Abstract
Chaperones and proteases share the ability to interact with unfolded proteins. Here we show that enzymatically inactive forms of the aspartic proteases HIV-1 protease and pepsin have inherent chaperone-like activity and can prevent the aggregation of denatured substrate proteins. In contrast to proteolysis, which requires dimeric enzymes, chaperone-like activity could be observed also with monomeric domains. The involvement of the active site cleft in the chaperone-like function was demonstrated by the inhibitory effect of peptide substrate inhibitors. The high structural similarity between aspartic proteases and the N-terminal double-ψ barrels of Cdc48-like proteins, which are involved in the unfolding and dissociation of proteins, suggests that they share a common ancestor. The latent chaperone-like activity in aspartic proteases can be seen as a relic that has further evolved to serve substrate binding in the context of proteolytic activity.
Keywords: aspartic proteases, chaperones, double-ψ barrel, AAA-ATPase, protein evolution
Molecular chaperones bind unfolded or nonnative polypeptides in order to prevent aggregation and to assist protein folding (Hartl and Hayer-Hartl 2002; Young et al. 2004). Their ability to bind unfolded polypeptides is shared by proteases, which hydrolyze their substrates after initial binding. This commonality raises the possibility that chaperones and proteases may have evolved from common, peptide-binding ancestors, and subsequently diverged in their functions and reaction mechanisms. Do the sequences or structures of chaperones and proteases contain information to support this assumption and have features of such a common ancestor still survived in modern-day proteins? While homology between chaperones and proteases cannot be detected on the sequence level, related structures are found in both classes of proteins (Wlodawer et al. 1989; Coles et al. 1999). Aspartic proteases and double-ψ barrel domains, which act as modulators of protein unfolding and dissociation in ATPases associated with various cellular activities (AAA-ATPases) (Gerega et al. 2005), adopt a similar fold and could therefore represent a homologous protease–chaperone pair.
Pepsin and HIV-1 (human immunodeficiency virus) protease are aspartic proteases that are classified in the MEROPS classification system of proteases (Rawlings and Barrett 1999) in the same clan (aa), but in different families (A1 and A2, respectively). Whereas viral proteases of family A2 are homodimers of the aspartic protease domain (Wlodawer et al. 1989), eukaryotic proteases of family A1 are bilobed enzymes, where each lobe consists of a single domain and contributes one catalytic aspartic acid residue to the active site. These domains are considered to be homologous and evolved by gene duplication of the single domain with subsequent gene fusion (Tang et al. 1978). Viral proteases are regarded to be homologous to the eukaryotic proteases because of a common fold, the active-site locations, active-site residues, and inhibitor affinities (Lin et al. 1994). However, sequence similarity between the two families is essentially undetectable. While the phylogeny within each family is clearly described (Dunn et al. 2002; Kageyama 2002), the direction of evolution between these families is still enigmatic. Tang et al. postulated a sequential evolution originating from a viral protease that was retained in the eukaryotic host and evolved to present-day aspartic proteases (Tang et al. 1978). In contrast to this, Rao et al. describe the evolution of viral proteases from the eukaryotic ones by gene deletion under the selective pressure of viral coding capacity (Rao et al. 1991). The idea of evolution from low-sequence specificity of the digestive eukaryotic proteases toward highly specific regulatory viral proteases would also corroborate the second scenario (Neurath 1984). Divergent evolution from a common monomeric precursor has also been proposed (Rao et al. 1991; Coles et al. 1999). But, the lack of prokaryotic aspartic proteases has been used to argue against that hypothesis. Provided that the potential last common ancestor had existed before life divided into the three domains, one would expect the existence of descendents in each of them.
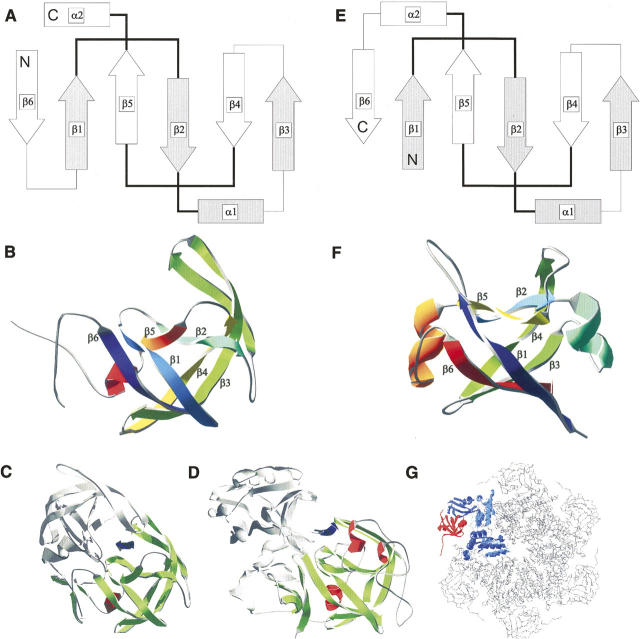
The fold of the aspartic proteases contains two topologically complex ψ-loop motifs as a result of the interleaved connection between two β-hairpins (Fig. 1) (Castillo et al. 1999). Strand 2 (β2) of one hairpin crosses underneath the loop of the other, giving the fold a pseudo-knotted structure and resembling in top view the Greek letter ψ. This remarkable arrangement is strikingly similar to the ψ-loops present in the double-ψ barrel protein family (Castillo et al. 1999). In double-ψ barrels, two symmetrically interleaved ββαβ-elements comprise the ψ-loops (Fig. 1). In the N-terminal domain of the valosine containing protein-like ATPase from Thermoplasma acidophilum (VAT-NN), the internal symmetry is reflected even on the sequence level (Coles et al. 1999). The double-ψ barrel fold comprises two different superfamilies, namely, the barwin-like endoglucanases and ADC-like proteins. Families of the ADC-like superfamily are aspartate–decarboxylase, formate/DMSO reductase (C-terminal domain), and Cdc48 N-terminal domain-like proteins. Members of the last family can be found as N-terminal domains of AAA-ATPases like NSF, p97, PEX1, and VAT, which act as protein unfoldases and as the N-terminal domain of Ufd1, which is involved in ubiquitin-dependent protein degradation (Park et al. 2005). N-terminal domains of AAA-ATPases provide substrate specificity and are involved in protein–protein interactions. For the N-terminal domain of VAT, a basic chaperone activity has been shown in vitro (Golbik et al. 1999). However, its native function seems to be substrate binding and regulation of its unfolding in the context of the entire AAA-ATPase (Gerega et al. 2005).
