Abstract
Amyloid diseases, including Alzheimer's and prion diseases, are each associated with unbranched protein fibrils. Each fibril is made of a particular protein, yet they share common properties. One such property is nucleation-dependent fibril growth. Monomers of amyloid-forming proteins can remain in dissolved form for long periods, before rapidly assembly into fibrils. The lag before growth has been attributed to slow kinetics of formation of a nucleus, on which other molecules can deposit to form the fibril. We have explored the energetics of fibril formation, based on the known molecular structure of a fibril-forming peptide from the yeast prion, Sup35, using both classical and quantum (density functional theory) methods. We find that the energetics of fibril formation for the first three layers are cooperative using both methods. This cooperativity is consistent with the observation that formation of amyloid fibrils involves slow nucleation and faster growth.
Keywords: amyloid energetics, Sup35, GNNQQNY, density functional theory
Elongated, unbranched protein fibrils, each formed from a different type of protein monomer, are associated with some 25 “amyloid” diseases, including Alzheimer's and prion diseases. Similar “amyloid-like” fibrils are often formed when normal soluble proteins are transferred to destabilizing solvents. The atomic structures of some amyloid-like fibrils (Makin and Serpell 2005; Nelson et al. 2005) have recently been revealed, but little is known about their energetics of formation, other than that their growth from monomers is slow until a nucleus is introduced (Lomakin et al. 1996).
Computationally, amyloid formation has previously been studied by molecular dynamics (MD) simulations. Pellarin and Caflisch (2006) suggest that the fibrillogenesis is mainly dependent on the relative stability of an “amyloid-competent state” of the monomer. According to these investigators, peptides predominantly in that state form fibrils readily and without intermediates. Conversely, in the amyloid-protected state, the kinetics of protein aggregation are slow. Nussinov et al. (Zheng et al. 2006) argue that aggregation is caused by maximizing the van der Waals interactions between side-chains, and by backbone hydrogen bonds. In addition, they argue that shape complementarity between neighboring molecules plays a key role. Ma and Nussinov (2006) attribute the driving force of aggregation to side-chain interactions that are mostly hydrophobic but note that polar side-chains can also form additional hydrogen bonds. Simulations support the proposal of Nelson et al. (2005) that the minimal nucleus seed for fibril formation consists of only three to four peptides. Larger oligomers were found not to disassociate quickly due to slow diffusion coefficients. While these previous studies have provided important insights into the process of fibril formation, they have employed nonpolarizable fixed charge models and thus do not report on the possible contribution of hydrogen bond polarization to the cooperativity of amyloid formation.
Here we show that the strength of hydrogen bonds between layers of fibrils increases nonlinearly up to four layers and then levels off. Thus, the hydrogen bonding interactions within the β-sheets of the amyloid structure are cooperative, with contributions to the energy of binding from several layers away within the fibril. This cooperativity of hydrogen bonding probably contributes to the well-established pattern of nucleation-dependent growth of amyloid fibrils.
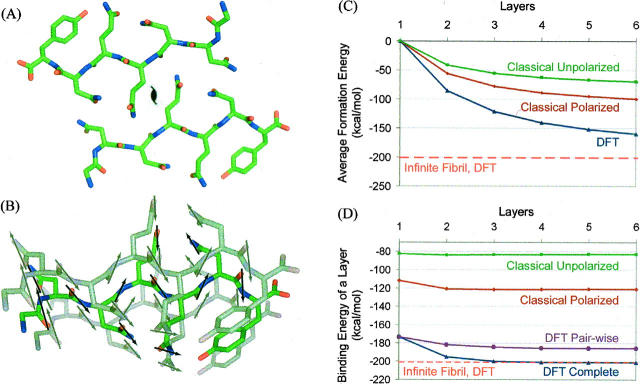
Our computations are based on the accurately known atomic structure of the heptameric peptide with amino acid sequence GNNQQNY from the yeast prion Sup35 (Fig. 1A; Nelson et al. 2005). Peptide molecules are extended and stacked as pairs into two β-sheets, with the stack forming the needle axis of the fibril. Each peptide forms 11 hydrogen bonds with both the peptides above and below it in the stacked sheet. Of these 11 hydrogen bonds, six are between backbone amide groups and five are between amide side-chains, bonding to the same side-chain above and below it. Two such sheets face each other, with their side-chains tightly interdigitating. That is, one layer of the fibril-like structure consists of two peptides (Fig. 1A).
Figure 1.
(A) One layer of two GNNQQNY molecules, viewed down the fibril axis, showing arrows for each amide dipole. (B) Three layers of one of the two β-sheets of the GNNQQNY fibril, showing the stacking of amide dipoles. (C) Energy per two-peptide layer in an n-layer fibril minus the self-energy of the layer, giving the stability of an n-layer fibril. The dashed line corresponds to the binding energy of one layer in an infinite fibril calculated using DFT. (D) Binding energy of a two-peptide layer to an n-layer fibril, showing the cooperativity of fiber formation. The dashed line corresponds to the binding energy of one layer in an infinite fibril calculated using DFT.
