Abstract
A key step in the signaling cascade responsible for activation of the transcription factor NF-κB involves Lys63-linked polyubiquitination of TRAF6. Covalent attachment of ubiquitin (Ub) to TRAF6, and subsequent poly(Ub) chain synthesis, is catalyzed by the hUev1a–hUbc13 heterodimer. hUbc13 is a catalytically competent E2 enzyme, and hUev1a is an E2-like protein that binds substrate Ub. The hUev1a–hUbc13 heterodimer is targeted to TRAF6 through interactions between hUbc13 and the N-terminal RING domain from TRAF6. Nuclear magnetic resonance (NMR) spectroscopy was used to determine the solution state structure of the RING domain from human TRAF6, and the interaction between hUbc13 and TRAF6 was characterized using NMR chemical shift mapping. The main-chain dynamics of the RING domain from TRAF6 were studied using 15N NMR relaxation. Analysis of the main-chain dynamics data indicates that residues within the α-helix and β-sheet of the RING domain are as rigid as regions of canonical secondary structure in larger proteins, consistent with the biological role of RING-domain E3 proteins, which requires that the E3 contain a recognition site for recruitment of E2 ubiquitin conjugation enzymes.
Keywords: TRAF6, E3 ubiquitin ligase, Ubc13, Uev1a, polyubiquitination, ubiquitin
TRAF proteins are key signal transducers for the TNF receptor superfamily and the interleukin-1/Toll-like receptors (Bradley and Pober 2001; Bodmer et al. 2002; Chung et al. 2002). TRAF-binding receptors recruit TRAF proteins and play important roles in the activation of cells, cell differentiation, immunity, and signaling for survival (Liu 2004; Watts 2005). TRAF2 and TRAF6 have been widely studied because of their involvement in the activation of NF-κB (Kobayashi et al. 2004; Chen 2005), a transcription factor that activates genes involved in the cell cycle, differentiation, apoptosis, and the immune response (Ghosh et al. 1998; Garg and Aggarwal 2002; Viatour et al. 2005).
The C-terminal MATH domain from TRAF2 and TRAF6 is involved in interactions with the cytoplasmic domains of TNF receptors (Wu and Arron 2003). The structure of the MATH or TRAF domain from TRAF6 has been determined using X-ray crystallography and forms an eight-stranded antiparallel β-sandwich (PDB: 1LB6) (Ye et al. 2002). The structure of the C-terminal region of TRAF2 is a trimer with a “mushroom” shape, whose self-association is due in part to the coiled-coil region, which forms the stalk of the mushroom (PDB: 1CA4) (Park et al. 1999). Peptides from the TNF receptor have been shown to interact with the TRAF domain from TRAF2 (Park et al. 1999). The molecular basis for specificity differences between TRAF6 and other TRAF domains is believed to be due to different binding orientations of TNF receptor peptides to the TRAF domain from TRAF6 compared to peptide binding to TRAF2 (Ye et al. 2002).
The N-terminal region of TRAF6 is critical for signaling and contains three Zn2+ binding domains of unknown structure, with the first Zn2+-binding region containing a RING-domain consensus sequence (∼60 residues) (Regnier et al. 1995). Recently, TRAF6 has been identified as a substrate for covalent attachment of Lys63 poly(Ub) chains, and also functions as an E3 Ub ligase in conjunction with the E2 hUbc13 (∼150 residues), most likely through the N-terminal RING domain (Deng et al. 2000; Wang et al. 2001). TRAF6 and hUbc13 have been shown to be required for the activation of NF-κB through polyubiquitination of the NF-κB essential modulator NEMO (Zhou et al. 2004). A yeast two-hybrid assay has been used to demonstrate the interaction between the RING domain from TRAF6 and the E2 enzyme hUbc13 (Wooff et al. 2004).
The RING domain from TRAF proteins differs in comparison to the typical C3HC4 “Cross-Brace” motif (Takahashi et al. 1988; Barlow et al. 1994; Borden and Freemont 1996) in that the C-terminal Cys involved in Zn2+-ligation is an Asp residue (Regnier et al. 1995). Sequence differences that may have an impact on the RING-domain interaction with E2 enzymes include a conserved Trp residue involved in the E2–RING interface for c-Cbl–UbcH7 (Trp-408 in the c-Cbl RING domain) that is replaced by an exclusively polar residue (Ser) for the RING domain from TRAF6. Furthermore, a surface-exposed negatively charged residue in Zn2+ binding loop 1 from the RING domain of c-Cbl is substituted with a hydrophobic Met residue for TRAF6.
From a structural perspective, the structure of c-Cbl bound to UbcH7 determined by X-ray crystallography is one of the few known structures for E2–E3 complexes (Zheng et al. 2000), and may provide an example for the interaction between hUbc13 and the RING domain from TRAF6 (TRAF6-RD). RING domains have been found to associate as heterodimers (BRCA1–BARD1) (Brzovic et al. 2001), interact with an E2 as a single domain (c-Cbl–UbcH7) (Brzovic et al. 2001), and as single RING domains in the SCF E3 Ub-protein ligase complex (Zheng et al. 2002). For these cases, the RING domains share a common E2-binding motif. In particular, the E2-binding site is centered on a shallow hydrophobic groove that passes between the Zn2+-binding loops. For the c-Cbl–UbcH7 interaction, a key structural aspect of the binding site for the RING domain of the E2 enzyme is a hydrophobic patch on a loop joining helices α2 and α3 of UbcH7. The interaction between the RING domain from CNOT4 and UbcH5B deduced from NMR chemical shift mapping and subsequent model building is similar to that observed for c-Cbl/UbcH7 (Dominguez et al. 2004). These studies raise a fundamental question underlying the protein ubiquitination cycle in humans: It is not known how ∼80 E2 enzymes and ∼500 E3 ubiquitin ligases containing RING domains discriminate among each other to achieve different biological outcomes. Regulation of E2/E3 expression within tissue types and cellular localization may play a role in E2–E3 specificity. In addition, specificity can also be achieved directly at the molecular level through sequence differences within and at the periphery of E2–E3-binding sites. The latter contribution to specificity is beginning to be assessed through detailed structural and binding studies using X-ray crystallography and NMR spectroscopy (Zheng et al. 2000; Dodd et al. 2004; Dominguez et al. 2004; Kellenberger et al. 2005).
In this study, we have used high-resolution, solution state NMR spectroscopy to determine the solution structure of the C4HC2D RING domain from TRAF6 and interactions with its cognate E2, hUbc13. Additionally, the main-chain dynamics of the RING domain from TRAF6 were assessed using 15N NMR relaxation.
Results
Solution structure of the RING domain from TRAF6-RD
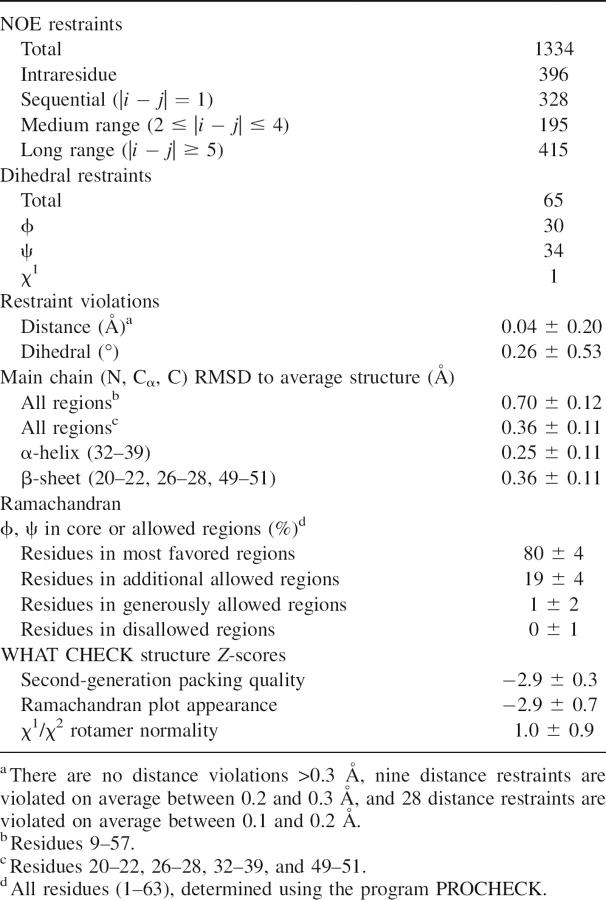
The solution structure of TRAF6-RD was determined using high-resolution, solution state 1H, 13C, 15N-NMR spectroscopy. The characteristics of the solution structures for TRAF6-RD are shown in Table 1. The main chain of the protein is well defined, superimposing with an RMSD of 0.7 Å over residues 9–57 (Table 1; Fig. 1). The side chains from the core of the protein, residues 9–57, are well defined, superimposing with an RMSD of 1.87 Å. Secondary structural elements include a short α-helix (residues 32–39) and a β-sheet (residues 20–22, 26–28, and 49–51), as determined using the program VADAR (Willard et al. 2003). These secondary structural elements contribute to the hydrophobic core of the protein (Fig. 1). There are two loops involved in binding two Zn2+ atoms (L1: residues 9–16; L2: residues 44–47). Residues whose side chains are directly involved in coordinating Zn2+ include Cys9, Cys12, Cys29, and Cys32 for one Zn2+ atom, and residues Cys24, His26, Cys44, and Asp47 for the second Zn2+ atom.
Table 1.
Structural statistics for TRAF6-RD
Figure 1.
Superposition of the ensemble of 50 NMR structures for TRAF6-RD. Traces through the Cα atoms of TRAF6-RD are shown in the ribbon representation and superimposed from residues 9–57. Key elements of secondary structure are labeled, including the Zn2+-binding loops L1 and L2 (residues 9–16 and 44–47, respectively), α-helix (residues 32–39), and β-sheet (20–22, 26–28, 49–51). Zn2+ atoms are shown as spheres.