Figure 1.
Structural comparison of double-ψ barrel proteins and aspartic proteases. Topology and structure of HIV-1 protease, pepsin, and Cdc48 N-terminal domain-like double-ψ barrels. Topology diagrams of (A) HIV-1 protease and a (E) double-ψ barrel. β-strands are represented as arrows, α-helices as bars, and loops as lines. Pseudo-symmetric units are depicted as filled and empty forms. The “ψ”-forming loops are in bold. The secondary structure elements are totally interleaved—elements of one symmetric unit make only contacts to elements of the other unit. Structures shown as ribbon diagrams. Order of secondary structure elements is color-coded from blue to red: (B) monomer of HIV-1 protease (5hvp) and (F) N-terminal double-ψ barrel domain of VAT (1cz4) from Thermoplasma acidophilum. (Bottom) Structural context of the domains: (C) One monomer of the HIV-1 protease dimer is colored and the substrate binding site is indicated by an arrow between both domains. (D) One domain of pepsin is colored and the substrate binding site is indicated by an arrow between both domains. (G) Structure of p97 (1r7r) representing the structural context of Cdc48 N-terminal domain-like double-ψ barrels. One monomer of the hexamer is colored with the N-terminal double-ψ barrel domain in red.
The double-ψ barrel can be converted topologically into the aspartic protease fold by a circular permutation, in which the N-terminal strand of the double-ψ barrel is relocated to the C terminus (Fig. 1). However, despite similar folds, there is no detectable sequence similarity between the two folds. The SCOP (Murzin et al. 1995) and CATH database (Orengo et al. 1997) therefore classify the double-ψ barrels and the aspartic proteases in two different folds, implying convergent evolution. Contrarily, Coles et al. suggested a scenario of divergent evolution of double-ψ barrels and aspartic proteases (Coles et al. 1999), in which their last common ancestor was a symmetrical single double-ψ barrel chaperone. From there on, the Cdc48 N-terminal domain-like proteins fused with AAA-ATPase domains and evolved specific substrate recognition and regulatory activity. The aspartic proteases underwent circular permutation and evolved protease activity. Further diversification occurred into viral and eukaryotic proteases.
In this work we asked whether aspartic proteases can be traced back to a chaperone ancestor that was lacking a proteolytically active site. In a sense, this evolutionary path has been probed in the opposite direction by the conversion of the chaperone cyclophilin into a serine protease by protein engineering on the basis of its peptide-binding capability (Quemeneur et al. 1998). In our studies we used the ability of chaperones to prevent aggregation of denatured proteins. We find that the enzymatically inactive aspartic proteases indeed have such an inherent chaperone-like activity, suggesting that they are evolutionarily linked to double-ψ barrels with which they share a common single-domain ancestor.
Results
Chaperone-like activity of pepsin
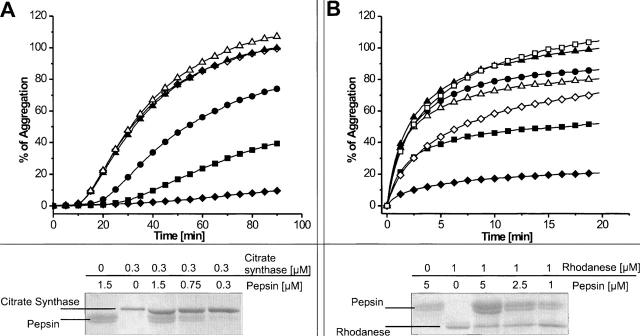
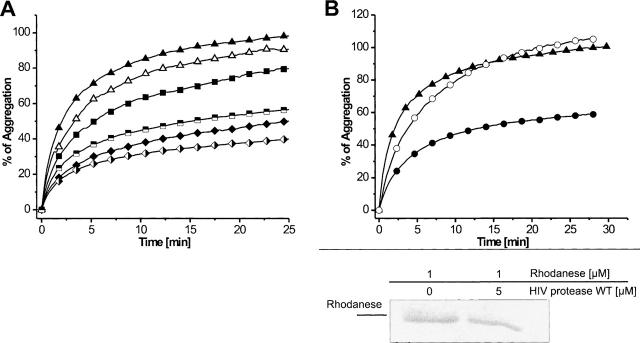
Chaperones are known to bind unfolded or nonnatively folded polypeptides. This feature can be used to test proteases for an intrinsic chaperone-like activity in vitro. If they are able to interact with denatured substrates, they might prevent protein aggregation ensuing from high concentrations of the denatured protein in the assay solution. Aggregation is caused either by thermal denaturation or by the inability of chemically denatured proteins to refold after dilution of the chaotropic denaturant. Protein aggregates cause an increase in attenuance, which can be prevented in the presence of chaperones. For pepsin, we took advantage of the fact that proteolytic activity is completely absent at pH 6.5 due to deprotonation of both catalytic aspartic acid residues (Lin et al. 1992). This allowed us to assess the ability of proteolytically inactive pepsin to prevent protein aggregation with minimal manipulation of the active site. The presence of pepsin reduced aggregation of thermally denatured citrate synthase (Fig. 2A) and chemically denatured rhodanese (Fig. 2B) effectively. With molar ratios of pepsin and citrate synthase of 1:1, 2.5:1, and 5:1, the aggregation could be reduced to 75%, 40%, and 10%, respectively. In the case of rhodanese, aggregation could be reduced to 85%, 50%, and 20%, with molar ratios of pepsin and rhodanese of 1:1, 2.5:1, and 5:1, respectively. A fivefold molar excess of bovine serum albumin (BSA) was used as a control for unspecific prevention of aggregation. To exclude remnant proteolytic activity of pepsin as a cause of this observation, the total protein amount of each sample was applied onto SDS-PAGE after the aggregation prevention assay. Protein amounts were evaluated by comparison of band intensities (Fig. 2). The band of pepsin occurs as a narrow doublet, which might be the result of two alternative cleavage sites of the presequence (Tang et al. 1973). The amount of citrate synthase was equal in all cases, and no degradation products could be detected. Proteolytic degradation during the assays can therefore be excluded and the aggregation protection effect can be assigned to a basic chaperone-like activity of pepsin.
