Abstract
Dissimilatory oxidation of thiosulfate in the green sulfur bacterium Chlorobium limicola f. thiosulfatophilum is carried out by the ubiquitous sulfur-oxidizing (Sox) multi-enzyme system. In this system, SoxY plays a key role, functioning as the sulfur substrate-binding protein that offers its sulfur substrate, which is covalently bound to a conserved C-terminal cysteine, to another oxidizing Sox enzyme. Here, we report the crystal structures of a stand-alone SoxY protein of C. limicola f. thiosulfatophilum, solved at 2.15 Å and 2.40 Å resolution using X-ray diffraction data collected at 100 K and room temperature, respectively. The structure reveals a monomeric Ig-like protein, with an N-terminal α-helix, that oligomerizes into a tetramer via conserved contact regions between the monomers. The tetramer can be described as a dimer of dimers that exhibits one large hydrophobic contact region in each dimer and two small hydrophilic interface patches in the tetramer. At the tetramer interface patch, two conserved redox-active C-terminal cysteines form an intersubunit disulfide bridge. Intriguingly, SoxY exhibits a dimer/tetramer equilibrium that is dependent on the redox state of the cysteines and on the type of sulfur substrate component bound to them. Taken together, the dimer/tetramer equilibrium, the specific interactions between the subunits in the tetramer, and the significant conservation level of the interfaces strongly indicate that these SoxY oligomers are biologically relevant.
Keywords: Chlorobium limicola f. thiosulfatophilum, thiosulfate oxidation, SoxY, crystal structure, sulfur binding
Dissimilatory oxidation of sulfur compounds is carried out by a variety of members of the Bacteria and Archaea. It is regarded as a primitive metabolism (Kelly 1987) that evolved to distinct sulfur-oxidizing pathways in phylogenetically diverse species (for reviews, see Friedrich 1998; Brüser et al. 1999). Thiosulfate is an important key intermediate in the sulfur cycle (Jørgensen 1990), which is reflected in the diversity of thiosulfate-oxidizing bacteria (Friedrich 1998). Two major thiosulfate-oxidizing pathways have been identified: (a) the “tetrathionate” pathway, which is mainly restricted to acidophilic thiobacilli and oxidizes thiosulfate to sulfate via tetrathionate as an intermediate (Kelly et al. 1997), and (b) the widely distributed periplasmic sulfur-oxidizing (sox) pathway found in neutrophilic, respiratory, and phototrophic Proteobacteria, and in green sulfur bacteria (Friedrich et al. 2001, 2005).
The best characterized thiosulfate-oxidizing systems to date are the Sox systems of Paracoccus versutus (Lu and Kelly 1983; Lu et al. 1985; Lu 1986) and Paracoccus pantotrophus (Friedrich et al. 2000; Rother et al. 2001). The P. pantotrophus system, for instance, combines four components, SoxYZ, SoxAX, SoxB, and SoxCD, which work together toward a full thiosulfate oxidizing activity that generates eight electrons and two sulfate molecules as end products (Friedrich et al. 2000). However, the functions of some components remain elusive. SoxYZ is regarded as the central protein complex of the Sox system because it binds reduced sulfur substrates via a thioether or a thioester bond at a conserved C-terminal cysteine of the SoxY subunit (Quentmeier and Friedrich 2001). Subsequently, SoxYZ is thought to present the activated thiosulfate to the other Sox enzymes. In addition to its sulfur-binding capacities, SoxYZ also appears to play a conditioning role in the thiosulfate-oxidizing activity of the Sox system through the redox state of the C-terminal cysteine of its SoxY subunit. It has already been shown that the reconstituted Sox system having reduced SoxYZ exhibits a substantially lower activity (Quentmeier and Friedrich 2001), while SoxYZ with SoxY disulfide-bridged subunits and SoxY-persulfide appears to increase the thiosulfate-oxidizing activity of the system (A. Quentmeier, P. Janning, and C.G. Friedrich, pers. comm.). The cytochrome c SoxAX complex has been proposed to mediate the binding event of thiosulfate to SoxYZ (Friedrich et al. 2001; Bamford et al. 2002), while the manganese-containing SoxB (Cammack et al. 1989) and the molybdoheme protein SoxCD (Wodara et al. 1997; Quentmeier et al. 2000) have been proposed to be implicated in completing the oxidation of the bound thiosulfate molecule to two sulfate molecules (Friedrich et al. 2001). Intriguingly, SoxCD is not present in the green sulfur bacteria Chlorobium limicola f. thiosulfatophilum (Verté et al. 2002) and Chlorobium tepidum (Eisen et al. 2002) and the phototrophic purple sulfur bacterium Allochromatium vinosum (Friedrich et al. 2005). In these bacteria, further oxidation of thiosulfate likely takes place in the cytoplasm by using a “reverse” sulfite reductase, DsrAB (Schedel et al. 1979; Pott and Dahl 1998; Eisen et al. 2002; Dahl et al. 2005), which is supposed to oxidize sulfide to sulfite, and by adenosine-5′-phosphosulfate (APS) reductase and ATP sulfurylase, which may catalyze the final oxidation step of sulfite to sulfate (Hipp et al. 1997; Eisen et al. 2002).
Despite a growing body of biochemical and physiological data, the molecular mechanism of the Sox system remains undeciphered, in particular with respect to the central SoxYZ complex. Thus far, most biochemical data have been generated using the heterodimeric SoxYZ protein. However, three lines of evidence strongly support the notion that the participating subunits SoxY and SoxZ can on their own form protomers. First, an apparently stand-alone “enzyme A” with an affinity for thiosulfate of 360 μM has been isolated as an essential protein of the thiosulfate-oxidizing system of Paracoccus versutus (Lu et al. 1985). Second, a “stand-alone” SoxZ crystal structure from Thermus thermophilus (PDB code 1v8h, unpubl.) has recently been determined, revealing an apparent dimer. Third, we have recently crystallized SoxY from Chlorobium limicola f. thiosulfatophilum as a tetramer (Stout et al. 2006). These results stress that ongoing and future biochemical studies of the SoxYZ complex will need to address SoxY and SoxZ individually, as well as in complex. Here, we report the high-resolution crystal structure of tetrameric SoxY whose subunits, despite the reducing crystal growth conditions (Stout et al. 2006), are remarkably linked via disulfide bridges. It also reveals other unexpected features and specific interactions leading to tetramer formation. Furthermore, we demonstrate the capacity of SoxY to bind reduced sulfur compounds and address the possible oligomeric states of SoxY using analytical chromatographic methods.
Results
Overall structure
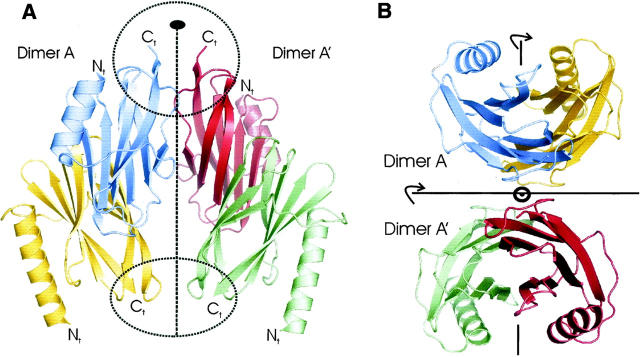
The SoxY crystal structure was determined by MIRAS and was refined to 2.15-Å resolution (Table 1). The structure reveals an αβ-protein consisting of an N-terminal α-helix and a β-sandwich domain. Two SoxY subunits are present in the asymmetric unit and interact extensively with each other, forming an extended β-sandwich structure with six and eight β-strands in the upper and lower sandwich layer, respectively. In turn, the resulting SoxY forms a tetramer with a second SoxY dimer related via a twofold crystallographic axis (Fig. 1A).
Table 1.