Zn2+ ligation for cysteine residues in TRAF6-RD
13Cα/Cβ chemical shifts from Cys residues were used to calculate the probabilities that these side chains are involved in Zn2+ ligation (Kornhaber et al. 2006). For site 1, the probabilities that Cys is coordinated to Zn2+ are 0.65, 0.97, 0.98, and 0.99 for Cys9, 12, 29, and 32, respectively. For site 2, the probabilities are 0.004 and 0.58 for Cys24 and 44, respectively. Thus, on the basis of these indirect chemical shift results, site 1 appears to be coordinated to Zn2+, whereas for site 2, only Cys44 has a >50% probability that it is coordinated to Zn2+. However, for site 2, two residues involved in Zn2+ ligation (His26, Asp47) are not cysteine residues and are not amenable to the above chemical shift analysis.
Titration of [U-15N; U-13C]-hUbc13 with TRAF6-RD
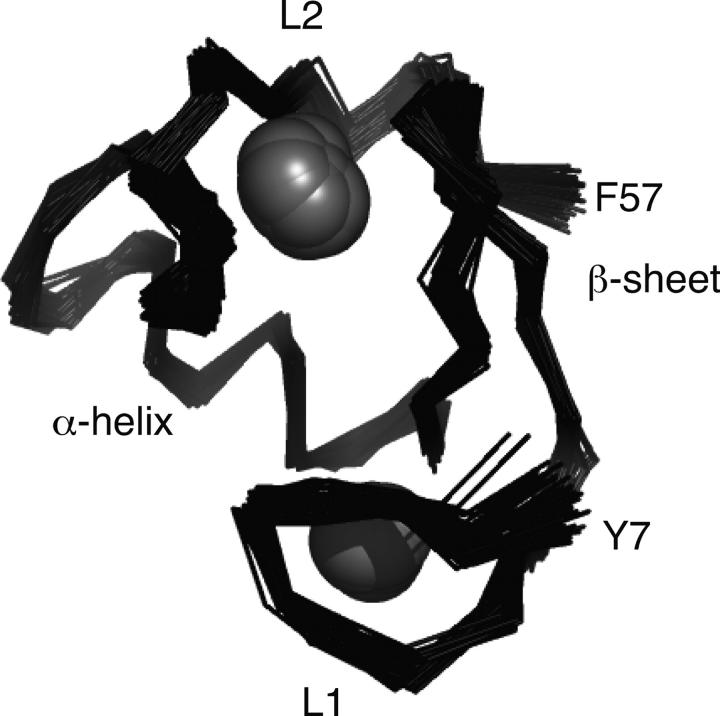
Backbone amide 1HN and 15N chemical shift changes for [U-15N; U-13C]-hUbc13 upon titration with the RING domain from TRAF6 are shown in Figure 2. Only 12 of 116 observable backbone amide 1HN–15N chemical shifts exhibit a significant change, suggesting that conformational changes in hUbc13 upon binding the RING domain from TRAF6 are minimal. The interaction between hUbc13 and TRAF6 is weak, given the high TRAF6/hUbc13 ratio that is required to perturb hUbc13 1HN–15N chemical shifts. Changes for the backbone amide 1HN and 15N chemical shifts of Ile101 were used to estimate a KD value of 1.9 ± 0.1 mM. Methyl group 1H and 13C chemical shift changes for [U-15N; U-13C]-hUbc13 upon titration with TRAF6-RD are shown in Figure 3. Residues Met64 and Ala98 are the only residues observed to undergo significant chemical shift changes, suggesting modest conformational changes for hUbc13 upon binding TRAF6-RD, consistent with the results for backbone amide 1HN and 15N chemical shift changes.
Figure 2.
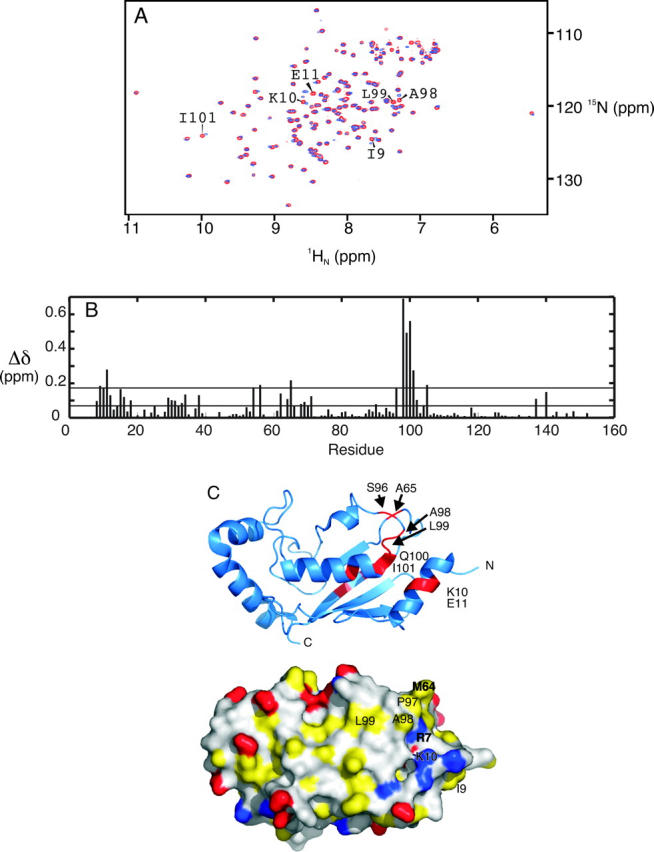
hUbc13 backbone amide chemical shift changes during titration with TRAF6-RD. (A) Superposition of hUbc13 1HN–15N HSQC NMR spectra collected for [TRAF6]/[hUbc13] ratios: (red) 0:1, (blue) 7.2:1. Various cross-peaks affected by complex formation are labeled according to residue number. (B) Per-residue plot of chemical shift perturbations for 7.2:1 [TRAF6]/[hUbc13]. The mean (0.07) and cutoff for one standard deviation from the mean (0.17) are included. (C) Cartoon (top) and surface (bottom) representations of hUbc13. For the cartoon representation, residues that experience chemical shift perturbation greater than one standard deviation from the mean upon titration of TRAF6 are red. The surface representation is colored with negatively charged atoms from Glu and Asp in red; positively charged atoms from Lys, His, and Arg in blue; and side-chain atoms from hydrophobic residues (Ala, Val, Leu, Ile, Phe, Tyr, Met, and Pro) in yellow. Atoms from remaining residues and those from the main chain are white.
Figure 3.
hUbc13 methyl group chemical shift changes during titration with TRAF6-RD. (A) Superposition of hUbc13 1H–13C HSQC NMR spectra collected for [TRAF6]/[hUbc13] ratios of 0:1 (black) and 7:1 (red). (B, inset) An expansion of the spectra around Met64; (C, inset) an expansion of the spectra around Ala98. For the [TRAF6]/[hUbc13] ratio of 7:1, some methyl group chemical shifts for TRAF6 arising from natural abundance 13C are observed (TRAF6-RD Met14 just below the label for hUbc13 Met64).
Titration of [U-15N]-TRAF6 with hUbc13
Backbone amide 1HN and 15N chemical shift changes for the [U-15N]-TRAF6 RING domain upon titration with hUbc13 are shown in Figure 4. Eight of 55 observable backbone amide 1HN–15N chemical shifts show a significant change upon binding of hUbc13. The uncharacterized residues that were not observed were either prolines, N-terminal residues that are presumed to be in rapid exchange with water, or exchange broadened (His42), or are partially/completely overlapped in the 2D 1H–15N HSQC NMR spectra. The small magnitude of the observed chemical shift differences suggests that conformational changes in TRAF6-RD upon binding hUbc13 are small.
Figure 4.
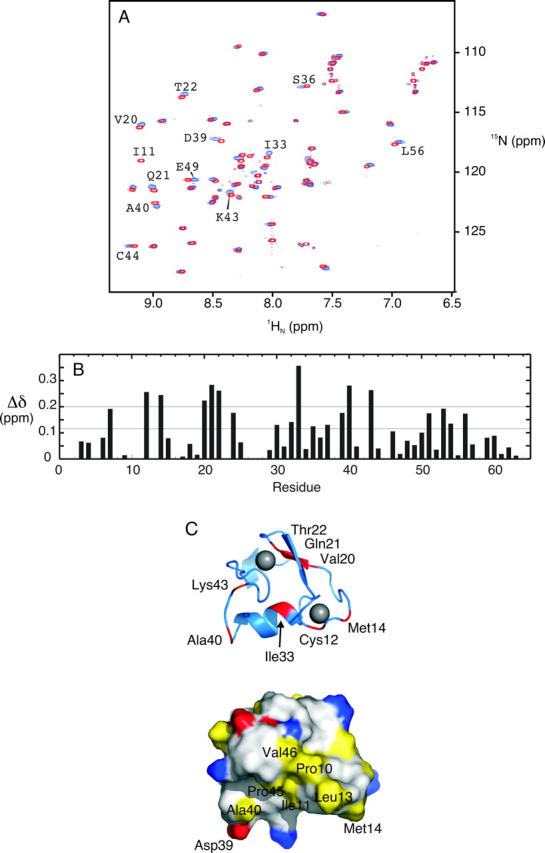
TRAF6-RD backbone amide chemical shift changes during titration with hUbc13. (A) Superposition of TRAF6-RD 1HN–15N HSQC spectra collected for TRAF6 alone (red) and ∼1:4 TRAF6:hUbc13 (blue), with various resonances labeled according to residue number. (B) Per-residue plot of chemical shift perturbation. The mean (0.11 ppm) and cutoff for one standard deviation from the mean (0.20 ppm) are included. (C) Cartoon (top) and surface (bottom) representation of the core residues (9–57) from the RING domain from TRAF6. For the cartoon representation, residues that experience chemical shift perturbation greater than one standard deviation from the mean upon titration with hUbc13 are red. Zn2+ ions are gray spheres. The surface representation is colored according to the scheme outlined in the legend to Figure 2.
