibits activity as a potent tumor repressor in cancer cell lines (Kim et al. 2003). CAR possesses an extracellular region that is comprised of two domains termed CAR–D1 and CAR–D2. CAR–D1 has been well characterized by structural biology and shown to be an immunoglobulin-like domain (Bewley et al. 1999; van Raaij et al. 2000; Jiang et al. 2004). CAR–D2 is also predicted to be an immunoglobulin-like domain (Bergelson et al. 1997), and its secondary structure has recently been characterized by NMR spectroscopy (Jiang and Caffrey 2005). In an effort to better understand the function of the extracellular domain of CAR, we have determined the solution structure of CAR–D2 by NMR spectroscopy.
Results and Discussion
The solution structure of CAR–D2, consisting of residues 142–235 of human CAR, was determined using information from a total of 488 NOEs, 46 H-bonds, and 232 dihedral restraints (Table 1). The structural statistics of a family of the 40 lowest energy structures and the final minimized mean structure are summarized in Table 1. The superposition of the backbone of the final 40 lowest energy simulated annealing structures is shown in Figure 1A as a stereo diagram. The final ensemble shows no NOE violations over 0.5 Å and no dihedral angle violations >10°. The RMSD of residues within elements of secondary structure relative to the mean is 0.69 Å for the backbone atoms and 1.23 Å for the heavy atoms. The RMSD of residues within the core structure, defined as residues 145–231, relative to the mean is 1.05 Å for the backbone atoms and 1.75 Å for the heavy atoms. A ribbon representation of the minimized mean structure of CAR–D2 is shown in Figure 1B. CAR–D2, which is a member of the immunoglobulin superfamily, forms a β-sheet sandwich motif with an overall length of ∼46 Å and width of ∼26 Å. With respect to the classical c-type immunoglobulin fold (Bork et al. 1994), one β-sheet consists of β-strands A, B, and F (residues 145–151, 156–163, and 206–215, respectively) and the other β-sheet consists of β-strands C, E, and G (residues 172–178, 196–201, and 220–230, respectively). Interaction between the β-sheets is stabilized by the presence of two disulfide bonds found between residues 146–223 and 162–212, which were identified by long-range NOEs. Interestingly, a lone helix encompassing residues 185–192 is found in the position of β-strand D of the c-type immunoglobulin fold, and therefore we will refer to this region as helix D. CAR–D2 does not exhibit a high degree of sequence identity to other immunoglobulin domains; thus, its relatively unique structure is not surprising. Note that in the native structure the N terminus of CAR–D2 would be attached to the C terminus of CAR–D1 and the C terminus of CAR–D2 would be attached to a transmembrane domain that anchors CAR in the extracellular milieu. A surface electrostatic representation of CAR–D2 shown in Figure 1C reveals that the charge is evenly distributed; however, the surface of helix D is relatively hydrophobic, and thus a potential interaction site for the cellular partners of CAR.
Table 1.
Structural statistics
Figure 1.
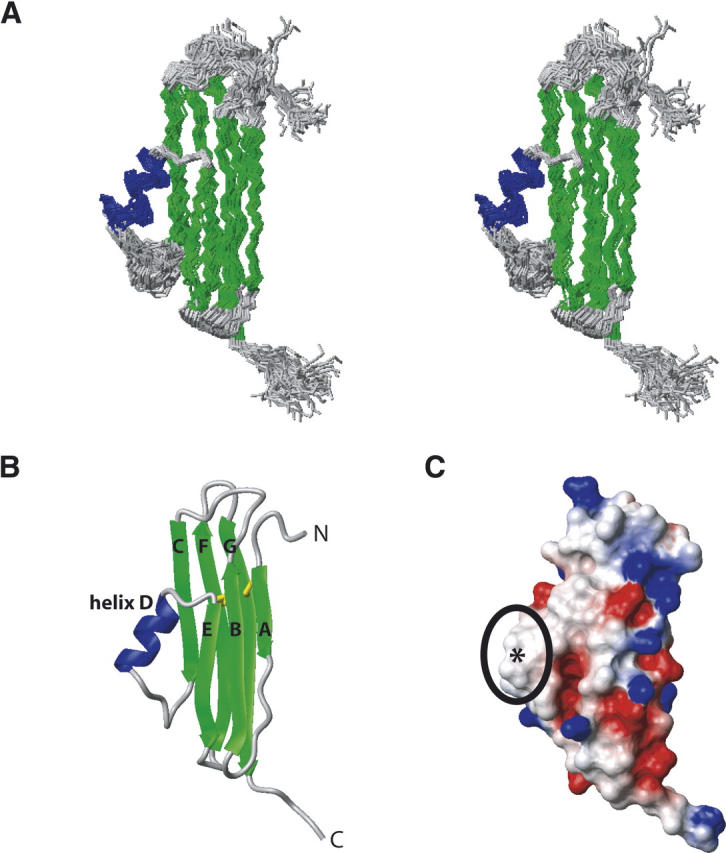
Solution structure of human CAR–D2. (A) Stereo diagram of the ensemble of 40 low-energy structures showing the superimposition of the backbone atoms. (B) Ribbon representation of the minimized mean structure. Disulfide bonds are shown in yellow. The β-strands and helix are labeled according to the classical c-type immunoglobulin fold (Bork et al. 1994). (C) Electrostatic map of the minimized mean structure. The location of helix D is highlighted. In A and B β-strands and the helix have been colored green and blue, respectively.