For the ab initio quantum mechanical calculations, we used the density functional theory (DFT). DFT is known to work well for hydrogen-bonded systems (Morozov et al. 2004 and references therein) and is much more feasible for large molecules or crystals than the traditional wave-function–based quantum chemistry methods (Parr and Yang 1989). It is implemented for both finite and infinite periodic systems, and with growing computer power and code parallelization, systems of >1000 atoms have been studied using DFT.
Results
The DFT result for the energy per layer in the infinite crystal is −201.1 kcal/mol. The energy per hydrogen bond for adhesion of peptide molecules to the amyloid can be estimated by dividing by the number of hydrogen bonds per layer (2 × 11 = 22), or −9.1 kcal/mol-of-hydrogen bond. This is somewhat larger than the hydrogen bond energy of ice, −6.7 kcal/mol-of-hydrogen bond (Eisenberg and Kauzmann 2005). Two pairs of peptides have a binding energy of just −7.8 kcal/mol-of-hydrogen bond, which is 1.3 kcal/mol smaller than the same energy for binding to the infinite crystal. Coordinates from the known atomic structure of GNNQQNY (Protein Data Bank [PDB] entry 1YJP) were used for all calculations; it should be noted that the interactions in the fibril will differ somewhat from the interactions in the crystal as the β-sheet in the fibril, unlike the flat sheets in the crystal, presumably has a twist.
To determine the origins of the increase in binding energy in the infinite crystal, two types of energy differences were computed. The first is the energy per monomer in a fibril of n layers (2n peptides), shown in Figure 1C. The second is the binding energy of a layer to a preexisting fibril of n layers, shown in Figure 1D. This second energy difference is more directly related to the cooperative effects in which we are interested.
The energy of adding a layer to a fiber with n layers was obtained by subtracting the energies of the one layer and n layer systems from that of the n + 1 layer fiber (Fig. 1D, blue lines). To isolate the electron density polarization from other contributions to the cooperativity of binding (such as differing numbers of H-bonds formed by edge strands), the binding energies between the added layer and each individual layer in the growing fiber were also computed separately and then summed (Fig. 1D, purple lines). The energy for adding a layer to a fiber clearly increases with increasing fiber length in both the classical and QM models up to a fiber length of four monomers and then levels off. In both the classical and QM models, the increase in binding energy results primarily from polarization of the electron density.
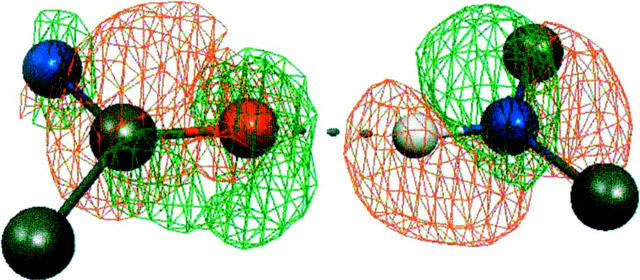
Figure 2 shows the electron density in a single O…H-N hydrogen bond, as determined by the DFT method. Upon polarization, the density shifts away from the hydrogen atom (red mesh) toward the oxygen atom (green mesh), indicating the formation of the hydrogen bond. In the classical electrostatic model, the increased polarization is reflected in the enhanced dipole moments of the amide groups in the zipper spine from about 3.5–4.5 Debye units.
Figure 2.
The DFT density change upon polarization for a single O…H-N hydrogen bond, with green mesh showing increased electron density around the oxygen atom, and red mesh showing diminished electron density around the hydrogen atom. Shown is the difference between the density in the infinite crystal and the density computed for isolated layers superimposed back onto the crystal structure.
Discussion
The calculations on seven-layer fibrils involve 1498 atoms, large for ab initio methods; the validity of these results is confirmed by the convergence of both the energy per monomer and the binding energy with increasing fiber length to the values for the infinite crystal (dashed lines), which are computed using a single layer with only 214 atoms with periodic boundary conditions to generate the crystal. It is notable that the crystalline symmetry here allows perhaps the first accurate computation of energies using ab initio quantum methods for a protein-like system.
The results with the classical and DFT methods are similar: In both, the energy per monomer increases nonlinearly for the first few layers of the growing fiber and then becomes linear for additional layers. Electron density polarization contributes significantly to this cooperativity (cf. the classical polarized to the unpolarized results, and the full DFT calculations to the pairwise layer results), and is illustrated in Figure 2. The stronger hydrogen bonding interactions in longer fibers may contribute to the nucleation growth kinetics observed experimentally.
Materials and Methods
Classical electrostatics method
In the classical electrostatics approach, each H-NCRHC=O amide group is represented by a point dipole μ with an initial dipole moment of 3.5 Debye units (Hol 1985). Each peptide contains 11 amide dipoles, whose poles form donors and acceptors for the H-bonds. The orientation of each dipole was defined using the atomic coordinates of the oxygen and nitrogen atoms for the negative and positive poles, respectively. The electric field E is summed from neighboring dipolar amide groups, and the binding energy is computed as the sum of μ·E. In “unpolarized” calculations (green lines), each dipole is undistorted by the presence of other dipoles.