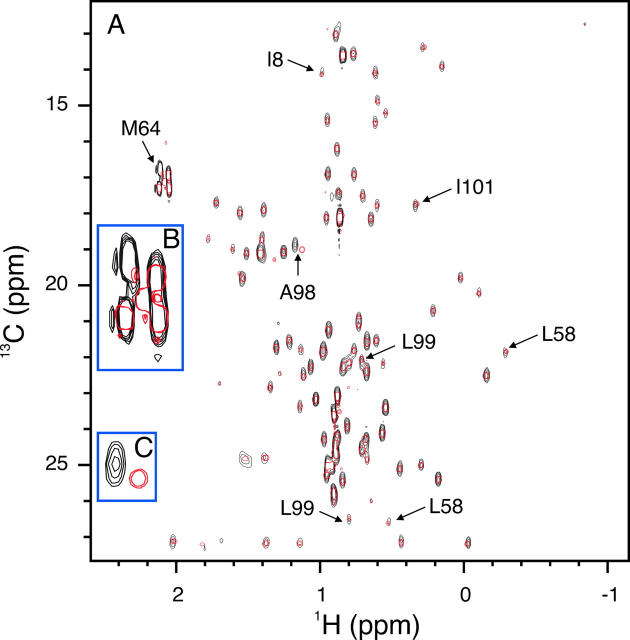
Main-chain dynamics: 15N-T1, T2, and NOE data
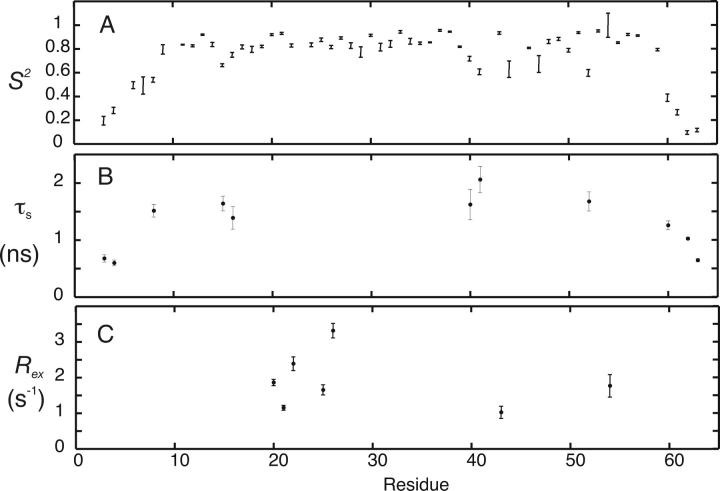
15N NMR relaxation data for 55 of a total of 63 residues were obtained. Uncharacterized residues are the same as those in the titration of [U-15N]-TRAF6-RD with hUbc13. The average R 1 for the core residues (9–57) is 2.2 ± 0.1 sec−1, and the average error is 0.03 sec−1 (Fig. 5A). The average R 2 for core residues is 7 ± 1 sec−1 (Fig. 5A) with an average error of 0.08 sec−1. The average NOE for the core residues is 0.71 ± 0.04 with an average error of 0.02 (Fig. 5B). The average core values of 15N-R 1, R 2, and NOE match the theoretical R 1 and R 2 values of 2.1 and 6.1 sec−1, respectively, and the NOE value of 0.74 determined using a τm value of 4.3, with S 2 = 0.85 and τf = 25 psec. The τm value was determined from the 15N R 2/R 1 ratio as previously described (Kay et al. 1989; Spyracopoulos et al. 2005). The τm value for TRAF6-RD is shorter than that for residues 1–63 of the RING domain from CNOT4 (5.7 nsec) (Houben et al. 2005). If the τm for TRAF6-RD is multiplied by the ratio of the viscosity of water at 20°C and 25°C (1.12) (Lide 2005), the resulting τm value for TRAF6-RD increases to 4.8 nsec, but is still shorter than 5.7 nsec for CNOT4. Given that these proteins are similar in size and structure, the larger τm value for CNOT4 may be due to self-association.
Figure 5.
Main-chain amide relaxation data for TRAF6-RD. (A) (•) 15N R 1 and (○) R 2and (B) {1H}-15N NOE. Elements of secondary structure include Zn2+-binding loops L1 and L2 (residues 9–16 and 44–47, respectively), α-helix (residues 32–39), and β-sheet (20–22, 25–28, 49–51).
Model-independent analysis of main-chain dynamics
S2 values were calculated for TRAF6-RD using five basic motional models on a per-residue basis as previously described (Fig. 6; Mandel et al. 1995; Spyracopoulos et al. 2005; Spyracopoulos 2006). The model-independent analysis yielded the following results: One residue was fit with model 1 (S 2 = 0.921 ± 0.006). Eleven residues were fit with model 2 (S 2, τf) with an average S 2 value of 0.79, and with τf values ranging from 19 psec to 2 nsec. Model 3 (S 2, R ex) was required to fit the relaxation data for three residues, with an average S 2 value of 0.88, and R ex parameters extending from 0.6 ± 0.2 sec−1 to 1.9 ± 0.1 sec−1. Model 4 (S 2, τf, R ex) was necessary to fit the relaxation data for 15 residues with an average S 2 of 0.89, τf values ranging from 9 psec to 2 nsec, and R ex parameters extending from 0.4 ± 0.1 sec−1 to 3.4 ± 0.2 sec−1. The necessity for R ex parameters to fit some of the relaxation data may be indicative of microsecond-to-millisecond time-scale conformational exchange phenomena at these main-chain amide sites. However, small R ex (<1.0 sec−1) values should be cautiously interpreted, and microsecond-to-millisecond timescale motions can be more rigorously assessed with specialized NMR experiments (see Discussion; Palmer et al. 2001; Wang and Palmer 2003), leaving residues 7, 20–22, 25–27, 43, 54, and 56 (R ex parameters >1 sec−1), as residues that may be involved in a genuine conformational exchange phenomenon. In addition, residues that were fit with models 3 or 4 were culled on the basis of R 1 R 2 values (see Materials and Methods). Thus, residues 20–22, 25, 26, 43, and 54 remain as residues that may be undergoing exchange. Residues not selected on the basis of R 1 R 2 were subjected to a second round of model selection using AIC with models 1 and 2. Following this selection, an additional two residues were fit with model 1, and an additional 10 residues were fit with model 2.
Figure 6.
Model-independent analysis for TRAF6-RD. (A) Plots of S 2 values determined from main-chain 15N NMR 〈R 1 R 2〉 relaxation measurements. (B) τs parameters from the model-independent analysis. (C) R ex parameters from the model-independent analysis. R ex parameters are shown only for residues selected on the basis of R1R2><R1R2>+σR1R2.
Twenty-four residues were fit using model 5 (Ss2, Sf2, τs) with an average S2 of 0.61 (S2 = Ss2, Sf2) and τs values ranging from 0.6 to 4.3 nsec. Some of these τs values are equal to the overall correlation time (residues 11, 19, and 44) and are likely to be failed models (d'Auvergne and Gooley 2006). In addition, if a residue was fit with model 5, the fit was only considered meaningful if the error in τs was <2.5σcutoff, with σcutoff = 0.115. The choice of this cutoff value is arbitrary; however, it is the largest τs error for flexible N- and C-terminal residues whose NOE was less than ∼0.56. Using this selection protocol, 11 residues were fit with model 5, and all of these residues had (NOE − σNOE) values <0.65. Model selection for residues not fit with model 5 using the above protocol was accomplished with AIC, and the requirement that R ex for models 3 and 4 was not 0. Six of these residues were fit with model 1, and nine were fit with model 2, bringing the total number of residues fit with model 1 to nine, and those with model 2 to 30. It should be noted that τs values should be cautiously interpreted because of the decoupling approximation (τs ≪ τm), and the fact that this model was developed in the absence of an analytical motional model.
Discussion
Solution structure of the RING domain from TRAF6
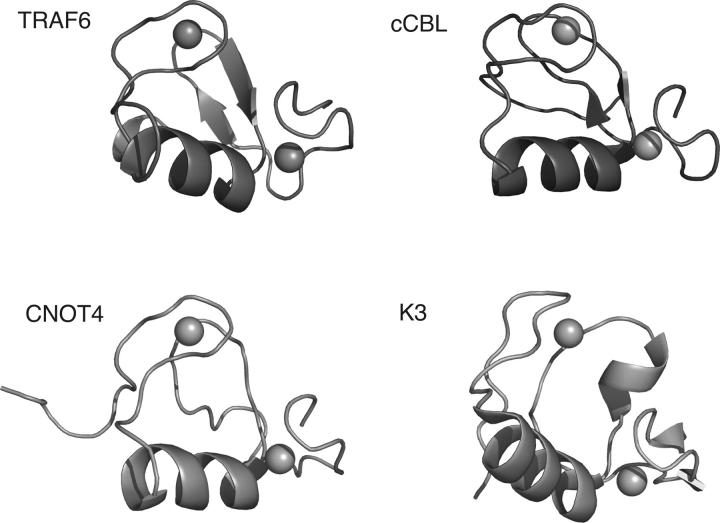
The topology for the RING domain from TRAF6 is similar to that observed for other RING domains (Fig. 7). Some representative examples of Zn2+ ligation using the canonical “cross-brace” motif (Takahashi et al. 1988; Barlow et al. 1994; Borden and Freemont 1996) include the RING domains from TRAF6, c-Cbl (1FBV) (Zheng et al. 2000), CNOT4 (1E4U) (Hanzawa et al. 2001), and K3 (1VYX) (Dodd et al. 2004). Interestingly, TRAF6 uses an Asp residue for Zn2+ ligation in place of Cys in the second Zn2+-binding site. For this residue, the χ1 angle was determined to be −60 ± 30° from the HNHB and HN(CO)HB experiments. For Asp47, the χ1 angle places the side chain in an orientation suitable for Zn2+ ligation. The structural features that are shared among the RING domains shown in Figure 7 include a short α-helix, and two Zn2+-binding loops. The overall fold of the C4HC2D RING domain from TRAF6 is most similar to the RING domains from CNOT4 (C4C4 RING) and c-Cbl (C3HC4 RING). However, there are differences in secondary structure. For example, CNOT4 lacks the β-sheet observed in TRAF6 and c-Cbl, and contains a poorly defined single helical turn in place of the N-terminal β-strand.