Refinement statistics
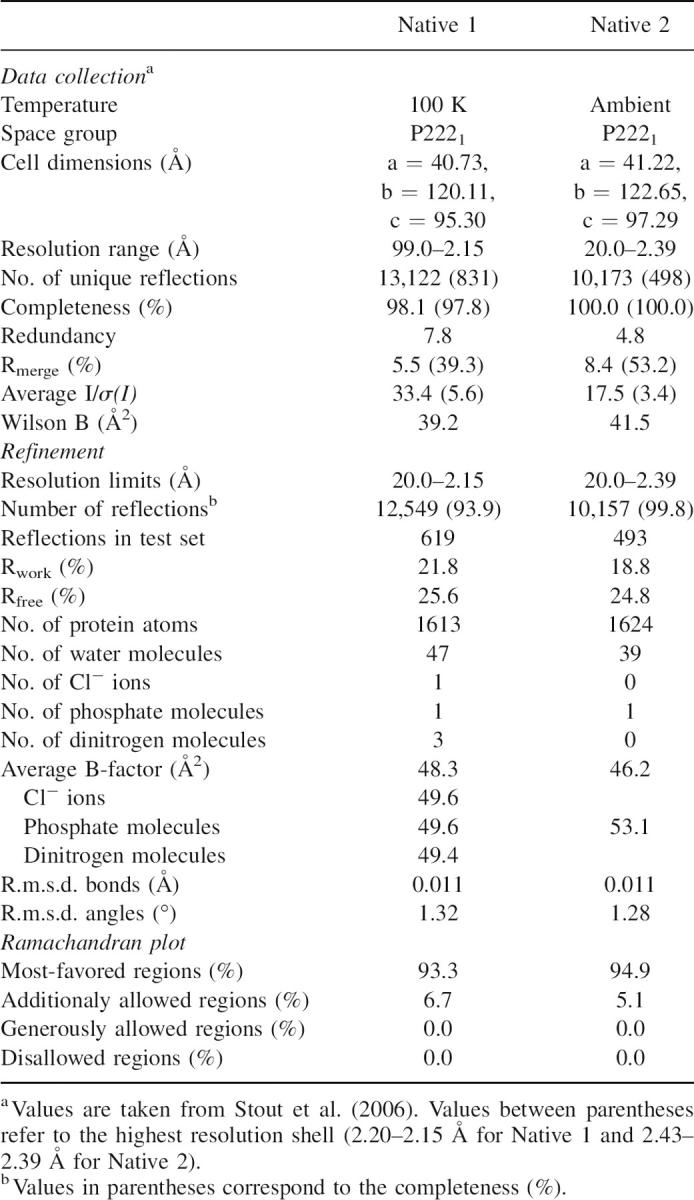
Figure 1.
(A) The tetramer viewed from the side, showing dimer A and dimer A′, which are related via a crystallographic twofold axis (dashed line with filled ellipsoid on top). Tetramerization occurs by means of two small interface patches at the distal ends of the oligomer (two dashed ellipsoids). (B) The SoxY tetramer viewed along the crystallographic twofold axis. The total symmetry consists of three orthogonal twofold axes: a twofold axis perpendicular to the page, and two twofold axes parallel to the page; this is referred as D2 symmetry.
Gel filtration experiments already indicated that recombinant, “as isolated” SoxY is present in solution as a 52-kDa tetramer (Stout et al. 2006). Interestingly, SDS-PAGE showed that the tetramer is built of dimer subunits that are covalently linked via a disulfide bridge, as demonstrated by reduction with β-mercaptoethanol (data not shown). Furthermore, untreated crystalline SoxY migrates identically on an SDS-polyacrylamide gel as nonreduced purified SoxY, proving that crystallized SoxY remains covalently linked via a disulfide bridge, despite the presence of 10 mM dithiothreitol (DTT) in the crystallization buffer (Stout et al. 2006). The site of covalent linkage was assigned to C120, which is the only cysteine residue in the mature SoxY (Fig. 2). Consequently, a total of two disulfide bridges are formed between the four cysteine residues in the tetramer. Unfortunately, no model could be built for the disulfide bridges due to the very poor quality of the electron densities of the C120 residues, indicating a highly flexible structural feature, which is in all likelihood correlated with the high glycine content of the GGCG(G) motif. Similar intersubunit disulfide bridges were also demonstrated for the SoxY subunit of P. pantotrophus (Quentmeier et al. 2003). From this point on, the term “oxidized SoxY” will be used to refer to SoxY being covalently linked via disulfide bridges.
Figure 2.
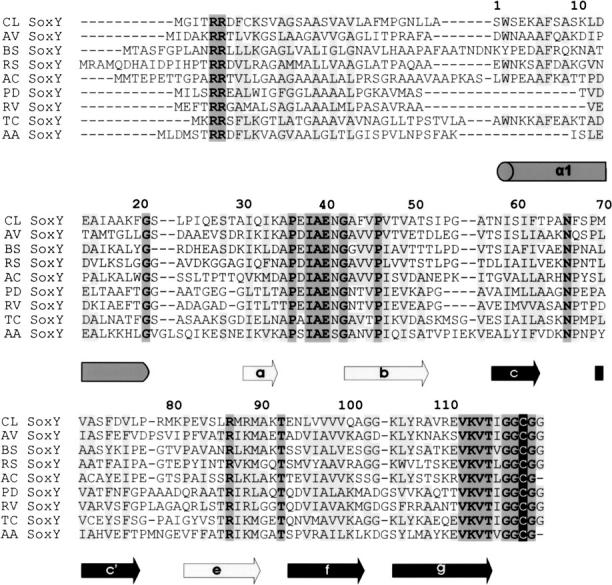
Sequence alignment of SoxY. The secondary structure of Chlorobium limicola f. thiosulfatophilum is indicated. From top to bottom, sequences from Chlorobium limicola f. thiosulfatophilum DSM249 (CL) (AAL68884), Allochromatium vinosum (AV) (ABE01369), Bradyrhizobium sp. Bta1 (BS) (ZP_00863129), Ralstonia solanacearum (RS) (ZP_00944480), Acidiphilium cryptum (AC) (ZP_01144909), Paracoccus denitrificans (PD) (CAB94380), Rhodovulum sulfidophilum (RV) (AAF99432), Thiomicrospira crunogena (TC) (YP_390873), and Aquifex aeolicus (AA) (NP_214241) are aligned. (Black highlighting) Strictly conserved C-terminal cysteine, (dark gray) all other identical residues (allowing for Asp/Glu replacements), (pale gray) identical residues in five or more sequences and conservative replacements in all sequences. The sequence alignment was executed in STRAP. Secondary structure elements indicated: helices (gray colored cylinders) and β-strands (arrows); β-strands a, b, and e (pale gray) and c, c′, f, and g (dark gray) constitute β-sheets I and II, respectively.
SoxY monomer
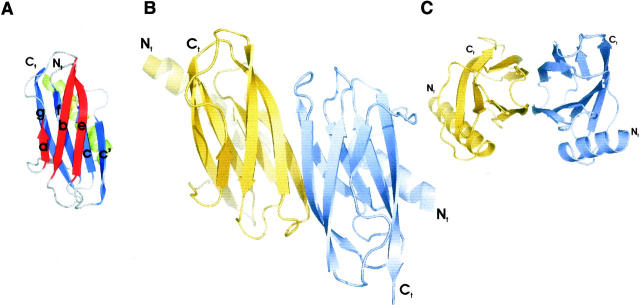
The 13-kDa SoxY monomer consists of an N-terminal α-helix (S1-F20) packed against an Ig-like domain (I31-I117). A loop (G21-T30) covering one side of the β-sandwich domain connects the α-helix to the Ig-like domain (Fig. 3A). Variations exist in the 3D-topology of Ig-domains, and distinct Ig-sets have been defined on the basis of their topological differences (Williams and Barclay 1988; Bork et al. 1994; Harpaz and Chothia 1994). The Ig-like fold we observe in SoxY is an s-type Ig-domain based on the classification scheme of Bork et al. (1994) (Fig. 3A). The core of SoxY is built up of hydrophobic residues from both β-sheets. This is a key signature of the Ig-like fold (Halaby et al. 1999; Kister et al. 2002). In contrast to the core residues, the residues of the β-sheets pointing to the solvent are mainly hydrophilic, resulting in alternating polar/nonpolar sequence patterns of the β-strands, which is a typical sequence feature in antiparallel β-structures (Mandel-Gutfreund and Gregoret 2002).
Figure 3.