Figure 2.
Chaperone-like activity of pepsin. Aggregation assays with thermally denatured citrate synthase (A) and chemically denatured rhodanese (B) in the absence of pepsin (▴); an equal molar amount (•), 2.5-fold (▪), or fivefold (♦) molar excess of pepsin; a 2.5-fold molar excess of pepsin inhibited with pepstatin (B, □); a fivefold molar excess of pepsin inhibited with pepstatin (⋄); a fivefold molar excess of BSA (▵). (Bottom) SDS-PAGE analysis of the assay solutions to show protein composition and to exclude remnant proteolytic activity of pepsin.
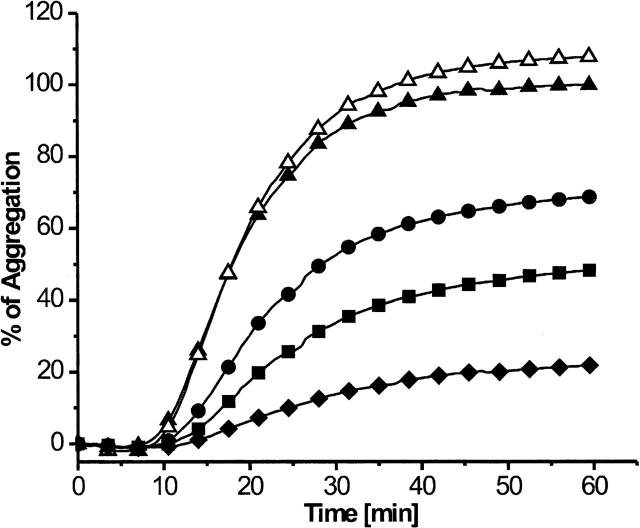
Pepsin consists of two domains with different thermal stabilities (Privalov et al. 1981). The N-domain of pepsin is likely to be unfolded in the assays at 50°C, suggesting that the C-domain alone causes the chaperone-like effect. To test this, we performed aggregation prevention assays with the pepsin C-domain that we generated from native pepsin by limited proteolysis with proteinase K. At pH 7.3, the N-domain is unfolded and therefore protease sensitive, whereas the C-domain is folded and protease resistant (Lin et al. 1993). The C-domain of pepsin has no proteolytic activity by itself. Chaperone-like activity was tested with thermally denatured citrate synthase. Our data show that the C-domain of pepsin prevented aggregation of citrate synthase to 70%, 50%, and 20% when applied in one-, 2.5-, and fivefold molar excess, respectively (Fig. 3). The results show that the C-domain of pepsin is sufficient to elicit this effect and probably also responsible for chaperone-like activity of full-length pepsin at 50°C.
Figure 3.
Chaperone-like activity of the pepsin C-domain. Aggregation assay with thermally denatured citrate synthase: Citrate synthase in the absence of the pepsin C-domain (▴); an equal molar amount (•), 2.5-fold (▪), or fivefold (♦) molar excess of pepsin C-domain; a fivefold molar excess of BSA (▵).
To test whether the active site of pepsin, which is located between the ψ-loops of each domain, participates in the interaction with nonnative substrate proteins, we performed aggregation prevention assays with pepsin that was preincubated with the active-site inhibitor pepstatin. As substrates, we used thermally denatured citrate synthase at 50°C and chemically denatured rhodanese at 25°C. Chaperone-like activity was largely abolished now (Fig. 2). Only in the case of rhodanese, a diminished chaperone-like activity could still be observed with a fivefold molar excess of pepsin; aggregation increased here from 20% to 70% in the presence of pepstatin. The aspartic protease inhibitor pepstatin is known to be a specific competitive inhibitor of the active site of pepsin that causes only minor conformational changes in the overall structure of the protein, which has been shown by structural investigations of pepsin–pepstatin complexes (Fujinaga et al. 1995).
Inhibition of chaperone-like activity at 50°C might seem surprising at first glance, since we consider the N-domain to be unfolded and the C-domain alone is unable to bind pepstatin tightly. However, the presence of pepstatin stabilizes the native fold of pepsin and increases the melting temperature of the N-domain (Privalov et al. 1981). Thus, the use of the active-site inhibitor pepstatin shows clearly the participation of the proteolytic site in binding of nonnative substrate proteins.
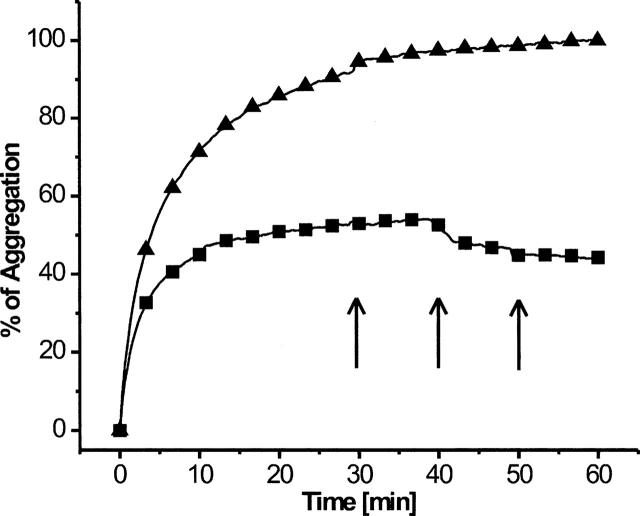
To estimate the stability of the interaction between pepsin and rhodanese, complexes were allowed to form, and after reaching a plateau of aggregation, pepstatin was added in three distinct steps. A reinstated aggregation of released rhodanese was expected if pepstatin could displace rhodanese from pepsin. However, this was not the case. Instead, the level of attenuance remained unchanged after each addition of pepstatin once the plateau was reached, indicating that the pepsin–rhodanese complexes were stable once they had formed (Fig. 4). A similarly strong association between a chaperone and its substrate has been observed with small heat-shock proteins (sHsps), in which case, dissociation relies on other ATP-dependent chaperones (Mogk et al. 2003).