Figure 7.
Comparison of the structure of TRAF6-RD to typical RING-domain structures. The main-chain atoms from the RING domains of c-Cbl, CNOT4, and herpesvirus K3 are shown in the cartoon representation. Zn2+ atoms are shown as spheres. Secondary structure was rendered using the default algorithm within Pymol.
There are significant differences in the structure of the RING domain from Kaposi's sarcoma-associated herpesvirus K3 compared to that from TRAF6. While K3 and TRAF6 use the canonical cross-brace motif, K3 contains a short β-sheet within the N-terminal Zn2+-binding loop that is not observed in other RING domains. In addition, the C-terminal strand from the β-sheet is replaced with a single α-helical turn. In spite of the differences between K3 and TRAF6, both of these RING domains interact with Ubc13 (Dodd et al. 2004).
The surface of the RING domain from c-Cbl, shown by crystallography (Zheng et al. 2000) to be involved in the interaction with UbcH7, is centered on a shallow groove between the Zn2+-binding loops. This region contains a hydrophobic strip that is composed of residues Pro10, Ile11, Pro45, and Val46 in TRAF6 (Fig. 4C), appears to be a common feature of the RING domains, and occurs on the surface of CNOT4 as well. A notable difference between the RING domains from TRAF6 and c-Cbl is that Met14 replaces Glu in TRAF6, and may be an important determinant of E2–E3 specificity (see below, the discussion regarding chemical shift mapping data).
Titration of [U-15N; U-13C]-hUbc13 with TRAF6-RD
As shown in Figure 2C, hUbc13 residues experiencing a significant change in chemical shift upon binding the RING domain from TRAF6 are clustered around the N-terminal helix of hUbc13, and the loop connecting helices α2 and α3. Interestingly, the chemical shifts from the methyl groups of Met64 and Ala98 are the only ones from hUbc13 observed to experience significant changes in chemical shift in 2D 1H–13C HSQC NMR spectra upon binding of the RING domain from TRAF6 (Fig. 3). Changes in chemical shift for the backbone amides and methyl groups from hUbc13 upon binding of TRAF6 indicate that the binding region includes the central region from the α1-helix, the N-terminal end of helix α3, the loop connecting α2 and α3, and the loop connecting β3 and β4. However, it should be pointed out that these chemical shift differences are indicative of changes in chemical environment, and not necessarily direct structural interactions.
Residues Met64, Pro97, Ala98, and Leu99 are partly/completely exposed to solvent, clustered together on the surface of hUbc13, and may be directly involved in the interaction with the RING domain from TRAF6. Interestingly, an analysis of 190 protein sequences and 211 structures identified a conserved motif on the N-terminal helix of E2 enzymes: xRϕxx−x, where x is any residue, ϕ is hydrophobic and packed into the core of the E2, and − is a negatively charged residue (Winn et al. 2004). In the case of hUbc13, this motif is found on helix α1, and ϕ corresponds to Ile8. The sequence conservation of the N-terminal motif has been suggested to be critical for E1 binding (Winn et al. 2004). This hypothesis is supported in this study, with the hUbc13-binding region (Met64, Pro97, Ala98) for the RING domain from TRAF6 being similar to that observed for the E3–RING E2 interaction between c-Cbl and UbcH7 (Phe63, Pro97, Ala98). The main-chain amide chemical shift changes observed for helix α1 of hUbc13 upon TRAF6 binding may reflect transmission of structural changes through the close packing between residues comprising the E3 binding site and helix α1, rather than a direct interaction between the RING domain and helix α1 hUbc13. For example, Ile8 is buried and interacts with Tyr62, a residue involved in direct contacts with the loop connecting α2 and α3, and the loop connecting β2 and β3.
Titration of [U-15N]-TRAF6 with hUbc13
As shown in Figure 4, TRAF6-RD residues Ile11, Cys12, Leu13, and Met14 form part of the first Zn2+-binding loop, and the main-chain amide chemical shifts for these residues are observed to either disappear or shift upon binding hUbc13. Residues Ile11, Leu13, and Met14 from the first, or N-terminal, Zn2+-binding loop from TRAF6 are hydrophobic and surface-exposed. On this basis, it is possible that these residues may be involved in binding through interactions with residues Met64, Pro97, Ala98, and Leu99 of hUbc13. This result suggests that the mode of binding between hUbc13 and the RING domain from TRAF6 may be different from that observed for the c-Cbl/UbcH7 interaction. In particular, residues from the C-terminal, or second, Zn2+-binding loop of the RING domain from c-Cbl are directly involved in interactions with hydrophobic residues (Pro97, Ala98, and Phe63) on the surface of UbcH7. However, given the structural ambiguity inherent in chemical shift mapping, and the weak affinity of TRAF6-RD for hUbc13, the results presented herein indicate that TRAF6-RD and hUbc13 interact through similar surfaces as observed in other E2–E3 complexes, and the binding is not necessarily different (Zheng et al. 2000; Zhang et al. 2005).
Biological relevance of the TRAF6-RD hUbc13 interaction
The crystallographically determined structures of the E2–E3 complexes formed between c-Cbl (RING-E3) and UbcH7 (E2) (Zheng et al. 2000), and between the CHIP homodimer (U-box E3) and hUev1a–hUbc13 (Uev/E2) (Zhang et al. 2005) indicate that the RING and U-box domains constitute key binding components in these E2–E3 interactions and that overall, the RING-E2 and U-box–E2 interactions are structurally similar. It should be noted that U-box domains are similar in structure to RING domains, but lack the ability to bind metal ions (Aravind and Koonin 2000; Zhang et al. 2005). Interestingly, the U-box domain is the minimal E3 component required to interact with hUbc13 (Zhang et al. 2005). On the basis of these structural studies and biochemical studies involving the interaction between hUbc13 and TRAF6 (Deng et al. 2000; Wooff et al. 2004; Zhou et al. 2004; Andersen et al. 2005), it is reasonable to expect the RING domain from TRAF6 to interact with hUbc13. Currently, the nature of the interaction between intact TRAF6 and hUbc13 is not known. From the chemical shift titration data presented herein, the interaction appears too weak (K D ∼ 2 mM) to be biologically relevant. For example, the E2 UbcH5B shows comparable chemical shift changes upon binding the RING domain from CNOT4 at a significantly lower E3:E2 ratio (2:1), indicative of stronger binding (Dominguez et al. 2004). It is possible that the region of TRAF6 adjacent to the RING domain, which is composed of two TRAF-type Zn2+-binding domains, may be involved in interactions with hUbc13, and these interactions may serve to increase the affinity of intact TRAF6 for hUbc13. However, the affinities of intact TRAF6, or the TRAF6 RING domain and surrounding regions of the protein for hUbc13, are not currently known. Finally, it has recently been reported that human TRAF6 can be degraded by ubiquitination through the activity of its own RING domain, suggesting that the RING domain can interact with E2 enzymes other than hUbc13 that are involved in degradation (He et al. 2006). Furthermore, the protein regions surrounding the RING domain may play important roles in determining the biological function of a given TRAF protein (He et al. 2006), and perhaps the E2 enzymes with which TRAF6 can interact.
Model-independent analysis of main-chain dynamics
Solution state NMR relaxation measurements can be used to assess protein main-chain dynamics by allowing for the calculation of order parameters, or S 2 values, for main-chain 1H–15N pairs (Palmer 2001). S 2 values give the magnitude of spatial restriction for a main-chain 1H–15N bond vector, and can be used to assess regions of flexibility in proteins and determine changes in protein dynamics upon interaction with ligands or upon unfolding (Palmer 2001; Spyracopoulos and Sykes 2001; Homans 2005; Spyracopoulos 2005; Jarymowycz and Stone 2006).
The S 2 values from the model-independent analysis indicate that, in general, for motions on the fast picosecond–nanosecond timescale, most of the TRAF6-RD residues within the α-helix, β-sheet, and some of the residues in the Zn2+-binding loops are as rigid as elements of canonical secondary structure in larger proteins, and many residues within the termini and some within the Zn2+-binding loops are flexible on the subnano-to-nanosecond timescale. That the core residues of TRAF6-RD are mostly rigid is consistent with known biological roles of RING-domain E3s. That is, this small domain serves as a rigid platform for recognition of E2 ubiquitin conjugation enzymes (Figs. 2C, 4C).