(A) SoxY monomer showing the secondary structure elements. Respectively, 16.4% and 36.9% of the modeled residues are present in α-helices and β-strands. The remaining residues are part of the loops and turns connecting the secondary structure elements. The N-terminal α-helix (S1-F20) is connected via a loop to a β-sandwich domain (I31-G122) having seven antiparallel β-strands: β-strands a (I31-K34), b (A43-T51), c (N58-T63), c′ (M70-L77), e (P82-M90), f (E94-A102), and g (K105-T116). Nomenclature of the β-strands was taken from Bork et al. (1994). β-strands a, b, and e (red) form β-sheet I. β-strands c, c′, f, and g (blue) constitute β-sheet II. Seven turns are also identified. E40-A43 and L77-M80 are type II β-turns, A102-K105 is a type II′ β-turn, and I53-A56 and T63-N66 are type III β-turns (Venkatachalam 1968). P36-A39 and N66-P69 are also cataloged as turns because the Cα(i)–Cα(i+3) distance is <7 Å (Rose et al. 1985). (B) The SoxY dimer viewed along the twofold noncrystallographic symmetry axis showing the extended β-sandwich. The two monomers are shown in different colors. (C) Side view of the SoxY dimer showing the extended β-sandwich consisting of two continuous β-sheets.
SoxY dimer
The two monomers within the asymmetric unit interact strongly. They are related to each other via a twofold noncrystallographic axis, perpendicular to the β-sheets, and use one edge of their Ig-like domains to form an extended β-sandwich dimer: β-sheet I consists of strands a, b, and e of subunit A and B; β-sheet II is built up of strands c, c′, f, and g of both subunits (Fig. 3B). Both β-sheets exhibit a continuous hydrogen-bonding network, in contrast to canonical extended β-sandwiches. The latter have one continuous (sheet II) and one discontinuous (sheet I) β-sheet separated in the middle by water molecules (Richardson and Richardson 2002) that solvate the free amide and carbonyl peptide groups that are too distant from each other to form direct intersubunit hydrogen bonds. In the dimer interface of SoxY, however, a total of 12 direct intersubunit hydrogen bonds are present between the main chains of β-strands c′ and e and their equivalent β-strands of the opposite monomer (Supplemental Table 1).
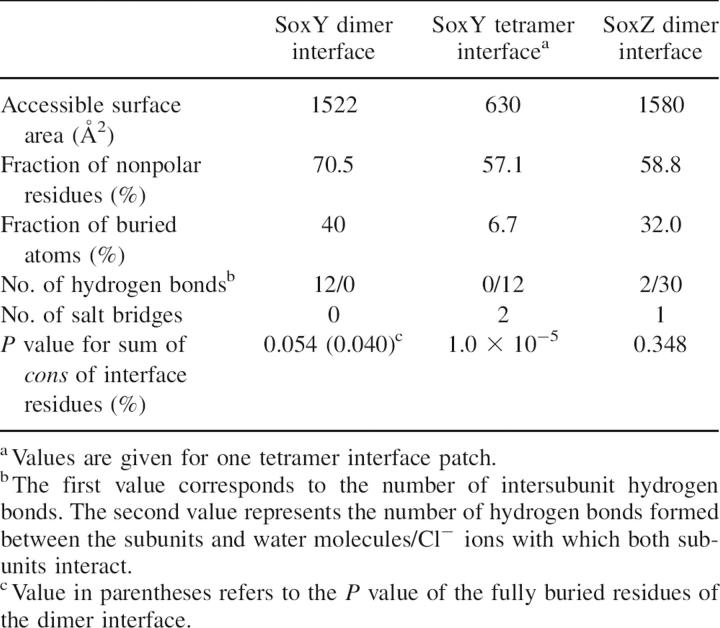
Examination of the SoxY dimer interface indicates that the structural and physicochemical characteristics are similar to those of other stable protein–protein interfaces in biological protein complexes (Table 2). The dimer interface buries a surface of 1522 Å2, a typical value for standard-size protein–protein interfaces (1200–2000 Å2) (Lo Conte et al. 1999; Bahadur et al. 2004). The dimer interface has a substantial fraction (40%) of the completely buried interface atoms and is mainly hydrophobic (70.5%). The former value is in accordance with the mean percentage of completely shielded interface atoms (34%–36%) (Lo Conte et al. 1999; Bahadur et al. 2003). The considerable burial of hydrophobic surface is assumed to be a key feature of stable protein–protein interfaces (Young et al. 1994; Jones and Thornton 1996; Tsai et al. 1997). The bulk of the hydrophobic residues in the dimer interface is located at the center and constitutes the inward-pointing residues of edge β-strands c′ and e of both monomers. They are flanked at the bottom and at the top by the two layers of intersubunit hydrogen bonds (12 in total) that are part of the β-ladder network of the two extended β-sheets as mentioned above. Hydrogen bonds are thought to play an important role in the specificity of protein–protein binding (Xu et al. 1997a,b) since they are mainly formed by complementary electrostatic, polar, or charged atom groups of both proteins.
Table 2.
Overview of the structural, physicochemical, and conservation parameters of the SoxY dimer and tetramer interface, and of the SoxZ dimer interface
A dops score of 78% indicated that the multiple sequence alignment had sufficient discriminative power to make a distinction between conserved and nonconserved residues. The significance of the conservation of the dimer interface residues could therefore be statistically assessed by a bootstrap analysis (see Materials and Methods). Estimated P values of 5.4% and 4.0% for the total dimer interface (20 residues) and the core of the dimer interface (10 residues) suggest that the central zone of dimer interface is significantly conserved.
SoxY tetramer
In contrast to the considerable extent of the dimerization interface, contacts that stabilize the tetramer occur at the distal ends of the tetramer by virtue of two small interface patches, separated by a solvent-filled space between the two SoxY dimers (Fig. 1A). Two strongly conserved protein regions of both SoxY subunits contribute to the tetramer interface: the a-b-loop connecting β-strands a and b (P36-G42) and the beginning of β-strand b (A43-P46), R111-I117 of β-strand g, and the equivalent regions of the symmetry-related subunit. The core of the interface is built of the strictly conserved peptide sequences E37-E40 of the a-b loop of both subunits, exhibiting a β-like conformation. These short peptides are oriented in an antiparallel manner but do not interact via direct intersubunit backbone hydrogen bonds. Instead, six ordered water molecules and two ordered chloride ions intercalate between the peptides and set up an elaborate hydrogen-bonding network involving 14 water molecule/Cl− ion–protein hydrogen bonds and seven mutual water molecule/Cl− ion hydrogen bonds (Fig. 4B). For this purpose, the two chloride ions and four water molecules are organized as a hexagon at the center of the interface, while the remaining two water molecules lie next to the hexagon in the same plane. These chloride ions appear to be an artifact of crystal cryo-cooling at 100 K, since they are replaced by water molecules in a room-temperature crystal structure (data not shown). In contrast, the four water molecules of the hexagon are present in both crystal structures.
Figure 4.
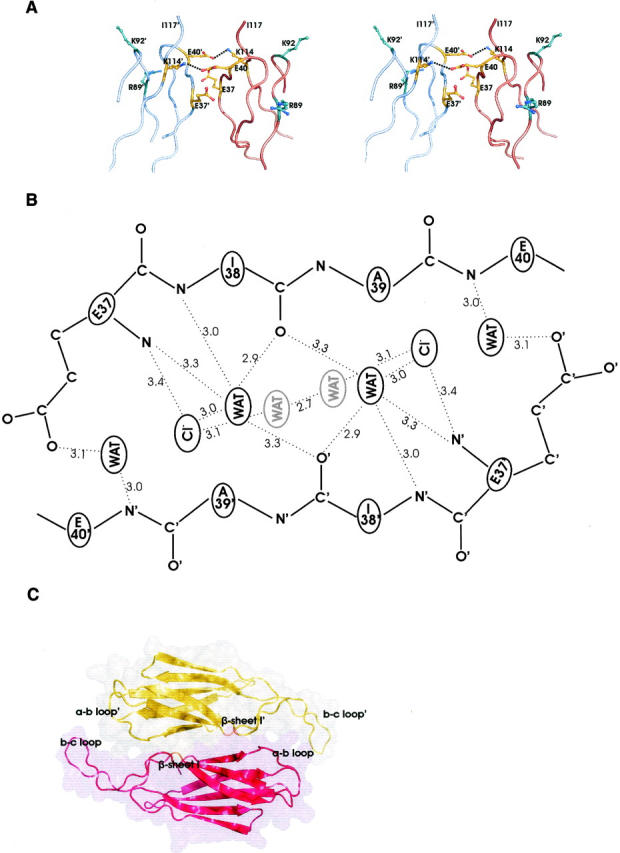
(A) The tetramer interface patch with the two conserved salt bridges (dashed lines) between E40-K114′ and E40′-K114 defining both sides of the patch. E37′ and E37 form hydrogen bonds with E40 and E40′, respectively. K114, K114′, and the differently colored K92, K92′, R89, and R89′ are potential candidates to interact with a modified, sulfur substrate-bound cysteine. (B) Schematic overview of the hydrogen bonds at one tetramer interface patch. The four central water molecules and the two chloride ions form a hexagon. From this top view, the waters in gray are the lower vertexes, the chloride ions constitute the middle vertexes, and the waters in black are the top vertexes. (C) Space-filling representation of the SoxZ dimer, with both monomers differently colored. Dimerization occurs at β-sheets I of both monomers. Contacts also exist between their a-b and b-c loops.