Figure 4.
Stability of complexes between pepsin and denatured polypeptide. Aggregation of rhodanese in the absence (▴) or with a 2.5-fold molar excess (▪) of pepsin. Arrows indicate addition of pepstatin during the measurement.
Isolation of complexes of pepsin and denatured citrate synthase or rhodanese
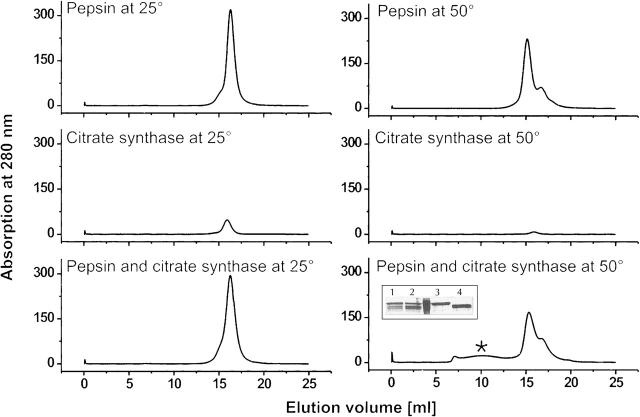
To obtain information on the stoichiometry of the interaction between pepsin and substrate proteins, we set out to isolate complexes of pepsin and denatured citrate synthase. Complex formation was promoted by incubation of pepsin and citrate synthase at 50°C for 90 min. An equal sample mixture incubated at room temperature, and each protein by itself, served as controls. Without prior heating, pepsin and citrate synthase eluted as separate proteins on a gel-size exclusion column (Fig. 5, left). In contrast, heating the samples resulted in a broad peak at higher molecular weights in the elution profile (Fig. 5, right). Neither pepsin nor citrate synthase alone showed a comparable peak. Additionally, SDS-PAGE analysis of these new peak fractions revealed that they contained both proteins: pepsin, and citrate synthase (Fig. 5, inset, bottom, right). Equivalent results have been obtained using rhodanese in place of citrate synthase (Supplemental Fig. S1). Further analysis of the complexes between pepsin and citrate synthase with static laser-light scattering revealed that the complexes do not adhere to a defined stoichiometry and are heterogeneous in size between 500 and 2000 kD (data not shown), consistent with the broad shape of the peak. The finding of an undefined stoichiometry is not surprising, since proteases may not be restricted to the binding of hydrophobic patches, and thus could bind multiple times to an extended protein chain or to small conglomerates. The occurrence of an undefined stoichiometry concurs with similar results obtained from aggregation prevention studies with the AAA-ATPase Cdc48 (Thoms 2002) and with sHsps (Mogk et al. 2003). The small shift to lower elution volumes with pepsin alone when incubated at 50°C can be explained by the above-mentioned partial heat denaturation of the N-domain; the shoulder is caused by still undenatured pepsin. In conclusion, these results indicate that pepsin interacts stably only with unfolded, but not with native citrate synthase or rhodanese, and thereby keeps them in solution.
Figure 5.
Complex formation of pepsin with denatured citrate synthase. UV-profiles of gel-size exclusion chromatography runs. The asterisk (*) (bottom, right) indicates the additional peak occurring when pepsin and citrate synthase are incubated together at 50°C. The inset shows SDS-PAGE analysis of the run with the mixture of citrate synthase and pepsin. Loaded samples are: fraction at 10 mL elution (1), fraction at 15 mL elution volume (2), references citrate synthase (3), and pepsin (4).
Chaperone-like activity of HIV-protease
HIV-1 protease forms homodimers by a four-stranded β-sheet with two strands from each subunit (comprising amino acids P1, Q2, I3, of the first and T96, L97, N98, F99 of the second strand; these dimerization strands are not considered in the topology diagram of Fig. 1). HIV-1 protease was proteolytically inactivated by the introduction of the mutation D25N, thereby removing the catalytic aspartic acid residue. In addition, HPR-DS carries the mutations Q2C, C95A, and L97C, which now positions two cysteine residues within the dimerization strands in such a way that they form an intramolecular disulfide bond. This, in turn, distorts the orientation of the dimerization strands and results in monomeric proteins that are also catalytically inactive (Louis et al. 2003). To confirm the formation of the disulfide bridges, the number of free cysteines per protein monomer was determined and found to be as expected (data not shown). Both HIV-1 protease mutants were subjected to aggregation prevention assays with chemically denatured rhodanese at 25°C and were found to be active (Fig. 6A). The dimeric HPR-M reduced aggregation to 80% and 50% with a molar excess of 2.5 and 5, respectively. The monomeric HPR-DS reduced aggregation to 60% and 40% with 2.5- and fivefold molar excess, respectively. In analogy to our experiments with pepsin, we tried to show the participation of the active site by preincubation of the protein with the HIV-1 protease inhibitor Ac-Leu-Val-Phe-Aldehyd. However, in this case, chaperone-like activity was not affected (data not shown). We reasoned that the mutation of the catalytic aspartate to asparagine might have caused the lack of sensitivity toward this inhibitor. Therefore, we decided to test wild-type HIV-1 protease in aggregation-prevention assays. This turned out to be possible because the protease was proteolytically inactive toward denatured rhodanese, presumably due to the lack of cleavage-recognition sites (Fig. 6B). Aggregation of rhodanese was reduced to 60% with a fivefold molar excess of protease and preincubation with Ac-Leu-Val-Phe-Aldehyd abolished this effect completely (Fig. 6B). This indicates that HIV-1 protease, like pepsin, shows chaperone-like activity as both monomer and dimer, with the involvement of the binding residues in the proteolytic site.
Figure 6.