In the “polarized” calculations (red lines), each dipole becomes polarized by its neighbors. The gain of dipole–dipole energy created by mutual polarization of the dipoles is calculated iteratively. At each iteration, a new moment μ for each dipole is calculated using the initial electric field (calculated above) and the isotropic polarizability of the amide group, for which a value of 2.56 Å3 was used (Krimm 2001). The permanent plus induced dipole moment μ is equal to the sum of the initial dipole moment μ0 plus the product of the polarizability α of the amide group and the local electric field E (μ = μ0 + αE). This creates a larger dipole moment and hence a larger electric field for each iteration. The electric field typically converges by 10 iterations. The energy required to polarize each dipole was calculated by a harmonic approximation as (μ − μ0)2/2α (Coulson and Eisenberg 1966) and was subtracted from the sum of the dipole–dipole energies to obtain the net energy gain. The calculation was repeated for two, three, four, and five layers. Neighboring layers are related by translation of the unit cell in the GNNQQNY crystal.
Quantum mechanical method
In the quantum DFT-based approach, ground-state energies were computed (Fig. 1, blue lines) using coordinates from the relaxed crystal structure for fiber fragments with one to seven layers. The plane-wave implementation of DFT (VASP software suite) (Kresse and Hafner 1993) was used to simulate both infinite GNNQQNY fibrils and fragments with finite numbers of layers. The crystal structure was fully relaxed to obtain the electron density and the ground-state energy.
Finite fibrils were treated with the strands retaining their crystalline structure; structural optimization of each individual finite fibril yielded the same degree of cooperativity with somewhat lower absolute binding energies. We used Vanderbilt ultrasoft pseudopotentials and the Perdew-Wang 91 exchange-correlation functional. For fibrils composed of a finite number of layers (up to seven), we simulated systems periodic in the directions normal to the fibril axis, while along this axis the identical replicas were separated by a “vacuum” spacer with thickness triple the length of the lattice constant (close to 15 Å).
For the “pairwise layer” calculations, a similar vacuum spacer was used to compute energies of the added layer with each individual layer in the preexisting fiber, two, three, etc., lattice constants in the fibril axis direction away from this added layer. For the infinite β-sheet, the true periodicity of the crystal was imposed, and the result was converged with respect to the sampling of Brillouin zone along the fibril axis direction.
Acknowledgments
We thank HHMI and NIH for support.
Footnotes
Reprint requests to: David Baker, HHMI, Department of Biochemistry, University of Washington, Seattle, WA 98195, USA; e-mail: dabaker@u.washington.edu; fax: (206) 685-1792.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062609607.
References
- Coulson C.A. and Eisenberg, D. 1966. Interactions of H2O molecules in ice: The dipole moment of an H2O molecule in ice. Proc. R. Soc. Lond. 291: 445. [Google Scholar]
- Eisenberg D. and Kauzmann, W. 2005. The structure and properties of water. Oxford Classic Texts, Oxford, UK.
- Hol W. 1985. Effects of the α-helix dipole upon the functioning and structure of proteins and peptides. Adv. Biophys. 19: 133–165. [DOI] [PubMed] [Google Scholar]
- Kresse G. and Hafner, J. 1993. Ab initio molecular dynamics for liquid metals. Phys. Rev. B 47: 558–561. [DOI] [PubMed] [Google Scholar]
- Krimm S. 2001. A polarizable electrostatic model of the N-methylacetamide dimer. J. Comput. Chem. 22: 1933–1943. [Google Scholar]
- Lomakin A., Chung, D.S., Benedek, G.B., Kirschner, D.A., and Teplow, D. 1996. On the nucleation and growth of amyloid β-protein fibrils: Detection of nuclei and quantitation of rate constants. Proc. Natl. Acad. Sci. 93: 1125–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B. and Nussinov, R. 2006. Simulations as analytical tools to understand protein aggregation and predict amyloid conformation. Curr. Opin. Chem. Biol. 10: 445–452. [DOI] [PubMed] [Google Scholar]
- Makin O.S. and Serpell, L.C. 2005. Structures for amyloid fibrils. FEBS J. 272: 5950–5961. [DOI] [PubMed] [Google Scholar]
- Morozov A., Kortemme, T., Tsemekhman, K., and Baker, D. 2004. Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations. Proc. Natl. Acad. Sci. 101: 6946–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R., Sawaya, M.R., Balbirnie, M., Madsen, A.Ø., Riekel, C., Grothe, R., and Eisenberg, D. 2005. Structure of the cross-β spine of amyloid-like fibrils. Nature 435: 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr R. and Yang, W. 1989. Density functional theory of atoms and molecules. Oxford University Press, Oxford, UK.
- Pellarin P. and Caflisch, A. 2006. Interpreting the aggregation kinetics of amyloid peptides. J. Mol. Biol. 360: 882–892. [DOI] [PubMed] [Google Scholar]
- Zheng J., Ma, B., Tsai, C.J., and Nussinov, R. 2006. Structural stability and dynamics of an amyloid-forming peptide GNNQQNY from the yeast prion sup-35. Biophys. J. 91: 824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]