Both sides of one tetramer interface patch are defined by charged residues: E37, E40, and K114, and their symmetry-related residues (Fig. 4A). E40 is positioned with its carboxyl group between the carboxyl group of E37′ and NZ of K114′ of the symmetry-related monomer and forms a strictly conserved salt bridge with K114′ (3.8 Å). Moreover, it is within hydrogen-bonding distance of OE2 of E37′ (2.8 Å). This type of electrostatic interaction, consisting of an identical charge pair (E40–E37′) partially compensated by an opposite charge (K114′), occurs frequently at protein–protein interfaces (Xu et al. 1997b).
The buried surface area per tetramer interface patch is 630 Å2. In contrast to the dimer interface that buries a substantial fraction of interface atoms, only a small fraction (6.7%) of interface atoms is completely shielded from solvent in the tetramer interface. This can be explained by the hydration and the relatively hydrophilic nature of the interface (nonpolar area fraction of 57.1%), which in fact resembles that of solvent-exposed protein surfaces.
By analogy to the dimer interface, a bootstrap procedure was performed to assess the average conservation of the tetramer interface. The low P value (0.001%) points to a highly conserved interface region on the protein surface.
Location of the disulfide bridges and the potential sulfur-binding site
Although the reactive C-terminal cysteines could not be modeled, the position of the disulfide bridges can be deduced based on the quaternary structure of SoxY, which brings two symmetry-related C120 residues in close proximity to one another to allow formation of a disulfide bridge. The last modeled residues of both β-strands g, I117, and I117′, protrude from the tetramer interface into the solvent (Fig. 4A). It is therefore likely that the intersubunit disulfide bond between the C120 residues is also exposed to the solvent, residing at the top of each tetramer interface patch.
Quentmeier et al. (2003) showed that P. pantotrophus SoxY also forms an intersubunit disulfide bridge with a second SoxY molecule by means of a C-terminal cysteine, and suggested that the bridge activates the cysteines for sulfur substrate binding via a thiol/disulfide exchange reaction. The SoxY structure reveals several residues on, or in the vicinity of, the tetramer interface patch that may electrostatically stabilize the resulting adduct after sulfur substrate binding. The conserved residues K114 and K114′, which are part of the charge triads flanking the tetramer interface patches, and the conserved residues R89 and R89′ are good candidates to interact with the negatively charged S-thiocysteinesulfonate. The nonconserved residues K92 and K92′ are also potential candidates for such an interaction (Fig. 4A).
SoxZ dimer interface
SoxZ, like SoxY, is a β-sandwich protein exhibiting a single s-type Ig-like domain (pdb code 1v8h, unpubl.). Apparently, SoxZ crystallizes as a dimer, but, in contrast to SoxY dimers, SoxZ does not dimerize by means of its edge strands. Instead, the two SoxZ monomers within the dimer are stacked onto each other using their three-stranded β-sheets, making additional contacts between their extended b-c and a-b loops (Fig. 4C). The dimer interface buries a total surface area of 1580 Å2 and is less hydrophobic (58.8%) compared with the dimer interface of SoxY (Table 2). A rather limited number of electrostatic interactions between the monomers is present at the interface: two hydrogen bonds and one salt bridge. In addition, 11 water molecules reside at the interface, which form hydrogen bonds (30 in total) with both monomers. Furthermore, in contrast to the high conservation levels of the SoxY interfaces, the SoxZ interface appears to be evolutionarily less significant, as inferred by a P value of 0.348.
Because of the limited number of direct electrostatic interactions and the apparent evolutionary insignificance of this interface, the possibility that this is an artifact of the process of crystallogenesis has to be considered. It is therefore not possible to draw conclusions about the quaternary structure of SoxZ in solution. Nevertheless, this crystal structure of a stand-alone SoxZ, as for the SoxY structure, argues against SoxYZ being an obligate complex, i.e., a complex that is necessarily formed during folding because the SoxY and SoxZ polypeptide chains would depend on each other for their structural integrity. A potential interaction between SoxY and SoxZ is discussed below.
Sulfur binding
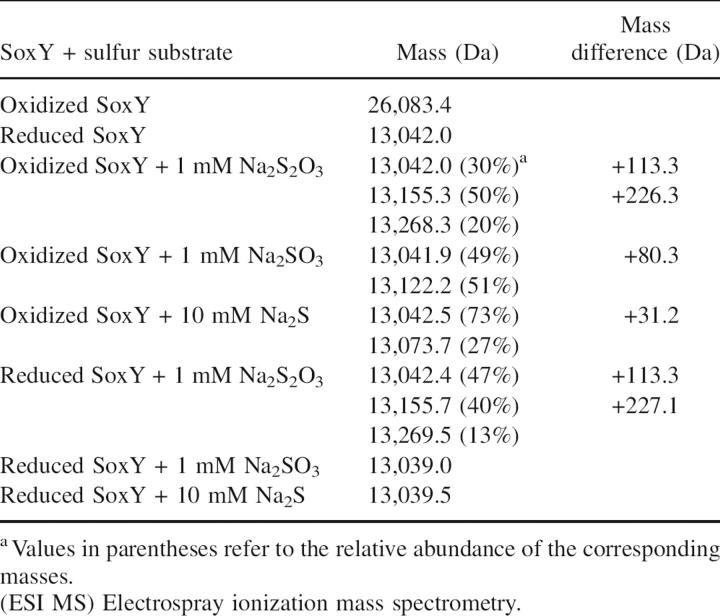
The heterodimeric Paracoccus pantotrophus SoxYZ was shown to undergo drastic changes in its quaternary structure, becoming a heterotetramer upon modification of its C-terminal cysteine with N-ethyl maleimide (Quentmeier et al. 2003). We investigated the effect of sulfur substrate binding on the oligomerization of Chlorobium limicola f. thiosulfatophilum SoxY. For this purpose, oxidized and reduced SoxY were incubated with thiosulfate, sulfite, and sulfide. Reaction mixtures were analyzed using electrospray ionization mass spectrometry (ESI MS), and the results of the binding reactions with different sulfur compounds are summarized in Table 3. The theoretical masses of reduced and oxidized SoxY were calculated to be 13,042.2 Da and 26,082.2 Da, respectively.
Table 3.
Masses measured by ESI MS of the reaction mixtures of oxidized and reduced SoxY with Na2S2O3, Na2SO3, and Na2S
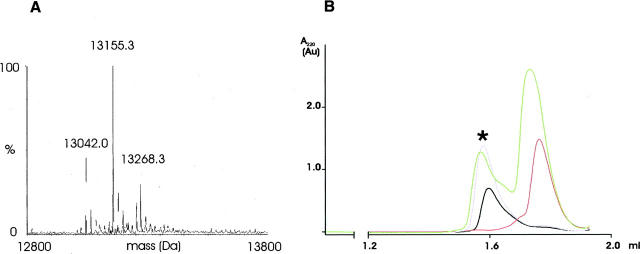
The observed masses of 13,155.3 Da and 13,155.7 Da for oxidized and reduced SoxY, respectively, incubated with 1 mM thiosulfate, are both 113.3 Da heavier than reduced SoxY and point to the addition of S2O3 2− (+112 Da) (Fig. 5A). Minor peaks of 13,268.2 and 13,269.5 Da, having an extra mass of 226.2 and 227.3 Da, respectively, were also observed. These additional masses can be explained in two ways. They might correspond to a covalently linked tetrathionate (S4O6 2−; 224 Da), although in this case addition does not proceed via a sulfur–sulfur bond. Alternatively, there is also the possibility that, in addition to the covalently bound molecule, a second thiosulfate molecule is bound noncovalently. Incubation of oxidized SoxY with 1 mM sulfite resulted in a peak of 13,122.2 Da, including an extra mass of 80.3 Da for SO3 2− (80 Da). Adding 10 mM Na2S to oxidized SoxY resulted in a complete reduction of the disulfide bridge, as inferred by the dominant peak of 13,042.5 Da. Only a minor fraction had an extra mass of 31.2 Da, pointing to the addition of sulfide (32 Da). Binding experiments of reduced SoxY with 1 mM sulfite and 10 mM sulfide were also conducted. However, no masses were detected that can correspond to a SoxY-sulfite and a SoxY-sulfide adduct. MALDI MS was performed on the peptides of tryptic digest reaction mixtures of reduced and oxidized SoxY with thiosulfate. Peptide masses of 773 Da and 1129 Da for a perfectly cleaved C-terminal peptide (VTIGGCGG) and a C-terminal peptide with one miscleavage (EVKVTIGGCGG), respectively, were detected at low intensity, strongly indicating the covalent binding of thiosulfate. Concerning a possible tetrathionate adduct, no masses were observed that correspond to C-terminal peptides covalently linked with tetrathionate.