Residues 7, 20–22, 25–27, 43, 54, and 56 require R ex parameters (>1 sec−1) to properly fit their relaxation data. The majority of these residues involve the β-sheet, a key structural element that contributes to hydrophobic core packing (Val20, Thr22). These results indicate that the hydrophobic core of TRAF6-RD may be involved in a conformational exchange phenomenon on the microsecond-to-millisecond timescale. The method of Bracken and coworkers was used to identify residues undergoing conformational exchange (Kneller et al. 2002). Using this method, R ex terms of 2.1 ± 0.1, 1.5 ± 0.1, 1.2 ± 0.2, 1.2 ± 0.1, 1.9 ± 0.2, 1.4 ± 0.2, and 2.5 ± 0.2 sec−1 are calculated for residues 20–22, 25, 26, 43, and 54, respectively. In comparison, using previously recorded main-chain 15N NMR relaxation data for ubiquitin (Tjandra et al. 1995) (BMRB accession number 6470), we estimate Rex values of 1.3 ± 0.3, 2.5 ± 0.2, 0.7 ± 0.2, 0.6 ± 0.2, and 0.6 ± 0.2 sec−1 for residues 23, 25, 27, 28, and 70, respectively, as previously noted (Spyracopoulos 2006). These estimates compare favorably with the observation that residues 23, 25, 55, and 70 from ubiquitin are involved in a chemical exchange process, as determined through 15N NMR R 1ρ relaxation measurements (Massi et al. 2005).
Residues that display nanosecond timescale internal motions include loop residues Ala15 and Leu16 in Zn2+-binding site 1; residues Ala40 and Gly41 at the C-terminal end of the α-helix; as well as N-terminal residues Leu3, Gly4 (cloning artifact), and Glu8; C-terminal residues Asn60, Ala62, and Lys63; and residue Leu52.
Conclusions
The common structural features of RING domains from various organisms and proteins include a short α-helix, and two Zn2+-binding loops that position the Zn2+ atoms in similar orientations with respect to the α-helices for the different proteins. This common architecture suggests that different RING domains may bind their cognate E2 proteins similarly. The RING domain from human TRAF6 is most similar in structure to the RING domain from c-Cbl, and the chemical shift mapping experiments in this study indicate that the binding interface between TRAF6-RD and hUbc13 is similar to that between c-Cbl and UbcH7. Given the involvement of TRAF6 in activation of NF-κB through Lys63-linked polyubiquitination, it is surprising that TRAF6-RD binds hUbc13 weakly. This weak binding may be due to the fact that only a fragment of TRAF6 was used in the binding studies. However, on the basis of other studies involving E2–E3 interactions, the RING domain constitutes a key component of the E2–E3 interaction. In addition, recent biochemical studies indicate that TRAF6 can be degraded by ubiquitination through the RING domain, raising the intriguing possibility that hUbc13 may not be the cognate E2 for TRAF6.
Analysis of the main-chain dynamics of TRAF6-RD through 15N NMR relaxation measurements indicates that the α-helical and β-sheet secondary structural elements for this small zinc-binding protein are as rigid as those found in larger proteins. This observation is consistent with the biological role of RING-domain E3s, which requires in part that these proteins provide a recognition site for binding of E2 ubiquitin conjugation enzymes.
Materials and Methods
Protein purification
TRAF6-RD
The amino acid sequence of the RING domain from human TRAF6 was identified using the PROSITE database (Hulo et al. 2006) from the ExPASy World Wide Web server (Gasteiger et al. 2003), and the sequence was aligned to that from the RING domain from c-Cbl using the program CLUSTAL W (Thompson et al. 1994). On the basis of the sequence alignment to the RING domain from c-Cbl and its structure (1FBV), residues 67–124 from human TRAF6 (TRAF6-RD) were subcloned into pGEX6P-1 as a GST fusion protein. Overexpression and purification of [U-15N, U-13C] TRAF6-RD was performed in a similar fashion to hUbc13K92R with the exception that M9 minimal medium was supplemented with 10 μM ZnCl2, and with exclusion of DTT and EDTA from all buffers. In addition, cleavage of the GST fusion protein resulted in the inclusion of five N-terminal residues (Gly-Pro-Leu-Gly-Ser) for TRAF6-RD from a cloning artifact; thus residue 6 of TRAF6-RD corresponds to residue 67 of intact TRAF6. Unlabeled TRAF6-RD was overexpressed and purified similarly to the protein hMms2 (Lewis et al. 2006), with the exclusion of DTT and EDTA from all buffers.
hUbc13
Expression and purification of unlabeled and [U-15N, U-13C]-hUbc13K92R was conducted in a similar fashion to that previously described for the protein hMms2 (Spyracopoulos et al. 2005; Lewis et al. 2006).
NMR spectroscopy
All NMR spectra were obtained using Varian Unity INOVA 500, 600, or 800 MHz NMR spectrometers. NMR experiments for chemical shift assignment, structure determination, and backbone 15N relaxation measurements of TRAF6-RD were conducted at 25°C. NMR experiments for chemical shift assignment of hUbc13 and hUbc13/TRAF6-RD titrations were carried out at 30°C. NMR samples for assignment of hUbc13 and assignment and structure determination of TRAF6-RD were 600 μL for standard 5-mm NMR tubes and 300 μL for SHIGEMI microcell NMR tubes, and contained 9:1 H2O/D2O with 50 mM phosphate (pH 7.5), 150 mM NaCl for TRAF6-RD, 250 mM NaCl for hUbc13, 1 mM DTT, 1 mM DSS, 3 μL of 100× stock protease inhibitor cocktail I for hUbc13 (Calbiochem catalog no. 539,131), and 6 μL of 25× stock EDTA-free protease inhibitor for TRAF6-RD (Roche catalog no. 11873580001), with ∼0.5 mM hUbc13 and ∼0.3–0.5 mM TRAF6-RD. For hUbc13/TRAF6-RD titrations, protein concentrations and buffers differed somewhat from the above conditions (see relevant section below).
Chemical shift assignment for hUbc13
The main-chain atoms of hUbc13 were unambiguously assigned using a combination of the HNCACB (Wittekind and Mueller 1993; Muhandiram and Kay 1994) and CBCA(CO)NNH (Grzesiek and Bax 1992a; Muhandiram and Kay 1994) experiments. Side-chain atoms were assigned using the (H)CCTOCSY(CO)NNH and H(CC)TOCSY(CO)NNH experiments (Grzesiek et al. 1993; Logan et al. 1993; Lyons and Montelione 1993; Gardner et al. 1996), and the HCCH-TOCSY experiment (Bax et al. 1990; Kay et al. 1993). All spectra were processed using the program NMRPipe (Delaglio et al. 1995), and chemical shift assignment was accomplished using the SPARKY 3 NMR software (University of California, San Francisco). Chemical shift assignments for hUbc13 have been deposited in the BMRB repository under accession number 15092.
Chemical shift assignment for TRAF6-RD
Main-chain chemical shift assignment for TRAF6-RD•2Zn2+ was performed using the SmartNotebook software package (http://www.bionmr.ualberta.ca/bds) and the following NMR experiments: 15N-HSQC (Zhang et al. 1994), 13C-HSQC, CBCA(CO)NH (Muhandiram and Kay 1994), HNCACB (Muhandiram and Kay 1994), HACA(CO)CANH (Löhr and Rüterjans 1995), HNCO (Grzesiek and Bax 1992b; Muhandiram and Kay 1994), HN(CO)HB (Grzesiek et al. 1992), and HNN(CO,CA) (Szyperski et al. 1995; Panchal et al. 2001). The main-chain 15N and 1HN atoms for His42 were not observed in 1HN-detected spectra, presumably because of line broadening, and were unassigned. Side-chain assignments were carried out using the HNHA (Kuboniwa et al. 1994), HNHB (Archer et al. 1991), 15N-DIPSI-HSQC (Zhang et al. 1994), HCCH-TOCSY (Bax et al. 1990; Kay et al. 1993), (H)CCTOCSY(CO)NNH, and H(CC)TOCSY(CO)NNH (Montelione et al. 1992; Grzesiek et al. 1993; Logan et al. 1993; Lyons and Montelione 1993; Gardner et al. 1996) set of NMR experiments. Aromatic side-chain assignments were obtained using the aromatic 2D 1H–13C HSQC (Pervushin et al. 1998), (HB)CB(CGCD)HD, and (HB)CB(CGCDCE)HE (Yamazaki et al. 1993) pulse sequences. NOE distance restraints were derived from 3D 15N-NOESY-HSQC (Zhang et al. 1994) and 13C-NOESY-HSQC experiments. The 3D 15N-NOESY-HSQC was collected at 500 MHz with spectral widths of 4500 (F 1: 1H indirect, 96 real points), 1200 (F 2: 15N indirect, 32 real points), and 5500 Hz (F 3: 1HN acquisition, 352 real points) and a mixing time of 150 msec. The 3D 13C-NOESY-HSQC was collected at 800 MHz with spectral widths of 7998 (F 1: 1H indirect, 120 real points), 4800 (F 2: 13C indirect, 32 real points), and 8292 Hz (F 3: 1HN acquisition, 512 real points), and a mixing time of 150 msec. NMR restraints and chemical shift assignments for TRAF6-RD have been deposited in the BMRB repository under accession number 15014.
Structure determination for TRAF6-RD
Structures were generated using the program CYANA 2.1 (Güntert et al. 1997; Herrmann et al. 2002; Güntert 2004) with the “noeassign” automatic assignment protocol (Jee and Güntert 2003). NOEs were calibrated automatically with the CYANA standard procedure, with upper bounds set to 6 Å. Chemical shift assignments obtained from NMRView/SmartNotebook were exported with pseudoatom name correction as required, for proper calibration of NOEs in CYANA. Eight rounds of structure generation and refinement (100 structures/round) were performed. A total of 1164 unambiguous proton–proton restraints were used for the final round. The “ramaaco” protocol within the CYANA “noeassign” routine was used to generate main-chain dihedral restraints based on the allowed and most favored regions of the Ramachandran plot.