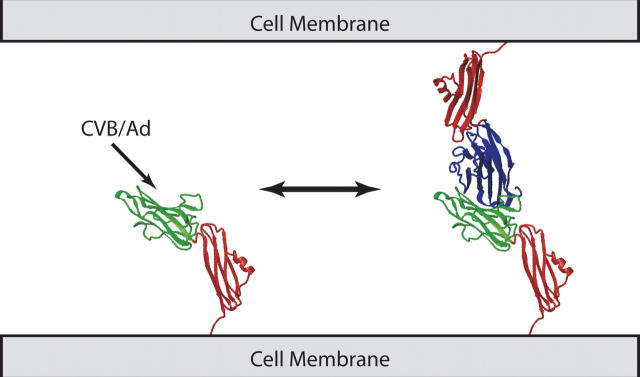
A model for CAR structure and function is presented in Figure 2. The structure of the CAR–D1 is taken from van Raaij et al. (2000), and the structure of CAR–D2 is taken from the work presented herein. As mentioned above, the N terminus of CAR–D2 is attached to the C teriminus of CAR–D1 and the C terminus of CAR–D2 is attached to a transmembrane domain. In this model, CAR is shown to exist in an equilibrium between monomeric and dimeric states. In the monomeric state, CAR is a receptor for coxsackievirus and adenovirus via contacts with CAR–D1 (Bewley et al. 1999; He et al. 2001). In the dimeric state, CAR plays a role in cell adhesion by crosslinking adjacent cells via dimerzation of CAR–D1, which has been deduced from analytical ultracentrifugation and X-ray crystallography studies in vitro (van Raaij et al. 2000) and homophilic association of CAR observed in vivo (Cohen et al. 2001). The CAR association is shown to be reversible, based on the relatively weak self-association of CAR–D1 in vitro (van Raaij et al. 2000; Jiang et al. 2004).
Figure 2.
Model for the mechanism of CAR. The CAR–D1 domains are colored blue and green and the CAR–D2 domains are colored red. The structure of the CAR–D1 monomer and dimer are taken from van Raaij et al. (2000). The structure of CAR–D2 represents the work presented herein. The relative orientation of the CAR domains to one another is arbitrarily chosen and not based on experimental data. The binding site of CVB and Ad on CAR–D1 is deduced from the work of Bewley et al. (1999) and He et al. (2001).
At present, the cellular role of CAR–D2 is not yet apparent. However, one possible role would be to place CAR–D1 in the correct orientation and distance from the membrane for interactions with cellular partners. Another potential role could consist of direct interactions between CAR–D2 (perhaps via the hydrophobic helix D) and cellular partners. In summary, the present NMR study represents the first high-resolution structural characterization of CAR–D2. Together, the structures of CAR–D1 and CAR–D2 lend insight into the role of CAR in cell adhesion, as well as the entry mechanisms of CVB and Ad, thereby allowing for the development of structure-based antivirals.
Materials and methods
Human CAR–D2 (residues 142–235) was prepared as previously described (Jiang and Caffrey 2005). For the NMR experiments, the experimental conditions were 1 mM CAR–D2 in 100 mM PO4/pH 6.0 and 10% D2O. NMR spectra were recorded at 25°C on Bruker DRX 600 and AVANCE 800 MHz spectrometers equipped with cryogenic triple resonance probes. Spectra were processed by NmrPipe and visualized with NmrDraw (Delaglio et al. 1995). Backbone assignments have been previously described (Jiang and Caffrey 2005). 3D 13C- and 15N-edited NOESY-HSQC (mixing times = 100 and 120 msec, respectively) were acquired for structural restraints. NOEs were manually assigned and classified as: strong (1.8–2.7 or 1.8–2.9 Å for the NOE of NH), medium (1.8–3.3 or 1.8–3.5 Å for NOE of NH), weak (1.8–5.0 Å), or very weak (1.8–6.0 Å). For distances involving methyl protons, 0.5 Å was added to the upper limit to account for the higher apparent intensity of methyl protons. ψ, ϕ, and χ1 torsion angle restraints were derived from database analysis of chemical shifts (N, HN, Cα, Cβ, C′, Hα) using the programs TALOS and SHIFTOR (Cornilescu et al. 1999; Neal et al. 2006). Minimum error ranges for the torsion angle restraints were set to ±40°. H-bonds, which were identified by the 13C backbone chemical shifts, NOE patterns, and nonexchangeable protons, were incorporated for the regions of secondary structure as two restraints per H-bond where rNH—O = 1.5–2.8 Å and rN—O = 2.4–3.5 Å. Structures were calculated by simulated annealing in torsion angle space starting from an extended strand, followed by conventional simulated annealing, using the program CNS (Brunger et al. 1998). The program was adapted to incorporate a conformational database (Kuszewski et al. 1996). A family of the 40 lowest energy structures was chosen and a minimized mean structure was calculated. The overall quality of the final structures was assessed using the program PROCHECK (Laskowski et al. 1993). Figures were generated using the program MOLMOL (Koradi et al. 1996).