Figure 5.
(A) Reconstructed electrospray mass spectrum (ESI MS) of oxidized SoxY treated with 1 mM Na2S2O3. (B) Analytical gel filtration chromatograms of SoxY. Experimental procedures are described in the Materials and Methods. (Black line) “As isolated” (oxidized) SoxY, (pink line) “as isolated” SoxY + 10 mM sodium thiosulfate, (red line) “as isolated” SoxY + 10 mM DTT, (green line) DTT-reduced SoxY + 10 mM sodium thiosulfate. Chromatograms of “as isolated” and DTT-reduced SoxY treated with sodium sulfide, sodium sulfite, and N-ethylmaleimide are similar to the chromatogram of DTT-reduced SoxY. The chromatograms of SoxY treated with thiosulfate and loaded on the same column pre-equilibrated with 100 mM sodium thiosulfate are identical to that of the tetrameric “as isolated” SoxY. (Asterisk) Fraction of the sample containing “as isolated” SoxY and 10 mM sodium thiosulfate collected at the elution volume of the tetramer and subsequently analyzed by ESI MS.
Analytical gel filtration experiments showed that treatment with different reduced sulfur components affects the oligomeric state of SoxY (Fig. 5B). Oxidized, “as isolated” SoxY (elution volume [Ve] = 1.60 mL) and “as isolated” SoxY incubated with thiosulfate (Ve = 1.58 mL) eluted as a tetramer, while SoxY treated with DTT (Ve = 1.76 mL), sulfite (Ve = 1.73 mL), sulfide (Ve = 1.76 mL), and glutathione (Ve = 1.74 mL), respectively, appears to dissociate into dimers. Incubation of reduced SoxY with sulfite (Ve = 1.74 mL), sulfide (Ve = 1.74 mL), and NEM (Ve = 1.72 mL) did not affect the dimeric state of SoxY (data not shown). Conversely, reduced SoxY incubated with thiosulfate shifted partially its oligomeric state from dimer (Ve = 1.73 mL) to tetramer (Ve = 1.57 mL). However, MS analysis of the product eluted as a tetramer (Fig. 5B, asterisk) revealed that SoxY was mainly present in the disulfide-bonded form. An identical reaction of oxidized SoxY incubated with thiosulfate was therefore also subjected to analytical gel filtration using the same buffer but spiked with Na2S2O3. SoxY again eluted as a tetramer, and ESI MS of the corresponding eluate showed that SoxY was now mainly present as a SoxY-thiosulfate adduct. Based on these data, it can be concluded that the chemical composition of the adhered molecule conditions the oligomeric state of SoxY. When treated with different reductants, SoxY appears to dissociate into dimers, while when incubated with thiosulfate it is predisposed to form tetramers.
Discussion
We have presented structural evidence of a SoxY tetramer consisting of two dimeric β-sandwiches that interact with each other by means of two small interface patches at the tips of the dimers. We have analyzed the structural and physicochemical properties of the dimer and tetramer interface and have assessed the statistical significance of the residue conservation of both interfaces. Finally, we have investigated the effect of binding of sulfur substrates on the oligomerization.
Friedrich and coworkers (2000) isolated SoxY from Paracoccus pantotrophus in complex with SoxZ as a heterodimer. To date, most biochemical data are offered for the Sox system using the SoxYZ heterodimer (Rother et al. 2001; Friedrich et al. 2001; Quentmeier and Friedrich 2001; Quentmeier et al. 2003). It is generally believed that this protein is a stable, obligate complex that is active only in the form of a heterodimer. However, a combination of structural and biochemical data suggest that SoxYZ is not an obligate heterodimer, and that the individual proteins may exist on their own. First, biochemical evidence, provided by the reconstitution of a fully active Sox system using an apparently stand-alone thiosulfate-binding protein “enzyme A” (Lu and Kelly 1983; Lu et al. 1985), is indicative of a possible biological function for this individual constituent. Second, the SoxY structure presented here also argues against the point of view that SoxYZ is an obligate complex in which both proteins are dependent on each other for their folding and structural integrity. Moreover, a recently determined SoxZ structure, showing an apparent dimer, indicates that this protein is also stable on its own (pdb 1v8h, unpubl.). A plethora of unprecedented structural data is provided, supplemented with biochemical data, that together point to biologically relevant interactions between SoxY subunits within the tetramer.
A first strong argument supporting the biological relevance of the dimer and the tetramer interfaces resides in their levels of conservation. Several studies state that protein–protein interfaces are significantly more conserved than their respective solvent-exposed surfaces (Valdar and Thornton 2001b; Caffrey et al. 2004; Mintseris and Weng 2005) and biologically irrelevant crystal contact points (Valdar and Thornton 2001a). Our bootstrap analyses are in line with these results, indicating that the core of the dimer interface (P value = 0.040) and of the tetramer interface (P value = 1.0 × 10−5) are statistically conserved. Concerning the conservation of the tetramer interface, we draw attention to the fact that these residues are in the vicinity of the sulfur substrate-binding cysteine and may also be involved in transient interactions with other components of the Sox system. Hence, the evolutionary pressure on these residues may be higher than on other surface residues since their combination is likely to be optimal for interactions with the different Sox enzymes.
Another convincing body of data covers gel filtration experiments and the structural and physicochemical analyses of the dimer interface and the tetramer interface. Analytical gel filtration demonstrated that reduced SoxY elutes as a dimer, indicating that the dimer on its own constitutes a stable protomer. Furthermore, the dissection of the dimer interface highlighted three major properties that strongly argue for the SoxY dimer as a specific protein–protein complex. First, a considerable surface area (1522 Å2) is buried at this dimer interface. Second, the dimer interface has a distinct central core of hydrophobic residues. Several studies (Miller 1989; Tsai et al. 1997; Bahadur et al. 2003) have emphasized the presence of such a hydrophobic center in medium and large protein–protein interfaces, strengthening the basic paradigm that hydrophobicity is a major stabilizing factor in protein–protein association (Chothia and Janin 1975). Third, the considerable number of hydrogen bonds (12 in total) also argues for a biologically relevant protein–protein interface. These hydrogen bonds not only make a considerable energetic contribution to protein–protein binding, but could also imprint binding specificity as a consequence of the electrostatic complementarity of the hydrogen bond donor and acceptor groups (Xu et al. 1997a,b). All hydrogen bonds at the dimer interface are part of the hydrogen bond ladder pattern across the two extended β-sheets. This arrangement of two rows of hydrogen bond donor and acceptor groups is specific for β-sandwiches and likely restricts potential binding partners to proteins having a similar arrangement of donor and acceptor groups.