For structure calculations within the CYANA software package, zinc atoms were included within the residue library. To conduct torsion angle dynamics, zinc atoms were chained to the protein with virtual linkers. Conservative distance restraints between the atom pairs Zn–S, S–Cβ, (His Nδ1/Nɛ2)–S, and (Asp Oδ1/Oδ2)–Zn for the Cys4 and Cys2/Asp/His-binding sites were derived from previously published structural studies in similar systems (Wang et al. 2003; Blair et al. 2005; Kellenberger et al. 2005; Sakharov and Lim 2005).
For His and Asp, there are two side-chain nitrogen and oxygen atoms, respectively, that can participate in zinc ligation. Thus, ambiguous restraints to zinc were defined to both side-chain nitrogen atoms (Nδ1, Nɛ1) of His26, and both side-chain oxygen atoms (Oδ1, Oδ2) of Asp47.
The 50 structures with the lowest target function value obtained from CYANA were refined in explicit solvent using XPLOR-NIH 2.13 (Linge et al. 2003; Schwieters et al. 2003; Nabuurs et al. 2004). Distance restraints were converted from CYANA to the XPLOR format, with the addition of 1 Å to restraints involving methyl groups or pseudoatoms (Güntert et al. 1991). For main-chain ϕ and ψ dihedral angles with a maximum range of 45° in the final ensemble of structures generated with CYANA, 30 ϕ and 34 ψ dihedral restraints were also included during water refinement. Main-chain dihedral angles were constrained to ±20° if their distribution within the final ensemble of structures calculated within CYANA was <20°. One χ1 dihedral restraint, derived experimentally from the HNHB and HN(CO)HB experiments, was included for Asp47, and this restraint contributes to positioning the side-chain oxygen atoms proximal to the zinc atom.
The force field used within the XPLOR-NIH program was modified to take into account charge transfer and polarization effects in zinc complexes based on recent molecular dynamics simulations of zinc bound to Cys and/or His in proteins (Sakharov and Lim 2005). Parameter set A in (Sakharov and Lim (2005) was used, with the charges for zinc, and sulfur from Cys, set to +1.24 and −0.61, respectively.
The PROCHECK (Morris et al. 1992; Laskowski et al. 1993), WHAT CHECK (Hooft et al. 1996), and VADAR (Willard et al. 2003) software packages were used to evaluate the quality of the structures. Fitting was performed using the McLachlan algorithm (McLachlan 1982) as implemented in the program ProFit (http://www.bioinf.org.uk/software/profit/). Coordinates for TRAF6-RD have been deposited in the RCSB protein data bank as entry 2JMD.
hUbc13/TRAF6-RD titrations
All hUbc13/TRAF6-RD titrations were conducted in the absence of DTT. For titration of [U-15N; U-13C]-hUbc13 with TRAF6-RD, three aliquots of TRAF6 were titrated into ∼0.2 mM [U-15N; U-13C]-hUbc13. Protein concentrations were determined by amino acid analysis, and the TRAF6-RD/Ubc13 molar ratios were calculated to be 0.6, 2.2, and 7.2. For titration of [U-15N]-TRAF6-RD with hUbc13, one titration point was acquired with ∼0.1 mM [U-15N]-TRAF6 and ∼0.4 mM hUbc13. In addition, the buffer composition differed from that described in the section entitled “NMR spectroscopy” with 50 mM TRIS replacing phosphate buffer, and 200 mM NaCl instead of 150 mM.
2D 1H–15N-HSQC NMR spectra were acquired for each titration point. Chemical shift perturbations for each resonance were calculated using the equation
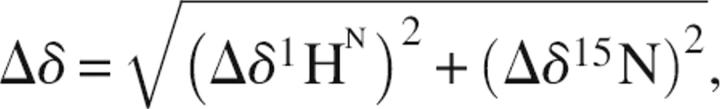 |
where Δδ1HN is the change in backbone amide proton chemical shift, and Δδ15HN is the change in backbone amide nitrogen chemical shift. Chemical shift changes that were greater than one standard deviation from the mean were considered significant (residues 9–11, 54, 56, 65, 96, 98–101, and 105 for hUbc13; and residues 12, 14, 20–22, 33, 40, and 43 for TRAF6-RD).
For titration of [U-15N; U-13C]-hUbc13 with TRAF6, the chemical shift changes for the backbone amide 1HN and 15N chemical shifts for Ile101 were calculated as described above. These chemical shift changes were fit to 1:1 protein–ligand binding as previously described using the program Xcrvfit in order to extract a macroscopic dissociation constant (K D) (http://www.bionmr.ualberta.ca/bds/software) (Williams et al. 1985; McKenna et al. 2003).
Main-chain 15N relaxation measurements
2D 1H–15N sensitivity-enhanced HSQC NMR spectra for measurement of 15N-R 1, 15N-R 2, and {1H}-15N NOE data were acquired at 600 MHz as previously described (Farrow et al. 1994; Spyracopoulos et al. 2005). Spectral processing and analysis were accomplished with the programs NMRPipe (Delaglio et al. 1995) and SPARKY, in a similar fashion to that previously reported (Spyracopoulos et al. 2005). Main-chain 15N-R 1 and 15N-R 2 values were obtained by nonlinear least squares fits of the 15N–1HN cross-peak intensities to a two-parameter exponential decay of the form I(t) = I 0 exp(−tRi), where Ri (i = 1, 2) is the longitudinal or transverse relaxation rate, using the program Mathematica (Wolfram 1996; Spyracopoulos 2006). Uncertainties in the measured Ri data were obtained from the covariance matrix of the nonlinear least squares fits. Uncertainties in the {1H}-15N NOE values were estimated from the base-plane noise in the 2D 1H–15N HSQC NMR spectra recorded in the presence and absence of proton saturation.
A Lipari-Szabo model-independent analysis (Lipari and Szabo 1982a,b) was conducted for TRAF6-RD with the program Mathematica (Wolfram 1996), using an approach similar to that recently described (Spyracopoulos et al. 2005; Spyracopoulos 2006). The model-independent analysis was carried out under the assumption that the rotational tumbling of TRAF6-RD is isotropic. This assumption is based on the observation that the normalized principal axes of the inertia tensor have values of 1:0.9:0.8 for the structured core domain (residues 9–58) of TRAF6-RD. 15N-R 1 and 15N-R 2 data indicate that residues 1–7 and 59–63 are flexible; therefore, these residues were excluded from the inertia tensor calculation. Additionally, hydrodynamic calculations using the program HYDRONMR (de la Torre et al. 2000) for the ensemble of structures of TRAF6-RD determined herein (including the flexible N- and C-terminal residues) indicate that, on average, the rotational tumbling is nearly isotropic (D∥/D⊥ = 1.1 ± 0.3).
Molecular graphics
Protein structure graphics were produced using the program Pymol (DeLano 2002).
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and the Alberta Heritage Foundation for Medical Research (AHFMR). L.S. is an AHFMR Medical Research Senior Scholar. We thank the Canadian National High Field NMR Centre (NANUC) for its assistance and use of the facilities. Operation of NANUC is supported by grants from the CIHR, the Natural Sciences and Engineering Research Council of Canada, and the University of Alberta. We thank Robert Boyko for assistance with SmartNotebook, and Logan Donaldson (York University) for CYANA to XPLOR conversion scripts. We also thank Deryk Webb for spectrometer maintenance, Yanni Batsiolas and Dean Schieve for computer support, and Lewis E. Kay for pulse sequences.
Footnotes
Reprint requests to: Leo Spyracopoulos, Department of Biochemistry, Room 416, Medical Sciences Building, University of Alberta, Edmonton, Alberta T6G 2H7, Canada; e-mail: leo.spyracopoulos@ualberta.ca; fax: (780) 492-0886.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062358007.
Abbreviations: E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; E3, ubiquitin protein ligase; HSQC, heteronuclear single quantum coherence; MATH domain, merpin and TRAF homology domain; NMR, nuclear magnetic resonance; NOE, nuclear Overhauser effect; RING, really interesting new gene; RMSD, root-mean-square deviation; R 1, longitudinal relaxation rate; R 2, transverse relaxation rate; TNF, tumor necrosis factor; TRAF, tumor necrosis receptor associated factor; TRAF6-RD, TRAF6-RING domain; Ub, ubiquitin.
References
- Andersen P.L., Zhou, H., Pastushok, L., Moraes, T., McKenna, S., Ziola, B., Ellison, M.J., Dixit, V.M., and Xiao, W. 2005. Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 170: 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. and Koonin, E.V. 2000. The U box is a modified RING finger—A common domain in ubiquitination. Curr. Biol. 10: R132–R134. [DOI] [PubMed] [Google Scholar]
- Archer S.J., Ikura, M.I., Torchia, D.A., and Bax, A. 1991. An alternative 3D NMR technique for correlating backbone 15N with side chain Hβ resonances in larger proteins. J. Magn. Reson. 95: 636–641. [Google Scholar]
- Barlow P.N., Luisi, B., Milner, A., Elliott, M., and Everett, R. 1994. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol. 237: 201–211. [DOI] [PubMed] [Google Scholar]
- Bax A., Clore, G.M., and Gronenborn, A.M. 1990. 1H–1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 88: 425–431. [Google Scholar]
- Blair D.E., Schuttelkopf, A.W., MacRae, J.I., and van Aalten, D.M.F. 2005. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. 102: 15429–15434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer J.L., Schneider, P., and Tschopp, J. 2002. The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 27: 19–26. [DOI] [PubMed] [Google Scholar]
- Borden K.L. and Freemont, P.S. 1996. The RING finger domain: A recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 6: 395–401. [DOI] [PubMed] [Google Scholar]
- Bradley J.R. and Pober, J.S. 2001. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20: 6482–6491. [DOI] [PubMed] [Google Scholar]
- Brzovic P.S., Rajagopal, P., Hoyt, D.W., King, M.C., and Klevit, R.E. 2001. Structure of a BRCA1–BARD1 heterodimeric RING–RING complex. Nat. Struct. Biol. 8: 833–837. [DOI] [PubMed] [Google Scholar]
- Chen Z.J. 2005. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7: 758–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.Y., Park, Y.C., Ye, H., and Wu, H. 2002. All TRAFs are not created equal: Common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 115: 679–688. [DOI] [PubMed] [Google Scholar]
- d'Auvergne E.J. and Gooley, P.R. 2006. Model-free model elimination: A new step in the model-free dynamic analysis of NMR relaxation data. J. Biomol. NMR 35: 117–135. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. 2002. The Pymol molecular graphics system. DeLano Scientific, San Carlos, CA.