Acknowledgments
The coordinates of the CAR–D2 minimized mean structure have been deposited in the PDB under the accession number 2NPL. This work was supported by the American Heart Association Scientist Development Grant 0030374Z to M.C. and grants to the UIC Structural Biology Center (NSF BIR 9601705 and NIH S10 RR15757).
Footnotes
Reprint requests to: Michael Caffrey, Department of Biochemistry and Molecular Genetics, University of Illinois at Chicago, Chicago, IL 60607, USA; e-mail: caffrey@uic.edu; fax: (312) 413-0353.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062643507.
References
- Abelmann W.. 1973. Viral myocarditis and its sequelae. Annu. Rev. Med. 24: 145–152. [DOI] [PubMed] [Google Scholar]
- Bergelson J., Cunningham, J., Droguett, G., Durt-Jones, E., Drithaivas, A., Hong, J., Horwitz, M., Crowell, R., and Finberg, R. 1997. Isolation of a common receptor for Coxsackie B viruses and Adenoviruses 2 and 5. Science 275: 1320–1323. [DOI] [PubMed] [Google Scholar]
- Bewley M., Springer, K., Zhang, Y.-B., Freimuth, P., and Flanagan, J. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286: 1579–1583. [DOI] [PubMed] [Google Scholar]
- Bork P., Holm, L., and Sander, C. 1994. The immunoglobulin fold. J. Mol. Biol. 242: 309–320. [DOI] [PubMed] [Google Scholar]
- Brunger A., Adams, P., Clore, G., DeLano, W., Gros, P., Grosse-Kunstleve, R., Jiang, J.-S., Kuszewski, J., Nilges, N., and Pannu, N., et al. 1998. Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta Crystallogr. D54: 905–921. [DOI] [PubMed] [Google Scholar]
- Cohen C., Shieh, J., Pickles, R., Okegawa, T., Hsieh, J., and Bergelson, J. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. 98: 15191–15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13: 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Hauwel M., Furon, E., and Gasque, P. 2005. Molecular and cellular insights into the coxsackievirus-adenovirus receptor: Role in cellular interactions in the stem cell niche. Brain Res. Brain Res. Rev. 48: 265–272. [DOI] [PubMed] [Google Scholar]
- He Y., Chipman, P., Howitt, J., Bator, C., Whitt, M., Baker, T., Kuhn, R., Anderson, C., Freimuth, P., and Rossman, M. 2001. Interaction of cosackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 8: 874–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T. and Kuwano, R. 2000. The coxsackievirus-adenovirus receptor protein as a cell adhesion molecule in the developing mouse brain. Brain Res. Mol. Brain Res. 14: 19–28. [DOI] [PubMed] [Google Scholar]
- Jiang S. and Caffrey, M. 2005. NMR assignment and secondary structure of the coxsackievirus and adenovirus receptor domain 2. Protein Pept. Lett. 12: 537–539. [DOI] [PubMed] [Google Scholar]
- Jiang S., Jacobs, A., Laue, T., and Caffrey, M. 2004. Solution structure of the coxsackievirus and adenovirus receptor domain 1. Biochemistry 43: 1847–1853. [DOI] [PubMed] [Google Scholar]
- Kim M., Sumerel, L., Belousova, N., Lyons, G., Carey, D., Krasnykh, V., and Douglas, J. 2003. The coxsackievirus and adenovirus receptor acts as a tumour suppressor in malignant glioma cells. Br. J. Cancer 88: 1411–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 52–55. [DOI] [PubMed] [Google Scholar]
- Kuszewski J., Gronenborn, A., and Clore, G. 1996. Improving the quality of NMR and crystallographic protein structures by means of a conformational database potential derived from structure databases. Protein Sci. 5: 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R., MacArthur, M., Moss, D., and Thorton, J. 1993. PROCHECK—A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Lukashok S. and Horwitz, M. 1998. New perspectives in adenoviruses. Curr. Clin. Top. Infect. Dis. 18: 286–305. [PubMed] [Google Scholar]
- Neal S., Berjanskii, M., Zhang, H., and Wishart, D. 2006. Accurate prediction of protein torsion angles using chemical shifts and sequence homology. Magn. Reson. Chem. 44: S158–S167. [DOI] [PubMed] [Google Scholar]
- Philipson L. and Pettersson, R. 2004. The coxsackie-adenovirus receptor—A new receptor in the immunoglobulin family involved in cell adhesion. Curr. Top. Microbiol. Immunol. 273: 87–111. [DOI] [PubMed] [Google Scholar]
- Tomko R., Xu, R., and Philipson, L. 1997. HCAR and MCAR: The human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. 94: 3352–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raaij M., Chouin, E., van der Zandt, H., Bergelson, J., and Cusack, S. 2000. Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 Å resolution. Structure 8: 1147–1155. [DOI] [PubMed] [Google Scholar]