In contrast to the extended dimer interface, the two dimers within the tetramer interact with each other via two small interface patches at the top and the bottom of the tetramer. At first sight, these patches can be interpreted as biologically irrelevant crystal contacts on the basis of their limited buried surface area (Bahadur et al. 2004) and their high level of hydration (Rodier et al. 2005). However, as mentioned above, this possibility is strongly contradicted by the high conservation level. Moreover, we propose that the apparent dependence of the dimer–tetramer equilibrium on the modification of the C-terminal cysteine—a phenomenon that was also shown for P. pantotrophus SoxYZ (Quentmeier et al. 2003)—is regulated at the tetramer interface. It has been stated that a considerable number of proteins, for which a change in activity or function is coupled to a change in oligomeric structure, form weak transient oligomers by means of a small interface (Nooren and Thornton 2003). The investigators reasoned that these proteins can easily stabilize or weaken their complexes by creating or breaking a limited number of interactions at these interfaces, making possible a dynamic response to a change in environment or to a covalent modification. In our case, the interface reveals a number of hydrogen bonds and four conserved salt bridges that are of less importance for the integrity of the tetramer than the disulfide bridges, as suggested by the gel filtration experiments. Treatment of SoxY with DTT, sulfide, sulfite, and thiosulfate, resulting in the reduction of the disulfide bridge, should have a similar destabilizing effect on the tetrameric complex. However, SoxY reduced with DTT, sulfide, and sulfite dissociates into dimers, while the SoxY-thiosulfate adduct remains a tetramer. This difference can be explained by the creation of an extra interaction between the negatively charged S-thiocysteinesulfonate and the tetramer interface that sufficiently stabilizes the tetrameric state of the SoxY-thiosulfate adduct. If so, a chemically similar S-cysteinesulfonate, resulting from the incubation of SoxY with sulfite, is also expected to stabilize the tetramer. Analytical gel filtration of SoxY treated with sulfite, however, contradicts this, thus indicating that other events in addition to a potential stabilization of the S-thiocysteinesulfonate moiety take place at the tetramer interface upon covalent binding of thiosulfate.
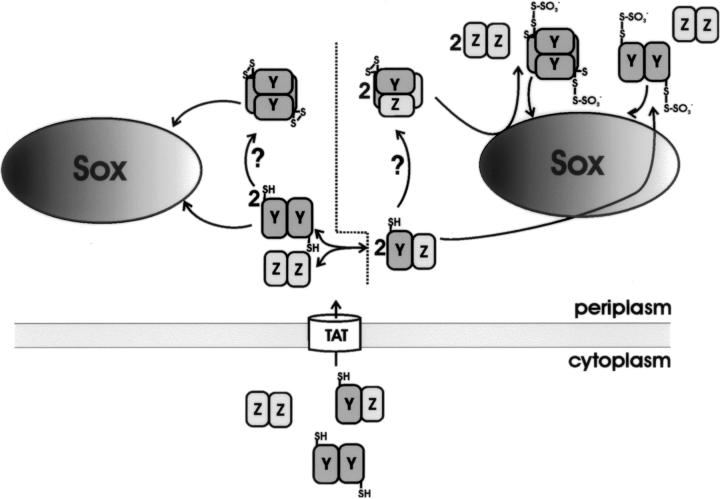
With the knowledge of the different SoxY and SoxZ dimer structures, we propose two potential modes for SoxYZ dimerization. Since SoxZ is a β-sandwich protein, it may be that SoxY interacts in a similar way with SoxZ as with a second SoxY monomer, forming an extended β-sandwich dimer. Thus, the hydrogen-bonding potential of both subunits would be combined with the creation of a hydrophobic center in the SoxYZ interface, thereby further stabilizing the SoxYZ complex. The SoxYZ complex may also be analogous to the SoxZ dimer where the two components also interact with each other via their β-sheets I. In both structures, the C terminus of SoxY would be in the vicinity of the extended b-c loop. This loop has a number of conserved charged residues that may play a role in the interaction with the other Sox components and/or in stabilizing the cysteine-sulfur substrate adduct (Friedrich et al. 2001). The presence of the intersubunit disulfide bridges between the SoxY subunits would, however, be possible only for the first model, whereas the SoxZ subunits in the second model would hinder two SoxY molecules to approach each other and to interact at the tetramer interface patch.
Although neither crystal structure gives any clues about how the transition from homodimers to heterodimers and vice versa proceeds, they are suggestive that such an exchange takes place. A generally accepted issue is that SoxZ, which lacks a signal peptide, needs SoxY for translocation to the periplasm (Friedrich et al. 2000). This implies that SoxYZ heterodimers need to be formed in the cytoplasm so that both proteins can be transported to the periplasm by the Tat-mechanism, a system capable of translocating folded proteins (Berks et al. 2003). Having arrived in the periplasm, these heterodimers become involved in thiosulfate oxidation, and it may be that they exchange their subunits at one or more steps during this process. The following two scenarios may occur: In one scenario, SoxYZ exchanges subunits, forming SoxY and SoxZ dimers (Fig. 6, left side). These SoxY dimers, being redox-active, could also assemble into SoxY tetramers with the closure of the intersubunit disulfide bridges, an event that is likely mediated via a thiol oxidoreductase system. Both SoxY protomers comprise the active sulfur substrate-binding species on which the other sulfur-oxidizing Sox enzymes act. One major drawback of this model is that it neglects SoxZ, which is believed to coordinate the sulfur substrate molecules (Quentmeier and Friedrich 2001). The second scenario takes SoxZ into account (Fig. 6, right side). It could be that SoxZ mediates the interaction between the heterodimer and the proteins responsible for disulfide bond closure and sulfur substrate addition, respectively, protecting in the meantime the disulfide bridge and the covalently linked sulfur substrate. An exchange from SoxYZ heterodimers to SoxY homodimers and tetramers could then happen after substrate binding, when SoxY offers its bound sulfur molecules to the other Sox enzymes. In both models, however, one SoxY protomer offers two covalently bound sulfur molecules, instead of one, at each encounter with another component of the sulfur-oxidizing system. This should in principle be a more efficient way of presenting sulfur substrate molecules to the Sox system.
Figure 6.
Schematic representation of the two proposed reaction models, taking the SoxYZ and SoxY homodimers and tetramers into account.
To test these hypotheses, we suggest that future research needs to address the role of homo-oligomeric SoxY and SoxZ proteins, and in particular to investigate the role of the intersubunit disulfide bridges between SoxY monomers. Moreover, we need to find out what the biological meaning is of the flexible balance between SoxY dimers and tetramers.
Materials and Methods
Expression of native SoxY, crystallization, data collection, and phasing
Expression, crystallization, data collection, and MIRAS phasing of the native SoxY and the Pt2+- and Hg2+-derivativatized SoxY crystals were performed as described previously (Stout et al. 2006). Initial polyalanine models of SoxY were built into experimental MIRAS electron density maps at 3-Å resolution after density modification (Stout et al. 2006).
Refinement and structural analysis
Each round of model building was performed in the graphics program Turbo-Frodo (Roussel and Cambillau 1992) and was followed by a refinement cycle consisting of the “slow cooling” simulated annealing protocol and the conjugated energy-minimization protocols implemented in the CNS software package (Brunger et al. 1998). Noncrystallographic-symmetry restraints were applied in the initial rounds of model building and structure refinement. Refmac5 (Murshudov et al. 1997) was used in the latter stages of structure refinement, in combination with TLS-refinement (Winn et al. 2001). The refinement steps of the room-temperature structure were similar to the latter stages of the structure solution of the “cryo”-structure, using a partially refined “cryo”-structure as a starting model. Ordered water molecules were added and refined using the ARP-water module of Refmac5. Furthermore, one Cl-anion, one phosphate anion, three dinitrogen molecules, and one phosphate anion were added in the “cryo”- and room-temperature structures, respectively. They were refined to structural moieties having electron densities without any residual features in the Fo-Fc maps, and B-factor values similar to those of other atoms in its immediate surrounding. Although the quality of the electron density is good for both structures, exceptions occur at the N and C termini, and at some solvent-exposed loops of both subunits. In subunit B the electron density at the residues E37-A43 and M90-A92 is weak for the “cryo”- and room-temperature structure. Residues S1-F7 and G118-G122 of subunit A and residues T116-G122 of subunit B were not modeled due to absence of electron density. The refinement statistics are presented in Table 1.
Structure analysis
Structure validation was performed with the program PROCHECK (Laskowski et al. 1993) and secondary structure was assigned using DSSP (Kabsch and Sander 1983). Hydrogen bonds were analyzed using the program HBPLUS (McDonald and Thornton 1994) using the default criteria of the program for hydrogen bond assignment. Salt bridges were identified when Asp or Glu side-chain carbonyl oxygen atoms were found to be within a 4.0-Å distance from the nitrogen atoms and Arg, Lys, and His side-chains. Accessible surface areas (ASA) were calculated with the programs ACCESS and ACCFMT (Lee and Richards 1971). Atoms losing 0.1 Å2 in a protein–protein interaction were identified as interface atoms, and atoms having 0 Å2 in a protein–protein interface were classified as buried interface atoms. Surface residues having >5% of their relative accessible surface area (RSA), calculated for each amino acid by Miller et al. (1987), were defined as exposed to the solvent. Fully buried interface residues have a maximum of 5% of their RSA exposed upon oligomerization. Residues having >5% RSA exposed in the interface are partially buried interface residues. Protein–protein interfaces were analyzed by the Protein–Protein Interaction server (www.biochem.ucl.ac.uk/bsm/PP/server/index.html). Figures were prepared with PyMOL.