Chaperone-like activity of HIV-1 protease. (A) Aggregation assay with chemically denatured rhodanese: Rhodanese in the absence of HIV-1 protease (▴); a 2.5-fold (▪) or fivefold (♦) molar excess of HPR-M, a 2.5-fold ( ) or fivefold (
) or fivefold ( ) molar excess of HPR-DS; a fivefold molar excess of BSA (▵). (B) Effect of HIV-1 protease inhibitor: Rhodanese in the absence (▴) or presence of a fivefold molar (•) excess of HPR-WT; a fivefold molar excess of HPR-WT inhibited with HIV-1 protease inhibitor (○). (B) (Bottom) SDS-PAGE analysis of the assay solutions to exclude remnant proteolytic activity of HPR-WT.
) molar excess of HPR-DS; a fivefold molar excess of BSA (▵). (B) Effect of HIV-1 protease inhibitor: Rhodanese in the absence (▴) or presence of a fivefold molar (•) excess of HPR-WT; a fivefold molar excess of HPR-WT inhibited with HIV-1 protease inhibitor (○). (B) (Bottom) SDS-PAGE analysis of the assay solutions to exclude remnant proteolytic activity of HPR-WT.
Chaperone assays with thermolysin and trypsin
To assess whether chaperone activity of proteases is a general phenomenon, we set out to test serine- amd metalloproteases. As an example for a eukaryotic serine protease, we opted for trypsin, which was proteolytically inactivated by substituting the active-site serine residue with alanine (trypsin S200A). Thermolysin is a prokaryotic Zn-protease that was inactivated with the Zn-chelator 1,10-phenanthrolin. By this treatment, calcium, which is required for the stability of thermolysin, is kept bound to the protein. In aggregation assays with thermally denatured citrate synthase and luciferase, or with chemically denatured rhodanese and luciferase, we could not observe any chaperone activity with either of these proteases (Supplemental Fig. S2).
Discussion
To exert their functions, proteases and molecular chaperones typically interact with nonnatively folded or extended regions of polypeptides as a first step in their respective reaction cycles. When proteolytic activity of the aspartic proteases pepsin and HIV-1 protease is abolished, an inherent chaperone-like activity surfaces. The occurrence of chaperone-like activity in proteolytically inactivated proteases is not universal as it might be assumed. We have tested the inactivated metalloprotease thermolysin and the serine protease trypsin as examples for other protease families in aggregation-prevention assays on chaperone-like activity and could not find any effect. Aggregation assays revealed that a single pepsin C-domain and a HIV-1 protease monomer are sufficient for the prevention of aggregation, i.e., binding and shielding of unstructured elements in denatured proteins similar to sHsps (Haslbeck et al. 2005). In contrast to this basic chaperone-like activity, proteolysis requires the interplay of two protease domains (on a single chain in pepsin and as homodimer in HIV-1 protease), thereby the substrate is bound in the interface of these two domains. Tight binding of the inhibitor pepstatin also requires a hydrogen-bond network in the domain interface and does not inhibit the C-domain alone. However, given the size of the deep groove (40 Å in length) (Davies 1990) and the known mode of interaction with peptides via both residue-specific and more generic hydrophobic interactions, it is entirely feasible that the C-domain provides protein–protein interactions of sufficient strength to prevent aggregation. Active-site inhibitors could compete with the chaperone-like activity, supporting our initial assumption that the active site is used for the interaction with denatured polypeptides. In an active protease, binding would be rapidly followed by the cleavage reaction. With pepsin, we did not observe a particular stoichiometry between chaperone and substrate; instead, the complexes appear to be of heterogeneous nature. We can demonstrate that aspartic proteases possess the ability to interact with nonnative polypeptides in a way that resembles the binding mode of molecular chaperones.
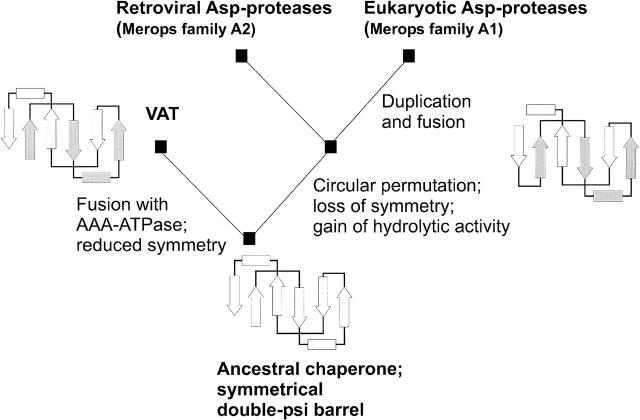
We explain the inherent chaperone-like effect of aspartic proteases in an evolutionary context, whereby the remnant chaperone-like activity has been inherited from a monomeric chaperone ancestor. Our findings support the evolutionary scenario suggested by Coles at al. (1999), which describes a divergent evolution of double-ψ barrels and aspartic proteases. According to this scenario (Fig. 7), the last common ancestor was a symmetrical single double-ψ barrel chaperone. From thereon, Cdc48 N-terminal domain-like proteins fused with a AAA-ATPase domain and can now be found in present-day proteins p97, Cdc48, VAT, or PEX1 as substrate-binding domains. These protein domains maintained their chaperone activity in the context of AAA-ATPases, as has been shown for VAT-N (Golbik et al. 1999). In parallel, circular permutation of the ancestor and acquisition of hydrolytic residues in the substrate-binding pocket led to the evolution of aspartic proteases. Further diversification occurred within the aspartic protease family into viral and eukaryotic proteases. Viral proteases have evolved high-substrate specificity under the selective pressure of virus assembly. Eukaryotic proteases evolved to highly efficient, but rather unspecific proteases of the digestive system. Gene fusion to obtain both domains on a single chain can be construed as advantageous in the sense of increased reaction efficiency.
Figure 7.
Scenario of Cdc48 N-terminal domain-like double-ψ barrel and aspartic protease evolution. The scenario originates with an ancestral chaperone showing a symmetrical double-ψ barrel fold. The left branch leads to AAA-ATPases by fusion with AAA-domains, e.g., in VAT. The right branch leads to circular permutation, loss of internal symmetry, and gain of hydrolytic activity. Further branching leads to the retroviral proteases like HIV-1 protease, and via gene duplication and fusion, to the eukaryotic aspartic proteases like pepsin.