- de la Torre J.G., Huertas, M.L., and Carrasco, B. 2000. HYDRONMR: Prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 147: 138–146. [DOI] [PubMed] [Google Scholar]
- Deng L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., Slaughter, C., Pickart, C., and Chen, Z.J. 2000. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361. [DOI] [PubMed] [Google Scholar]
- Dodd R.B., Allen, M.D., Brown, S.E., Sanderson, C.M., Duncan, L.M., Lehner, P.J., Bycroft, M., and Read, R.J. 2004. Solution structure of the Kaposi's sarcoma-associated herpesvirus K3 N-terminal domain reveals a novel E2-binding C4HC3-type RING domain. J. Biol. Chem. 279: 53840–53847. [DOI] [PubMed] [Google Scholar]
- Dominguez C., Bonvin, A.M.J.J., Winkler, G.S., van Schaik, F.M.A., Timmers, H.T.M., and Boelens, R. 2004. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutatgenesis, and docking approaches. Structure 12: 633–644. [DOI] [PubMed] [Google Scholar]
- Farrow N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Forman-Kay, J.D., and Kay, L.E. 1994. Backbone dynamics of a free and a phosphopeptide-complexed Src homology-2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003. [DOI] [PubMed] [Google Scholar]
- Gardner K.H., Konrat, R., Rosen, M.K., and Kay, L.E. 1996. An (H)C(CO)NH-TOCSY pulse scheme for sequential assignment of protonated methyl groups in otherwise deuterated 15N, 13C-labeled proteins. J. Biomol. NMR 8: 351–356. [DOI] [PubMed] [Google Scholar]
- Garg A. and Aggarwal, B.B. 2002. Nuclear transcription factor-κB as a target for cancer drug development. Leukemia 16: 1053–1068. [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R.D., and Bairoch, A. 2003. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31: 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., May, M.J., and Kopp, E.B. 1998. NF-κB and REL proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16: 225–260. [DOI] [PubMed] [Google Scholar]
- Grzesiek S. and Bax, A. 1992a. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114: 6291–6293. [Google Scholar]
- Grzesiek S. and Bax, A. 1992b. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J. Magn. Reson. 96: 432–440. [Google Scholar]
- Grzesiek S., Ikura, M., Clore, G.M., Gronenborn, A.M., and Bax, A. 1992. A 3D triple-resonance NMR technique for qualitative measurement of carbonyl-Hβ J couplings in isotopically enriched proteins. J. Magn. Reson. 96: 215–221. [Google Scholar]
- Grzesiek S., Anglister, J., and Bax, A. 1993. Correlation of backbone amide and aliphatic side chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. B101: 114–119. [Google Scholar]
- Güntert P. 2004. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278: 353–378. [DOI] [PubMed] [Google Scholar]
- Güntert P., Braun, W., and Wüthrich, K. 1991. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol. 217: 517–530. [DOI] [PubMed] [Google Scholar]
- Güntert P., Mumenthaler, C., and Wüthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273: 283–298. [DOI] [PubMed] [Google Scholar]
- Hanzawa H., de Ruwe, M.J., Albert, T.K., van Der Vliet, P.C., Timmers, H.T., and Boelens, R. 2001. The structure of the C4C4 ring finger of human NOT4 reveals features distinct from those of C3HC4 RING fingers. J. Biol. Chem. 276: 10185–10190. [DOI] [PubMed] [Google Scholar]
- He L.S., Wu, X.L., Siegel, R., and Lipsky, P.E. 2006. TRAF6 regulates cell fate decisions by inducing caspase 8-dependent apoptosis and the activation of NF-κB. J. Biol. Chem. 281: 11235–11249. [DOI] [PubMed] [Google Scholar]
- Herrmann T., Güntert, P., and Wüthrich, K. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319: 209–227. [DOI] [PubMed] [Google Scholar]
- Homans S.W. 2005. Probing the binding entropy of ligand–protein interactions by NMR. ChemBioChem. 6: 1585–1591. [DOI] [PubMed] [Google Scholar]
- Hooft R.W.W., Vriend, G., Sander, C., and Abola, E.E. 1996. Errors in protein structures. Nature 381: 272. [DOI] [PubMed] [Google Scholar]
- Houben K., Wasielewski, E., Dominguez, C., Kellenberger, E., Atkinson, R.A., Timmers, H.T.M., Kieffer, B., and Boelens, R. 2005. Dynamics and metal exchange properties of C4C4 RING domains from CNOT4 and the p44 subunit of TFIIH. J. Mol. Biol. 349: 621–637. [DOI] [PubMed] [Google Scholar]
- Hulo N., Bairoch, A., Bulliard, V., Cerutti, L., De Castro, E., Langendijk-Genevaux, P.S., Pagni, M., and Sigrist, C.J. 2006. The PROSITE database. Nucleic Acids Res. 34: D227–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarymowycz V.A. and Stone, M.J. 2006. Fast time scale dynamics of protein backbones: NMR relaxation methods, applications, and functional consequences. Chem. Rev. 106: 1624–1671. [DOI] [PubMed] [Google Scholar]
- Jee J. and Güntert, P. 2003. Influence of the completeness of chemical shift assignments on NMR structures obtained with automated NOE assignment. J. Struct. Funct. Genomics 4: 179–189. [DOI] [PubMed] [Google Scholar]
- Kay L.E., Torchia, D.A., and Bax, A. 1989. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 28: 8972–8979. [DOI] [PubMed] [Google Scholar]
- Kay L.E., Xu, G.Y., Singer, A.U., Muhandiram, D.R., and Forman-Kay, J.D. 1993. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J. Magn. Reson. B101: 333–337. [Google Scholar]
- Kellenberger E., Dominguez, C., Fribourg, B., Wasielewski, E., Moras, D., Poterszman, A., Boelens, R., and Kieffer, B. 2005. Solution structure of the C-terminal domain of TFIIH p44 subunit reveals a novel type of C4C4 ring domain involved in protein–protein interactions. J. Biol. Chem. 280: 20785–20792. [DOI] [PubMed] [Google Scholar]
- Kneller J.M., Lu, M., and Bracken, C. 2002. An effective method for the discrimination of motional anisotropy and chemical exchange. J. Am. Chem. Soc. 124: 1852–1853. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Walsh, M.C., and Choi, Y. 2004. The role of TRAF6 in signal transduction and the immune response. Microbes Infect. 6: 1333–1338. [DOI] [PubMed] [Google Scholar]
- Kornhaber G.J., Snyder, D., Moseley, H.N.B., and Montelione, G.T. 2006. Identification of zinc-ligated cysteine residues based on 13Cα and 13Cβ chemical shift data. J. Biomol. NMR 34: 259–269. [DOI] [PubMed] [Google Scholar]
- Kuboniwa H., Grzesiek, S., Delaglio, F., and Bax, A. 1994. Measurement of HN-Hα J couplings in calcium-free calmodulin using new 2D and 3D water flip-back methods. J. Biomol. NMR 4: 871–878. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S., and Thorton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–290. [Google Scholar]
- Lewis M.J., Saltibus, L.F., Hau, D.D., Xiao, W., and Spyracopoulos, L. 2006. Structural basis for non-covalent interaction between ubiquitin and the ubiquitin conjugating enzyme variant human MMS2. J. Biomol. NMR 34: 89–100. [DOI] [PubMed] [Google Scholar]
- Lide D.R. 2005. CRC handbook of chemistry and physics, 86th ed. CRC Press, Taylor and Francis Group, Boca Raton, FL.