Calculation of conservation scores
Homologs were searched for in the NCBI protein database, using psi-BLAST, and all sequences were aligned in STRAP (Gille and Frommel 2001) with SoxY as a structural template. Conservation scores (cons) between 0 and 1 were calculated for all positions in the alignment using the SCORECONS with default settings (Valdar and Thornton 2001b). The summations of the cons of the residues constituting the dimer and tetramer interfaces, respectively, were used as a measure for the interface conservation. Bootstrap experiments of 100,000 trials each were performed to evaluate the calculated conservation of the total dimer interface, the core of the dimer interface, and the tetramer, respectively. For each experiment, the H0 hypothesis was tested: The level of conservation of the interface set should not be higher than the average conservation of an equal number of surface residues drawn randomly without replacement. To exclude the effect of the residues of the other interface on the calculations, these residues were left out of the bootstrap analysis of the interface to be tested. The probability P that sets of randomly picked residues have a sum of cons score equal to or higher than the scores of the observed interfaces was estimated as p = tc/t, where tc is the number of trials consisting each of a set of residues that are at least as conserved as the observed set, and t is the total number of trials (Valdar and Thornton 2001b). If the probability P of H0 was <5%, the alternative hypothesis H1 was accepted, which states that the observed interface set is more conserved than a set of randomly picked residues.
Sulfur compound binding
The covalent binding of thiosulfate, sulfite, and sulfide was tested by incubating a 0.8 mg/mL SoxY solution, buffered in 50 mM Tris-HCl, pH 8.0, with 1 mM Na2S2O3, 1 mM Na2SO3, and 5 mM and 10 mM Na2S respectively, for 1 h. Subsequently, the reactions were desalted using Microspin concentrators (Millipore, MWCO 10 kDa). The samples were prepared for MS analysis by mixing 3 μL of a desalted SoxY solution (30 μM) with an equal volume of 50% acetonitrile, 0.1% formic acid. MS analyses were carried out on an electrospray ionization (ESI) hybrid quadrupole-time-of-flight (Q-TOF) mass spectrometer (Micromass), equipped with a nanoflow Z-spray ionization system. The MS spectra were transformed using the Masslynx 4.0 Software supplied with the mass spectrometer.
A trypsin digest of SoxY after thiosulfate binding was carried out using an enzyme to substrate ratio (mass/mass) of 1:55 for 20 h, at 37°C, in 100 mM ammonium bicarbonate, pH 8.0. The digest was spotted on a matrix assisted laser desorption ionization (MALDI) sample target plate loaded with a matrix consisting of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% trifluoroacetic acid. MS was performed in the positive and in the negative reflecton mode using a MALDI-TOF mass spectrometer (model 4700 Proteomic Analyzer, Applied Biosystems). Peptide mass spectra were obtained in a mass range of 700–2000 Da in the positive mode and of 600–1700 Da in the negative mode, respectively. Calibration was carried out in the default mode.
Analytical gel filtration
The oligomeric state of modified SoxY was determined by analytical gel filtration using a Superdex 200 PC 3.2/30 column (Amersham) installed on a SMART system (Amersham). For each run, 5–15 μg of protein was loaded onto the column, which was preequilibrated with 50 mM Tris-HCl, pH 8.0, 150 mM NaCl.
Protein Data Bank accession number
The coordinates of the SoxY Native1 and Native2 structures have been deposited in the RCSB Protein Data Bank, accession codes 2NNC and 2NNF.
Acknowledgments
This work was supported by the Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie en Vooruitgang in Vlaanderen (IWT). J.V.B. is indebted to the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen, (FWO-Flanders; grant G.0330.03). We gratefully acknowledge access to the EMBL beamline BW7A at the DORIS storage ring, DESY, Hamburg. J.S. also is indebted to Bjorn Vergauwen for suggestions and valuable discussions, to Terry Meyer for helpful discussions and critical reading of the manuscript, and to Maarten Aerts, Kjell Sergeant, and Kris De Vriendt for conducting the MALDI-TOF analysis and for assistance with the ESI MS measurements.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Jozef Van Beeumen or Savvas N. Savvides, Laboratory of Protein Biochemistry and Protein Engineering, Department of Biochemistry, Microbiology and Physiology, Ghent University, 9000 Ghent, B-Belgium; e-mail: jozef.vanbeeumen@ugent.be or savvas.savvides@ugent.be; fax: +32 9264 5338.
Abbreviations: PDB, RSCB protein data bank; ESI MS, electrospray ionization mass spectrometry; MALDI, matrix assisted laser desorption ionization; Ve, elution volume.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062633607.
References
- Bahadur R.P., Chakrabarti, P., Rodier, F., and Janin, J. 2003. Dissecting subunit interfaces in homodimeric proteins. Proteins 53: 708–719. [DOI] [PubMed] [Google Scholar]
- Bahadur R.P., Chakrabarti, P., Rodier, F., and Janin, J. 2004. A dissection of specific and non-specific protein–protein interfaces. J. Mol. Biol. 336: 943–955. [DOI] [PubMed] [Google Scholar]
- Bamford V.A., Bruno, S., Rasmussen, T., Appia-Ayme, C., Cheesman, M.R., Berks, B.C., and Hemmings, A.M. 2002. Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme. EMBO J. 21: 5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks B.C., Palmer, T., and Sargent, F. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47: 187–254. [DOI] [PubMed] [Google Scholar]
- Bork P., Holm, L., and Sander, C. 1994. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242: 309–320. [DOI] [PubMed] [Google Scholar]
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54: 905–921. [DOI] [PubMed] [Google Scholar]
- Brüser T., Lens, P., and Trüper, H.G. 1999. The biological sulfur cycle. In Environmental technologies to treat sulfur pollution (eds. P. Lens and L.H. Pol), pp. 47–86. IWA Publishing, London.