Circular permutation, which is needed to transform a double-ψ barrel topology into the topology of aspartic proteases, is a very common genetic event (Lindqvist and Schneider 1997). In the case of proteases, the selective advantage of a circularly permuted structure might lie in the location of the termini. In a ββαβ repeat like in VAT-NN, the ψ-loops are positioned opposite to the termini (Fig. 1). Dimerization involves the chain termini: by interaction of terminal dimerization strands in the case of HIV-1 protease or as domain fusion in the case of pepsin. A circular permutation positions the termini perpendicular to the ψ-loop region. This allows for spatial proximity of the ψ-loop regions of both domains where the active site is located upon dimerization.
Neither double-ψ barrels nor aspartic proteases are classified as superfolds (Orengo et al. 1994) that are found in many evolutionary unrelated protein families. It is believed that among superfolds, convergent evolution might have occurred because of the high content of supersecondary structure elements with a high folding propensity and particular advantages in the “fold competition” (Soding and Lupas 2003). For nonsuperfolds, scenarios of divergent evolution are considered more probable, since they reflect the parsimonious tendency of nature to reuse existing folds instead of reinventing them. Especially for such a complex pseudo-knotted fold topology as that of double-ψ barrels and aspartic proteases, a de novo invention in duplicate seems unlikely. Instead, the wide divergence of homologous proteins, including fold changes, has been illustrated by Murzin and Grishin (Murzin 1998; Grishin 2001). Thus, in proposing the evolutionary scenario in Figure 7, we used parsimony and known rules of molecular evolution to propose how aspartic proteases might have evolved from an ancestral chaperone by circular permutation, duplication and fusion, and loss of symmetry through divergence and neutral drift.
Materials and Methods
Materials
Mitochondrial citrate synthase from porcine heart (EC 4.1.3.7), bovine adrenal rhodanese (EC 2.8.1.1), porcine pepsin A (EC 3.4.23.1), thermolysin (EC 3.4.24.27), and BSA were purchased from Sigma. Proteinase K was purchased from Fermentas. Pepstatin and Ac-Leu-Val-Phe-Aldehyd HIV-1 protease inhibitor were purchased from Bachem.
Construction of HIV-1 protease mutants
Cloning was done according to standard procedures. pBD2-plasmid carrying the HIV-1 protease wild-type sequence, received as a gift from Felix Mueller-Sarnowski (Max von Pettenkofer-Institute, Munich, Germany), was used as template for a PCR. Three HIV-1 protease constructs were made by overlap-extension mutagenesis. The constructs carry the wild-type sequence (HPR-WT) or the mutations D25N (HPR-M) or Q2C, D25N, C95A, and L97C (HPR-DS). Terminal PCR primers introduced restriction sites for NdeI and HindIII. The products were ligated into pET30b expression vector (Novagen) and transformed into Escherichia coli C41 (DE3) cells for overexpression. Nucleotide sequences were confirmed by DNA sequencing.
Purification of HIV1-protease mutants
E. coli cells were grown at 37°C in Luria-Bertani medium to an optical density of 0.6 and induced with 1 mM isopropyl-β-D-thiogalactopyranoside. Cells were harvested 4 h after induction and lysed by French press. Inclusion bodies were washed multiple times by resuspending them in 50 mM Tris (pH 8), 50 mM NaCl, 5 mM EDTA, 2 M urea, and 0.5% Triton X-100 with subsequent centrifugation. After washing, they were dissolved in 50 mM Tris (pH 7.5) with 8 M urea and 10 mM dithiothreitol. The protein was purified under denaturing conditions with 8 M urea on a cation-exchange chromatography column (SP-Sepharose Fast Flow, 10 mL, GE Healthcare) with a linear gradient of increasing NaCl concentration from 0 to 500 mM within 10 column volumes. For refolding, the HIV-1 protease-containing fractions were pooled and dialyzed against 30 mM formic acid (pH 2.8). Refolding was done according to Louis et al. (2003). Briefly, the protein solution with 0.3 mg/mL in 30 mM formic acid (pH 2.8) was rapidly diluted with 5 vol of 10 mM sodium acetate (pH 6) and dialyzed overnight against 20 mM sodium phosphate buffer (pH 5.8) at ambient temperature. The amount of free-sulfhydryl groups per protease monomer was estimated by Ellman's reagent (Ellman 1959) after the protein concentration was quantified by light absorption at 280 nm using an extinction coefficient of ɛ(280 nm) = 12,780 M−1 cm−1.
Preparation of pepsin and pepsin C-domain
Porcine pepsin was purchased as lyophilized powder with lactose as stabilizer. It was dissolved in 50 mM 2-morpholin-4-ylethanesulfonic acid (MES) (pH 6.5), 100 mM NaCl, and lactose was removed by a NICK or PD10 desalting column (GE Healthcare). For preparation of the pepsin C-domain, pepsin powder was dissolved in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.3), 100 mM NaCl, and lactose was removed as described. The solution was kept at pH 7.3 for 2 h to let the N-domain denature, and subsequently, proteinase K was added in a mass ratio 1:1000 relative to pepsin and incubated at ambient temperature for 30 min. After adding phenylmethylsulfonylfluoride, proteinase K was separated from the pepsin C-domain by anion-exchange chromatography on an anion-exchange MonoQ-column (1 mL, GE Healthcare). Finally, the pooled fractions of pepsin C-domain were dialyzed overnight against 30 mM HEPES (pH 7.3), 100 mM KCl.
Preparation of thermolysin
Lyophilized thermolysin was dissolved in 30 mM HEPES (pH 7.3), 100 mM KCl, 2 mM 1,10-phenanthroline with a final concentration of 10 μM and dialyzed overnight at ambient temperature against the same buffer.