- Linge J.P., Williams, M.A., Spronk, C.A.E.M., Bonvin, A.M.J.J., and Nilges, M. 2003. Refinement of protein structures in explicit solvent. Proteins 50: 496–506. [DOI] [PubMed] [Google Scholar]
- Lipari G. and Szabo, A. 1982a. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 104: 4546–4559. [Google Scholar]
- Lipari G. and Szabo, A. 1982b. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 104: 4559–4570. [Google Scholar]
- Liu Y.C. 2004. Ubiquitin ligases and the immune response. Annu. Rev. Immunol. 22: 81–127. [DOI] [PubMed] [Google Scholar]
- Logan T.M., Olejniczak, E.T., Xu, R.X., and Fesik, S.W. 1993. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J. Biomol. NMR 3: 225–231. [DOI] [PubMed] [Google Scholar]
- Löhr F. and Rüterjans, H. 1995. A new triple-resonance experiment for the sequential assignment of backbone resonances in proteins. J. Biomol. NMR 6: 189–197. [DOI] [PubMed] [Google Scholar]
- Lyons B.A. and Montelione, G.T. 1993. An HCCNH triple-resonance experiment using carbon-13 isotropic mixing for correlating backbone amide and side-chain aliphatic resonances in isotopically enriched proteins. J. Magn. Reson. B101: 206–209. [Google Scholar]
- Mandel A.M., Akke, M., and Palmer, A.G. 1995. Backbone dynamics of Escherichia coli ribonuclease HI—Correlations with structure and function in an active enzyme. J. Mol. Biol. 246: 144–163. [DOI] [PubMed] [Google Scholar]
- Massi F., Grey, M.J., and Palmer, A.G. 2005. Microsecond timescale backbone conformational dynamics in ubiquitin studied with NMR R1ρ relaxation experiments. Protein Sci. 14: 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna S., Hu, J., Moraes, T., Xiao, W., Ellison, M.J., and Spyracopoulos, L. 2003. Energetics and specificity of interactions within Ub·Uev·Ubc13 human ubiquitin conjugation complexes. Biochemistry 42: 7922–7930. [DOI] [PubMed] [Google Scholar]
- McLachlan A.D. 1982. Rapid comparison of protein structures. Acta Crystallogr. A 38: 871–873. [Google Scholar]
- Montelione G.T., Lyons, B.A., Emerson, S.D., and Tashiro, M. 1992. An efficient triple-resonance experiment using carbon-13 isotropic mixing for determining sequence-specific assignments of isotopically-enriched proteins. J. Am. Chem. Soc. 114: 10975–10977. [Google Scholar]
- Morris A.L., Macarthur, M.W., Hutchinson, E.G., and Thornton, J.M. 1992. Stereochemical quality of protein structure coordinates. Proteins 12: 345–364. [DOI] [PubMed] [Google Scholar]
- Muhandiram D.R. and Kay, L.E. 1994. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. B 103: 203–216. [Google Scholar]
- Nabuurs S.B., Nederveen, A.J., Vranken, W., Doreleijers, J.F., Bonvin, A.M.J.J., Vuister, G.W., Vriend, G., and Spronk, C.A.E.M. 2004. DRESS: A Database of REfined Solution NMR Structures. Proteins 55: 483–486. [DOI] [PubMed] [Google Scholar]
- Palmer A.G. 2001. NMR probes of molecular dynamics: Overview and comparison with other techniques. Annu. Rev. Biophys. Biomol. Struct. 30: 129–155. [DOI] [PubMed] [Google Scholar]
- Palmer A.G., Kroenke, C.D., and Loria, J.P. 2001. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 339: 204–238. [DOI] [PubMed] [Google Scholar]
- Panchal S.C., Bhavesh, N.S., and Hosur, R.V. 2001. Improved 3D triple resonance experiments, HNN and HN(C)N, for HN and 15N sequential correlations in (13C, 15N) labeled proteins: Application to unfolded proteins. J. Biomol. NMR 20: 135–147. [DOI] [PubMed] [Google Scholar]
- Park Y.C., Burkitt, V., Villa, A.R., Tong, L., and Wu, H. 1999. Structural basis for self-association and receptor recognition of human TRAF2. Nature 398: 533–538. [DOI] [PubMed] [Google Scholar]
- Pervushin K., Riek, R., Wider, G., and Wüthrich, K. 1998. Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. J. Am. Chem. Soc. 120: 6394–6400. [Google Scholar]
- Regnier C.H., Tomasetto, C., Moog-Lutz, C., Chenard, M.P., Wendling, C., Basset, P., and Rio, M.C. 1995. Presence of a new conserved domain in CART1, a novel member of the tumor necrosis factor receptor-associated protein family, which is expressed in breast carcinoma. J. Biol. Chem. 270: 25715–25721. [DOI] [PubMed] [Google Scholar]
- Sakharov D.V. and Lim, C. 2005. Zn protein simulations including charge transfer and local polarization effects. J. Am. Chem. Soc. 127: 4921–4929. [DOI] [PubMed] [Google Scholar]
- Schwieters C.D., Kuszewski, J.J., Tjandra, N., and Clore, G.M. 2003. The XPLOR-NIH NMR molecular structure determination package. J. Magn. Reson. 160: 65–73. [DOI] [PubMed] [Google Scholar]
- Spyracopoulos L. 2005. Thermodynamic interpretation of protein dynamics from NMR relaxation measurements. Protein Pept. Lett. 12: 235–240. [DOI] [PubMed] [Google Scholar]
- Spyracopoulos L. 2006. A suite of Mathematica notebooks for the analysis of protein main chain 15N NMR relaxation data. J. Biomol. NMR 36: 215–224. [DOI] [PubMed] [Google Scholar]
- Spyracopoulos L. and Sykes, B.D. 2001. Thermodynamic insights into proteins from NMR spin relaxation studies. Curr. Opin. Struct. Biol. 11: 555–559. [DOI] [PubMed] [Google Scholar]
- Spyracopoulos L., Lewis, M.J., and Saltibus, L.F. 2005. Main chain and side chain dynamics of the ubiquitin conjugating enzyme variant human Mms2 in the free and ubiquitin-bound states. Biochemistry 44: 8770–8781. [DOI] [PubMed] [Google Scholar]
- Szyperski T., Braun, D., Fernandez, C., Bartels, C., and Wüthrich, K. 1995. A novel reduced-dimensionality triple-resonance experiment for efficient polypeptide backbone assignment, 3D CO HN N CA. J. Magn. Reson. B. 108: 197–203. [Google Scholar]
- Takahashi M., Inaguma, Y., Hiai, H., and Hirose, F. 1988. Developmentally regulated expression of a human “finger”-containing gene encoded by the 5′ half of the ret transforming gene. Mol. Cell. Biol. 8: 1853–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra N., Feller, S.E., Pastor, R.W., and Bax, A. 1995. Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. J. Am. Chem. Soc. 117: 12562–12566. [Google Scholar]
- Viatour P., Merville, M.-P., Bours, V., and Chariot, A. 2005. Phosphorylation of NF-κB and IκB proteins: Implications in cancer and inflammation. Trends Biochem. Sci. 30: 43–52. [DOI] [PubMed] [Google Scholar]
- Wang C.Y. and Palmer, A.G. 2003. Solution NMR methods for quantitative identification of chemical exchange in 15N-labeled proteins. Magn. Reson. Chem. 41: 866–876. [Google Scholar]
- Wang C., Deng, L., Hong, M., Akkaraju, G.R., Inoue, J., and Chen, Z.J. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412: 346–351. [DOI] [PubMed] [Google Scholar]
- Wang B., Alam, S.L., Meyer, H.H., Payne, M., Stemmler, T.L., Davis, D.R., and Sundquist, W.I. 2003. Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J. Biol. Chem. 278: 20225–20234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts T.H. 2005. TNF/TNFR family members in costimulation of T cell responses. Annu. Rev. Immunol. 23: 23–68. [DOI] [PubMed] [Google Scholar]
- Willard L., Ranjan, A., Zhang, H.Y., Monzavi, H., Boyko, R.F., Sykes, B.D., and Wishart, D.S. 2003. VADAR: A web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 31: 3316–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.C., Shelling, J.G., and Sykes, B.D. 1985. NMR approaches to the characterization of the interaction of metal ions with proteins NMR in the life sciences (eds. A.M. Bradbury and C. Nicolini), pp. 93–104. Plenum Press, New York.
- Winn P.J., Religa, T.L., Battey, J.N., Banerjee, A., and Wade, R.C. 2004. Determinants of functionality in the ubiquitin conjugating enzyme family. Structure 12: 1563–1574. [DOI] [PubMed] [Google Scholar]
- Wittekind M. and Mueller, L. 1993. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the α- and β-carbon resonances in proteins. J. Magn. Reson. B101: 201–205. [Google Scholar]
- Wolfram S. 1996. The Mathematica book, 3d ed. Wolfram Media/Cambridge University Press, Cambridge, England.
- Wooff J., Pastushok, L., Hanna, M., Fu, Y., and Xiao, W. 2004. The TRAF6 RING finger domain mediates physical interaction with Ubc13. FEBS Lett. 566: 229–233. [DOI] [PubMed] [Google Scholar]
- Wu H. and Arron, J.R. 2003. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays 25: 1096–1105. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Forman-Kay, J.D., and Kay, L.E. 1993. Two-dimensional NMR experiments for correlating 13Cβ and 1Hδ/ɛ chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J. Am. Chem. Soc. 115: 11054–11055. [Google Scholar]
- Ye H., Arron, J.R., Lamothe, B., Cirilli, M., Kobayashi, T., Shevde, N.K., Segal, D., Dzivenu, O.K., Vologodskaia, M., Yim, M., et al. 2002. Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418: 443–447. [DOI] [PubMed] [Google Scholar]
- Zhang O.W., Kay, L.E., Olivier, J.P., and Forman-Kay, J.D. 1994. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed-field gradient NMR techniques. J. Biomol. NMR 4: 845–858. [DOI] [PubMed] [Google Scholar]
- Zhang M.H., Windheim, M., Roe, S.M., Peggie, M., Cohen, P., Prodromou, C., and Pearl, L.H. 2005. Chaperoned ubiquitylation—Crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20: 525–538. [DOI] [PubMed] [Google Scholar]
- Zheng N., Wang, P., Jeffrey, P.D., and Pavletich, N.P. 2000. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102: 533–539. [DOI] [PubMed] [Google Scholar]
- Zheng N., Schulman, B.A., Song, L., Miller, J.J., Jeffrey, P.D., Wang, P., Chu, C., Koepp, D.M., Elledge, S.J., Pagano, M., et al. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709. [DOI] [PubMed] [Google Scholar]
- Zhou H., Wertz, I., O'Rourke, K., Ultsch, M., Seshagiri, S., Eby, M., Xiao, W., and Dixit, V.M. 2004. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature 427: 167–171. [DOI] [PubMed] [Google Scholar]