- Caffrey D.R., Somaroo, S., Hughes, J.D., Mintseris, J., and Huang, E.S. 2004. Are protein–protein interfaces more conserved in sequence than the rest of the protein surface? Protein Sci. 13: 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammack R., Chapman, A., Lu, W.P., Karagouni, A., and Kelly, D.P. 1989. Evidence that Protein-B of the thiosulfate-oxidizing system of Thiobacillus versutus contains a binuclear manganese cluster. FEBS Lett. 253: 239–243. [Google Scholar]
- Chothia C. and Janin, J. 1975. Principles of protein–protein recognition. Nature 256: 705–708. [DOI] [PubMed] [Google Scholar]
- Dahl C., Engels, S., Pott-Sperling, A.S., Schulte, A., Sander, J., Lubbe, Y., Deuster, O., and Brune, D.C. 2005. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum . J. Bacteriol. 187: 1392–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., Nelson, K.E., Paulsen, I.T., Heidelberg, J.F., Wu, M., Dodson, R.J., DeBoy, R., Gwinn, M.L., Nelson, W.C., Haft, D.H., et al. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. 99: 9509–9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C.G. 1998. Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 39: 235–289. [DOI] [PubMed] [Google Scholar]
- Friedrich C.G., Quentmeier, A., Bardischewsky, F., Rother, D., Kraft, R., Kostka, S., and Prinz, H. 2000. Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 182: 4677–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C.G., Rother, D., Bardischewsky, F., Quentmeier, A., and Fischer, J. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: Emergence of a common mechanism? Appl. Environ. Microbiol. 67: 2873–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C.G., Bardischewsky, F., Rother, D., Quentmeier, A., and Fischer, J. 2005. Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8: 253–259. [DOI] [PubMed] [Google Scholar]
- Gille C. and Frommel, C. 2001. STRAP: Editor for STRuctural Alignments of Proteins. Bioinformatics 17: 377–378. [DOI] [PubMed] [Google Scholar]
- Halaby D.M., Poupon, A., and Mornon, J.P. 1999. The immunoglobulin fold family: Sequence analysis and 3D structure comparisons. Protein Eng. 12: 563–571. [DOI] [PubMed] [Google Scholar]
- Harpaz Y. and Chothia, C. 1994. Many of the immunoglobulin superfamily domains in cell adhesion molecules ans surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol. 238: 528–539. [DOI] [PubMed] [Google Scholar]
- Hipp W.M., Pott, A.S., Thum-Schmitz, N., Faath, I., Dahl, C., and Trüper, H.G. 1997. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiol. 143: 2891–2902. [DOI] [PubMed] [Google Scholar]
- Jones S. and Thornton, J.M. 1996. Principles of protein–protein interactions. Proc. Natl. Acad. Sci. 93: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen B.B. 1990. The sulfur cycle of freshwater sediments: Role of thiosulfate. Limnol. Oceanogr. 35: 1329–1342. [Google Scholar]
- Kabsch W. and Sander, C. 1983. How good are predictions of protein secondary structure? FEBS Lett. 155: 179–182. [DOI] [PubMed] [Google Scholar]
- Kelly D.P. 1987. Sulfur bacteria first again. Nature 326: 830. [Google Scholar]
- Kelly D.P., Shergill, J.K., Lu, W.P., and Wood, A.P. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie van Leeuwenhoek 71: 95–107. [DOI] [PubMed] [Google Scholar]
- Kister A.E., Finkelstein, A.V., and Gelfand, I.M. 2002. Common features in structures and sequences of sandwich-like proteins. Proc. Natl. Acad. Sci. 99: 14137–14141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Macarthur, M.W., Moss, D.S., and Thornton, J.M. 1993. Procheck—A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Lee B. and Richards, F.M. 1971. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 55: 379–400. [DOI] [PubMed] [Google Scholar]
- Lo Conte L., Chothia, C., and Janin, J. 1999. The atomic structure of protein–protein recognition sites. J. Mol. Biol. 285: 2177–2198. [DOI] [PubMed] [Google Scholar]
- Lu W.P. and Kelly, D.P. 1983. Purification and some properties of two principal enzymes of the thiosulfate-oxidizing multi-enzyme system from Thiobacillus A2. J. Gen. Microbiol. 129: 3549–3564. [Google Scholar]
- Lu W.P., Swoboda, B.E.P., and Kelly, D.P. 1985. Properties of the thiosulfate-oxidizing multi-enzyme system from Thiobacillus versutus . Biochim. Biophys. Acta 828: 116–122. [Google Scholar]
- Lu W.P. 1986. A periplasmic location for the thiosulfate-oxidizing multi-enzyme system from Thiobacillus versutus . FEMS Microbiol. Lett. 34: 313–317. [Google Scholar]
- Mandel-Gutfreund Y. and Gregoret, L.M. 2002. On the significance of alternating patterns of polar and non-polar residues in beta-strands. J. Mol. Biol. 323: 453–461. [DOI] [PubMed] [Google Scholar]
- McDonald I.K. and Thornton, J.M. 1994. Satisfying hydrogen-bonding potential in proteins. J. Mol. Biol. 238: 777–793. [DOI] [PubMed] [Google Scholar]
- Miller S. 1989. The structure of interfaces between subunits of dimeric and tetrameric proteins. Protein Eng. 3: 77–83. [DOI] [PubMed] [Google Scholar]
- Miller S., Janin, J., Lesk, A.M., and Chothia, C. 1987. Interior and surface of monomeric proteins. J. Mol. Biol. 196: 641–656. [DOI] [PubMed] [Google Scholar]
- Mintseris J. and Weng, Z. 2005. Structure, function, and evolution of transient and obligate protein–protein interactions. Proc. Natl. Acad. Sci. 102: 10930–10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53: 240–255. [DOI] [PubMed] [Google Scholar]
- Nooren I.M. and Thornton, J.M. 2003. Structural characterisation and functional significance of transient protein–protein interactions. J. Mol. Biol. 325: 991–1018. [DOI] [PubMed] [Google Scholar]
- Pott A.S. and Dahl, C. 1998. Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiol. 144: 1881–1894. [DOI] [PubMed] [Google Scholar]
- Quentmeier A. and Friedrich, C.G. 2001. The cysteine residue of the SoxY protein as the active site of protein-bound sulfur oxidation of Paracoccus pantotrophus GB17. FEBS Lett. 503: 168–172. [DOI] [PubMed] [Google Scholar]
- Quentmeier A., Kraft, R., Kostka, S., Klockenkamper, R., and Friedrich, C.G. 2000. Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Arch. Microbiol. 173: 117–125. [DOI] [PubMed] [Google Scholar]
- Quentmeier A., Hellwig, P., Bardischewsky, F., Grelle, G., Kraft, R., and Friedrich, C.G. 2003. Sulfur oxidation in Paracoccus pantotrophus: Interaction of the sulfur-binding protein SoxYZ with the dimanganese SoxB protein. Biochem. Biophys. Res. Commun. 312: 1011–1018. [DOI] [PubMed] [Google Scholar]
- Richardson J.S. and Richardson, D.C. 2002. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. 99: 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F., Bahadur, R.P., Chakrabarti, P., and Janin, J. 2005. Hydration of protein–protein interfaces. Proteins 60: 36–45. [DOI] [PubMed] [Google Scholar]
- Rose G.D., Gierash, L.M., and Smith, J.A. 1985. Turns in peptides and proteins. Adv. Protein Chem. 37: 1–109. [DOI] [PubMed] [Google Scholar]
- Rother D., Henrich, H.J., Quentmeier, A., Bardischewsky, F., and Friedrich, C.G. 2001. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J. Bacteriol. 183: 4499–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel A. and Cambillau, C. 1992. Turbo-Frodo. Biographics, AFMB, Marseille, France.
- Schedel M., Vanselow, M., and Trüper, H.G. 1979. Siroheme sulfite reductase isolated from Chromatium vinosum. Purification and investigation of some of its molecular and catalytic properties. Arch. Microbiol. 121: 29–36. [Google Scholar]
- Stout J., De Smet, L., Panjikar, S., Weiss, M.S., Savvides, S.N., and Van Beeumen, J. 2006. Crystallization, preliminary crystallographic analysis and phasing of the thiosulfate binding protein SoxY from Chlorobium limicola f. thiosulfatophilum. Acta Crystallogr. 62: 1093–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.J., Lin, S.L., Wolfson, H.J., and Nussinov, R. 1997. Studies of protein–protein interfaces: A statistical analysis of the hydrophobic effect. Protein Sci. 6: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W.S.J. and Thornton, J.M. 2001a. Conservation helps to identify biologically relevant crystal contacts. J. Mol. Biol. 313: 399–416. [DOI] [PubMed] [Google Scholar]
- Valdar W.S.J. and Thornton, J.M. 2001b. Protein–protein interfaces: Analysis of amino acid conservation in homodimers. Protein Struct. Funct. Genet. 42: 108–124. [PubMed] [Google Scholar]
- Venkatachalam C.M. 1968. Stereochemical criteria for polypeptides and proteins V. Conformation of a system of three linked peptide units. Biopolymers 6: 1425–1436. [DOI] [PubMed] [Google Scholar]
- Verté F., Kostanjevecki, V., De, S.L., Meyer, T.E., Cusanovich, M.A., and Van Beeumen, J.J. 2002. Identification of a thiosulfate utilization gene cluster from the green phototrophic bacterium Chlorobium limicola . Biochemistry 41: 2932–2945. [DOI] [PubMed] [Google Scholar]
- Williams A.F. and Barclay, A.N. 1988. The immunoglobulin superfamily—Domains for cell surface recognition. Annu. Rev. Immunol. 6: 381–405. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Isupov, M.N., and Murshudov, G.N. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D57: 122–133. [DOI] [PubMed] [Google Scholar]
- Wodara C., Bardischewsky, F., and Friedrich, C.G. 1997. Cloning and characterization of sulfite dehydrogenase, two c-type cytochromes, and a flavoprotein of Paracoccus denitrificans GB17: Essential role of sulfite dehydrogenase in lithotrophic sulfur oxidation. J. Bacteriol. 179: 5014–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Lin, S.L., and Nussinov, R. 1997a. Protein binding versus protein folding: The role of hydrophilic bridges in protein associations. J. Mol. Biol. 265: 68–84. [DOI] [PubMed] [Google Scholar]
- Xu D., Tsai, C.J., and Nussinov, R. 1997b. Hydrogen bonds and salt bridges across protein–protein interfaces. Protein Eng. 10: 999–1012. [DOI] [PubMed] [Google Scholar]
- Young L., Jernigan, R.L., and Covell, D.G. 1994. A role for surface hydrophobicity in protein–protein recognition. Protein Sci. 3: 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]