Preparation of trypsin S200A mutant
pET3a-plasmid carrying the sequence of human trypsinogen was used as a PCR template for trypsin-protease constructs. This plasmid was a gift of Diethardt Mattanovich (University of Natural Resources and Applied Life Sciences, Vienna, Austria). The S200A mutation was introduced with the site-directed mutagenesis kit (Stratagene) and further transformed into E. coli BL21 (DE3) Gold cells for overexpression. Purification and refolding of trypsinogen S200A was done according to Hohenblum et al. (2004) with an additional chromatographic step after preparation of washed inclusion bodies. Cation-exchange chromatography (SP-Sepharose Fast Flow, 10 mL, GE Healthcare) was done under denaturing conditions in 8 M Urea and a linear gradient of increasing NaCl concentration for elution. Refolded trypsinogen S200A was processed to trypsin with enterokinase according to the manufacturer's instructions (Roche Diagnostics). After stopping with phenylmethylsulfonylfluoride, the protein solution was applied onto a Superdex G75 26/60 gel-size exclusion chromatography column (GE Healthcare) in 50 mM Tris (pH 8), 25 mM CaCl2 for final purification.
Aggregation-prevention assays
Protein aggregation was measured as the increase of attenuance at 340 nm in a Lambda 25 spectrophotometer (Perkin Elmer) equipped with a thermostated cell holder. Aggregation was promoted either by chemical or thermal denaturation. All signals were referred to the plateau attenuance, set as 100% aggregation, of a reference sample with denatured substrate protein alone. Chemical denaturation of rhodanese was done with 6 M guanidinium chloride and aggregation was initiated by rapid dilution in 100 vol of assay buffer, yielding a final concentration of rhodanese of 1 μM. For thermal denaturation, citrate synthase was incubated at 50°C with a concentration of 0.3 μM; however, in the presence of trypsin S200A, incubation for thermal denaturation was done at 43°C. Assays with pepsin were done in 50 mM MES (pH 6.5), 100 mM NaCl; with pepsin C-domain in 30 mM HEPES (pH 7.3), 100 mM KCl; with thermolysin in 30 mM HEPES (pH 7.3), 100 mM KCl, 2 mM 1,10-phenanthroline; with trypsin S200A in 50 mM Tris (pH 8), 25 mM CaCl2 in case of thermal denaturation and in 30 mM HEPES (pH 7.3), 100 mM KCl, 25 mM CaCl2 in case of chemical denaturation; with HIV1-protease in 20 mM sodium phosphate buffer (pH 5.8). To exclude that a decrease in attenuance was caused by proteolytic degradation of the substrate proteins, total protein was precipitated afterward with trichloroacetic acid. Neutralized samples were applied on SDS-PAGE. Band densities were compared after Comassie Brilliant Blue staining with samples lacking inactive protease. When competitive inhibitors were used, inhibitor stock solution (2 mg/mL in ethanol) was added to the assay-mixture 5 min prior to adding substrate, resulting in final concentrations of 35 μM for pepstatin and 62 μM for HIV-1 protease inhibitor Ac-Leu-Val-Phe-Aldehyd, if not otherwise stated.
Complex isolation by gel-size exclusion chromatography
A total of 10 μL of citrate synthase ammonium sulfate suspension (9 mg/mL yielding 3.6 μM final concentration) was incubated for 60 min with 500 μL 1 mg/mL pepsin (corresponding to 33 μM) in 50 mM MES (pH 6.5), 100 mM NaCl, either at ambient temperature or at 50°C. Samples were centrifuged to remove precipitation and supernatants were analyzed by gel-size exclusion chromatography (Superose 6, GE Healthcare) in 50 mM MES (pH 6.5), 100 mM NaCl. Aliquots of collected fractions were analyzed by SDS-PAGE, with subsequent silver staining. This experiment has been repeated with rhodanese in place of citrate synthase. The conditions matched exactly those that have been used with pepsin and citrate synthase, except that 20 μL of 6 mg/mL rhodanese were added to 500 μL of pepsin, yielding 7.2 μM of final concentration of rhodanese.
Electronic Supplemental Material
The Electronic Supplemental Material contains two figures. The first one shows the results of the complex isolation experiment with pepsin and rhodanese by gel-size exclusion chromatography. The second one shows aggregation prevention assays with proteolytically inactivated thermolysin and trypsin.
Acknowledgments
We thank Felix Mueller-Sarnowski (Max von Pettenkofer-Institute, Munich, Germany) for the pBD2-plasmid carrying the coding sequence of HIV-1 protease and Diethardt Mattanovich (University of Natural Resources and Applied Life Sciences, Vienna, Austria) for the plasmid with human trypsinogen.
Footnotes
Supplemental Material: see www.proteinscience.org
Reprint requests to: Jörg Martin, Department of Protein Evolution, Max-Planck-Institute for Developmental Biology, Spemannstrasse 35, D-72076 Tübingen, Germany; e-mail: joerg.martin@tuebingen.mpg.de; fax: 49-7071-601-349.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062478607.
References
- Castillo R.M., Mizuguchi, K., Dhanaraj, V., Albert, A., Blundell, T.L., and Murzin, A.G. 1999. A six-stranded double-ψ β barrel is shared by several protein superfamilies. Structure 7: 227–236. [DOI] [PubMed] [Google Scholar]
- Coles M., Diercks, T., Liermann, J., Groger, A., Rockel, B., Baumeister, W., Koretke, K.K., Lupas, A., Peters, J., and Kessler, H. 1999. The solution structure of VAT-N reveals a ‘missing link’ in the evolution of complex enzymes from a simple β α β β element. Curr. Biol. 9: 1158–1168. [DOI] [PubMed] [Google Scholar]
- Davies D.R. 1990. The structure and function of the aspartic proteinases. Annu. Rev. Biophys. Biophys. Chem. 19: 189–215. [DOI] [PubMed] [Google Scholar]
- Dunn B.M., Goodenow, M.M., Gustchina, A., and Wlodawer, A. 2002. Retroviral proteases. Genome Biol. 3: REVIEWS3006. [DOI] [PMC free article] [PubMed]
- Ellman G.L. 1959. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82: 70–77. [DOI] [PubMed] [Google Scholar]
- Fujinaga M., Chernaia, M.M., Tarasova, N.I., Mosimann, S.C., and James, M.N.G. 1995. Crystal-structure of human pepsin and its complex with pepstatin. Protein Sci. 4: 960–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerega A., Rockel, B., Peters, J., Tamura, T., Baumeister, W., and Zwickl, P. 2005. VAT, the Thermoplasma homolog of mammalian p97/VCP, is an N domain-regulated protein unfoldase. J. Biol. Chem. 280: 42856–42862. [DOI] [PubMed] [Google Scholar]
- Golbik R., Lupas, A.N., Koretke, K.K., Baumeister, W., and Peters, J. 1999. The janus face of the archaeal Cdc48/p97 homologue VAT: Protein folding versus unfolding. Biol. Chem. 380: 1049–1062. [DOI] [PubMed] [Google Scholar]
- Grishin N.V. 2001. Fold change in evolution of protein structures. J. Struct. Biol. 134: 167–185. [DOI] [PubMed] [Google Scholar]
- Hartl F.U. and Hayer-Hartl, M. 2002. Protein folding - molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295: 1852–1858. [DOI] [PubMed] [Google Scholar]
- Haslbeck M., Franzmann, T., Weinfurtner, D., and Buchner, J. 2005. Some like it hot: The structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 12: 842–846. [DOI] [PubMed] [Google Scholar]
- Hohenblum H., Vorauer-Uhl, K., Katinger, H., and Mattanovich, D. 2004. Bacterial expression and refolding of human trypsinogen. J. Biotechnol. 109: 3–11. [DOI] [PubMed] [Google Scholar]
- Kageyama T. 2002. Pepsinogens, progastricsins, and prochymosins: Structure, function, evolution, and development. Cell. Mol. Life Sci. 59: 288–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.Z., Fusek, M., Lin, X.L., Hartsuck, J.A., Kezdy, F.J., and Tang, J. 1992. Ph-dependence of kinetic-parameters of pepsin, rhizopuspepsin, and their active-site hydrogen-bond mutants. J. Biol. Chem. 267: 18413–18418. [PubMed] [Google Scholar]
- Lin X.L., Loy, J.A., Sussman, F., and Tang, J. 1993. Conformational instability of the N-terminal and C-terminal lobes of porcine pepsin in neutral and alkaline-solutions. Protein Sci. 2: 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.L., Lin, Y.Z., and Tang, J. 1994. Relationships of human-immunodeficiency-virus protease with eukaryotic aspartic proteases. Methods Enzymol. 241: 195–224. [DOI] [PubMed] [Google Scholar]
- Lindqvist Y. and Schneider, G. 1997. Circular permutations of natural protein sequences: Structural evidence. Curr. Opin. Struct. Biol. 7: 422–427. [DOI] [PubMed] [Google Scholar]
- Louis J.M., Ishima, R., Nesheiwat, I., Pannell, L.K., Lynch, S.M., Torchia, D.A., and Gronenborn, A.M. 2003. Revisiting monomeric HIV-1 protease - characterization and redesign for improved properties. J. Biol. Chem. 278: 6085–6092. [DOI] [PubMed] [Google Scholar]
- Mogk A., Schlieker, C., Friedrich, K.L., Schofeld, H.J., Vierling, E., and Bukau, B. 2003. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 278: 31033–31042. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. 1998. How far divergent evolution goes in proteins. Curr. Opin. Struct. Biol. 8: 380–387. [DOI] [PubMed] [Google Scholar]
- Murzin A.G., Brenner, S.E., Hubbard, T., and Chothia, C. 1995. Scop - a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247: 536–540. [DOI] [PubMed] [Google Scholar]
- Neurath H. 1984. Evolution of proteolytic-enzymes. Science 224: 350–357. [DOI] [PubMed] [Google Scholar]
- Orengo C.A., Jones, D.T., and Thornton, J.M. 1994. Protein superfamilies and domain superfolds. Nature 372: 631–634. [DOI] [PubMed] [Google Scholar]
- Orengo C.A., Michie, A.D., Jones, S., Jones, D.T., Swindells, M.B., and Thornton, J.M. 1997. CATH - a hierarchic classification of protein domain structures. Structure 5: 1093–1108. [DOI] [PubMed] [Google Scholar]
- Park S., Isaacson, R., Kim, H.T., Silver, P.A., and Wagner, G. 2005. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure 13: 995–1005. [DOI] [PubMed] [Google Scholar]
- Privalov P.L., Mateo, P.L., Khechinashvili, N.N., Stepanov, V.M., and Revina, L.P. 1981. Comparative thermodynamic study of pepsinogen and pepsin structure. J. Mol. Biol. 152: 445–464. [DOI] [PubMed] [Google Scholar]
- Quemeneur E., Moutiez, M., Charbonnier, J.B., and Menez, A. 1998. Engineering cyclophilin into a proline-specific endopeptidase. Nature 391: 301–304. [DOI] [PubMed] [Google Scholar]
- Rao J.K.M., Erickson, J.W., and Wlodawer, A. 1991. Structural and evolutionary relationships between retroviral and eukaryotic aspartic proteinases. Biochemistry 30: 4663–4671. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D. and Barrett, A.J. 1999. MEROPS: The peptidase database. Nucleic Acids Res. 27: 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. and Lupas, A.N. 2003. More than the sum of their parts: On the evolution of proteins from peptides. Bioessays 25: 837–846. [DOI] [PubMed] [Google Scholar]
- Tang J., Sepulved, P., Marcinis, J., Chen, K.C.S., Huang, W.Y., Tao, N., Liu, D., and Lanier, J.P. 1973. Amino-acid sequence of porcine pepsin. Proc. Natl. Acad. Sci. 70: 3437–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., James, M.N.G., Hsu, I.N., Jenkins, J.A., and Blundell, T.L. 1978. Structural evidence for gene duplication in evolution of acid proteases. Nature 271: 618–621. [DOI] [PubMed] [Google Scholar]
- Thoms S. 2002. Cdc48 can distinguish between native and non-native proteins in the absence of cofactors. FEBS Lett. 520: 107–110. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Miller, M., Jaskolski, M., Sathyanarayana, B.K., Baldwin, E., Weber, I.T., Selk, L.M., Clawson, L., Schneider, J., and Kent, S.B.H. 1989. Conserved folding in retroviral proteases - crystal-structure of a synthetic Hiv-1 protease. Science 245: 616–621. [DOI] [PubMed] [Google Scholar]
- Young J.C., Agashe, V.R., Siegers, K., and Hartl, F.U. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5: 781–791. [DOI] [PubMed] [Google Scholar]